- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring the Interactions between Peroxisomes and Chloroplasts by in situ Laser Analysis
Published: Vol 6, Iss 8, Apr 20, 2016 DOI: 10.21769/BioProtoc.1790 Views: 8085
Reviewed by: Tie LiuRu ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Isolation of Lysosome-Related Organelles from Stationary Phase and Iron-Overloaded Chlamydomonas reinhardtii Cells
Jiling Li and Huan Long
Nov 20, 2024 1767 Views

Isolation and Biophysical Characterization of Extracellular Vesicles From Hairy Root Cultures
Marisa Conte [...] Alfredo Ambrosone
Mar 5, 2025 2220 Views

Rapid Miniprep of Intact Chloroplasts from Arabidopsis thaliana Leaves
Brenda A. Carranza-Correa [...] Manuel Gutiérrez-Aguilar
May 20, 2025 2646 Views
Abstract
Quantitative analysis has been necessary for deeply understanding characteristic of organelles function. This is the detailed protocol for the quantification of the physical interaction between peroxisomes and chloroplasts taken by laser scanning microscopy described by Oikawa et al. (2015). To clarify the morphological interactions between both organelles, we measured the contact length between two organelles (interaction length) in the fluorescent microscope image by using image analysis software ImageJ. The result clearly revealed that the contact length in light condition is much longer than that in dark condition. In addition, the force of the morphological interaction was quantified utilizing intersection technology of femtosecond laser and atomic force microscope (AFM). When an intense femtosecond laser is focused near the interface of two organelles, the adhesion is broken by a force due to the laser. The adhesion strength in light and dark conditions was estimated from the force calibrated by AFM. The detailed procedure is described in Bio-protocol as another protocol entitled “Quantification of the adhesion strength between peroxisomes and chloroplasts by femtosecond laser technology” (Hosokawa et al., 2016). These methods can be applied to other physical interaction between different types of organelles such as nuclei, mitochondria, Golgi, and chloroplasts.
Keywords: PeroxisomeMaterials and Reagents
- Glass slide (super frost) (Matsunami Glass)
Note: Any types of glass slide suitable for fluorescence observation can be used. We attached black tape on the glass slide to envelop the sample with cover slip (see Figure 1D). - Cover slip (24 x 60 No.1, Thickness 0.12-0.17 mm) (Matsunami Glass)
- 10 ml disposable syringe (Terumo Medical Corporation)
- Arabidopsis thaliana (ecotype Columbia) expressing peroxisome-targeted GFP (GFP-PTS1) (Mano et al., 2002)
Note: 1-4 is shown in Figure 1A. - 1/3 x Murashige and Skoog salts (MS) medium (Wako Pure Chemical Industries, catalog number: 392-00591 ) containing vitamins, 1% sucrose, and MES buffer (pH 5.7) (Wako Pure Chemical Industries, catalog number: 341-01622 )
- Agar powder for plant growth (Funakoshi, catalog number: BA-10 )
- Distilled water
- KOH
- Plant culture medium (see Recipes)
Equipment
- Scissors and tweezers (shown in Figure 1A)
- 40x dry system objective lens (ZEISS, EC Plan-Neofluar®)
- Growth chamber for growing plants (100 µmol m-2 sec-1 white Light for 16 h and dark for 8 h, 22 °C)
- Black box for dark condition
- Confocal laser scanning microscope (ZEISS, model: LSM510 META )
Software
- NIH ImageJ software 1.46 (http://imagej.nih.gov/ij)
Note: ImageJ is also available on Mac OS X, Windows and Linux.
Procedure
- Preparation of leaf section
- Cut the rosette leaves of 3-week-old Arabidopsis plants in 1 cm diameter and placed in a syringe filled with water (Figure 1B).
- Plug the top of syringe by finger and pull plunger to deaerate the leaves (Figure 1B).
- Mount one piece of the deaerated-leaf section (Figure 1C) on slide glass with water and cover with cover glass (Figure 1D).
- Incubate the pretreated-leaf sample in the dark or light for 2 h in growth chamber.
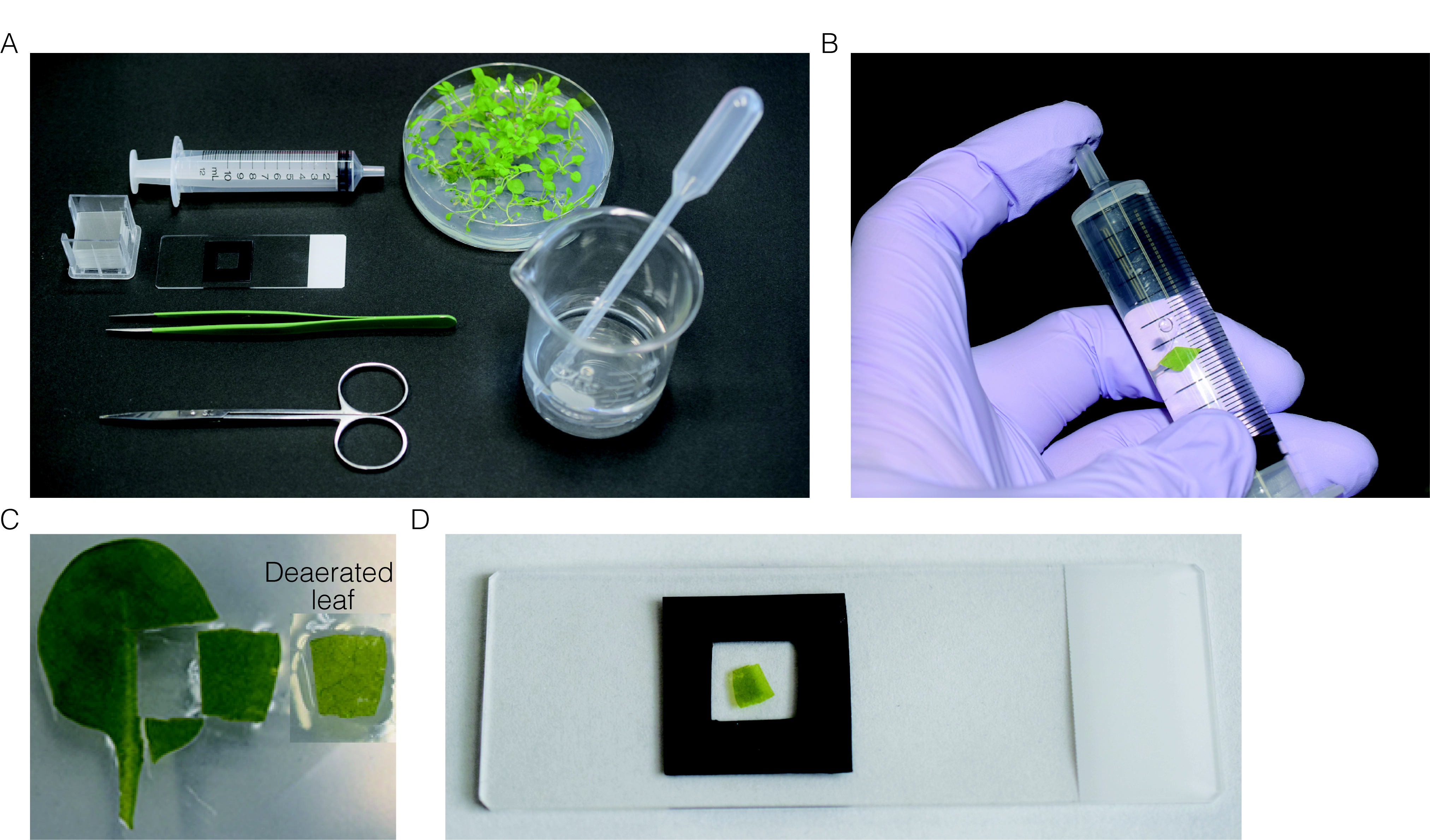
Figure 1. Preparation of leaf section for CLSM observation. A. All instruments used for preparation of leaf section. B. Deaeration of the leaves by pulling plunger while plugging the top of syringe. C. Leaf section used for observation. Before (left) and after (right) deaeration. D. Deaerated-leaf section mounted on the slide glass. - Taking organelles images by CLSM
- The images of organelles in leaf palisade mesophyll cells are focused and obtained through a 40x dry system objective lens using LSM 510 (Figure 4).
- To observe GFP fluorescence, a 488 nm Ar/Kr laser is used and the fluorescence is detected through an emission filter BP505-550. To detect the autofluorescence from chlorophyll, a 543 nm He/Ne laser is used and the signal is acquired through emission filters LP580.
- Obtain both fluorescence simultaneously and save the data as a single image of tif. file.
Note: The observation of organelles should be finished within 30 min after moving the sample from growth chamber, since the change of light condition affects the organelle motility and interaction.
- Quantification of the organelle interaction
- Launch an Image J software and open the obtained images.
- Set the scale of s: Analyze > Set Scale
- Set the measurements: Analyze > Set Measurements > check the column of Shape descriptors (Figure 2).
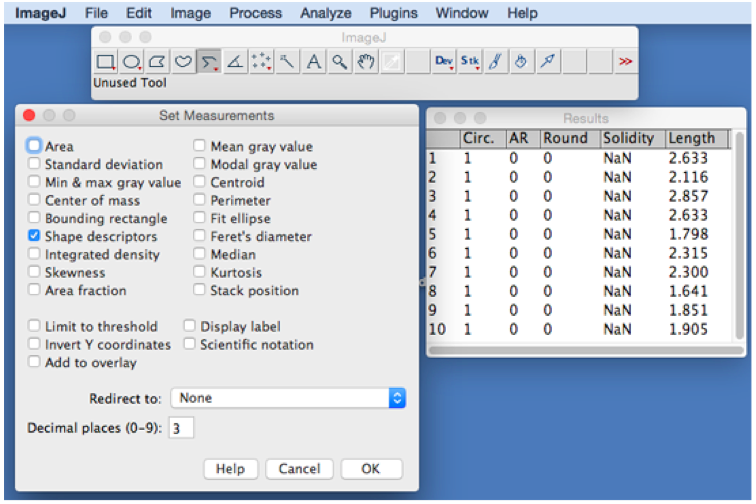
Figure 2. Analyze menu of ImageJ used to measure the length of segments lines. Choose Shape descriptors in Set Measurements window. Results of 10 interaction lengths are shown as an example of calculation after selecting Measure mode below Analyze menu. - Using line tool, draw a line connecting two intersection points between two organelles, or using segment tool, draw the curve line (red line in the right panel of Figure 3 and lower panel of Figure 4) for measuring.
Note: The image is magnified by zoom (in) function of ImageJ when drawing a line. Image > Zoom > In{+}.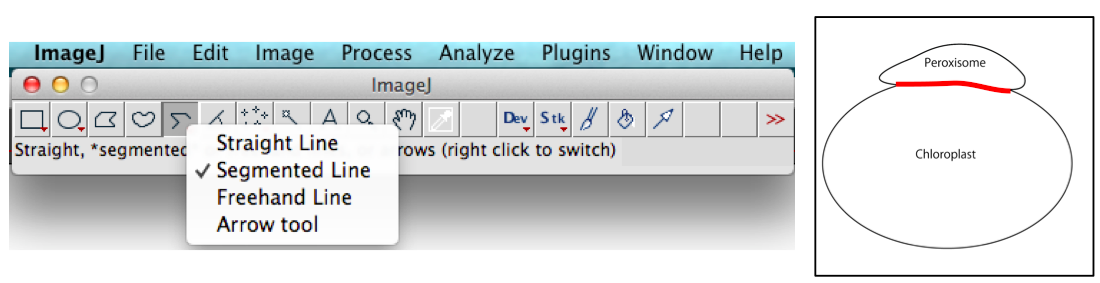
Figure 3. Segmented Line tool of ImageJ used to measure the interaction length (red line in the right panel) between a peroxisome and a chloroplast - Measure the length of lines: Analyze > Measure
Note: An average of at least 50 interactions between peroxisomes and chloroplasts should be measured to calculate the average length in independent three experiments (Figure 4, Table 1). Here, we show results of ten samples for measurements of interaction lengths between peroxisomes and chloroplasts.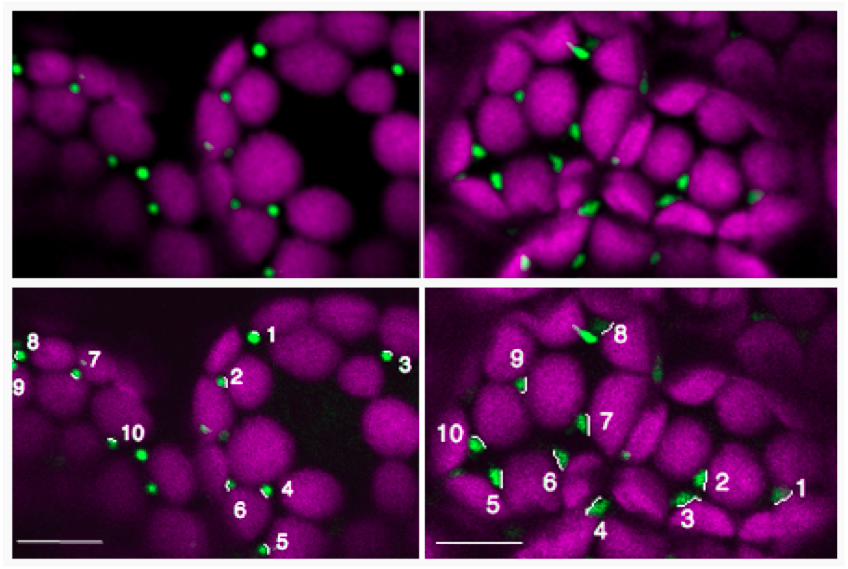
Figure 4. Measurements of the 10 selected-interaction lengths of membrane contact area between peroxisomes (green) and chloroplasts (magenta) in the dark (left panel) and light (right panel) at lower panels. Spherical peroxisomes in the dark change their shape to elongation in light, suggesting that their physiological interaction is enhanced in light. Upper panels are original images. Bars = 10 µm
Table 1. Results of measurements of the 10 selected-contact lengths between peroxisomes and chloroplasts in dark and light. The contact length in light condition was longer than that in dark condition. This fact suggests that the peroxisomes assist photorespiration by tightly contacting with the chloroplasts in photosynthesis.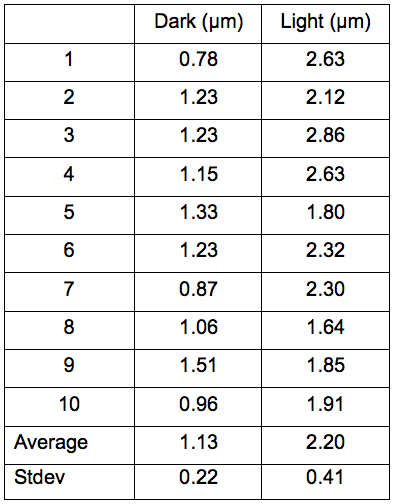
Recipes
- Plant culture medium
Dissolve 10 g sucrose, 1.53 g MS salt, and 0.5 g MES in 1 L pure water, and then adjust the pH to 5.7 with 1 M KOH
Add 8 g agar in the medium, and then autoclave
Acknowledgments
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) [KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas to M. N. (No. 22120007) and Y. H. (No. 22120010)].
References
- Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M. and Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol 43(3): 331-341.
- Hosokawa, Y., Iino, T., Oikawa, K., Mano, S., Yamada, K. and Nishimura, M., Quantification of the adhesion strength between peroxisomes and chloroplasts by femtosecond laser technology. Bio-protocol 6(11): e1834.
- Oikawa, K., Matsunaga, S., Man, S., Kondo, M., Yamada, K., Hayashi, M., Kagawa, T., Kadota, A., Sakamoto, W., Higashi, H., Watanabe, M., Mitsui, T., Shigemasa, A., Iino, T., Hosokawa, Y. and Nishimura, M. (2015). Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat Plants 1: 15035.
- Shibata, M., Oikawa, K., Mano, S. and Nishimura, M. (2014). Measurement of the number of peroxisomes. Bio-protocol 4(21): e1284.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Oikawa, K., Mano, S., Yamada, K., Hosokawa, Y. and Nishimura, M. (2016). Measuring the Interactions between Peroxisomes and Chloroplasts by in situ Laser Analysis. Bio-protocol 6(8): e1790. DOI: 10.21769/BioProtoc.1790.
Category
Plant Science > Plant cell biology > Organelle isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











