- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of the Adhesion Strength between Peroxisomes and Chloroplasts by Femtosecond Laser Technology
Published: Vol 6, Iss 11, Jun 5, 2016 DOI: 10.21769/BioProtoc.1834 Views: 10381
Reviewed by: Tie LiuJaroslav ĎurkovičDennis Nürnberg

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
This is the detailed protocol to quantify adhesion strength between peroxisomes and chloroplasts in plant leaf palisade mesophyll cells described by Oikawa et al. (2015). The quantification was performed by utilizing local explosion induced by focusing femtosecond laser pulses into a mesophyll cell under a confocal microscope. When an impulsive force generated by an explosion is loaded on the interface between a peroxisome and a chloroplast, the peroxisome is frequently detached from the chloroplast. The probability of a peroxisome detaching from a chloroplast was estimated (left-top of Figure 1). Next, the magnitude of the impulsive force was quantified by an atomic force microscope (AFM) cantilever (right-top of Figure 1). On the basis of these results, the pressure to break adhesion between a peroxisome and a chloroplast was quantified as an index of the adhesion strength (bottom of Figure 1). In this protocol, these procedures are summarized. As the local explosion is induced not only in the medium of the mesophyll cells but also in aqueous medium generally, this method could be applied to various adhesions between organelles and between cells around 1 to 100 μm in diameter (e.g., adhesions between mitochondria and chloroplasts, between nucleus and cell membrane, and between two cells with weak physical interaction). Additionally, we have evaluated the interaction between peroxisomes and chloroplasts from the interaction length between two organelles. This protocol has been presented in Bio-protocol as “Measuring the interactions between peroxisomes and chloroplasts by in situ laser analysis” (Oikawa et al., 2015).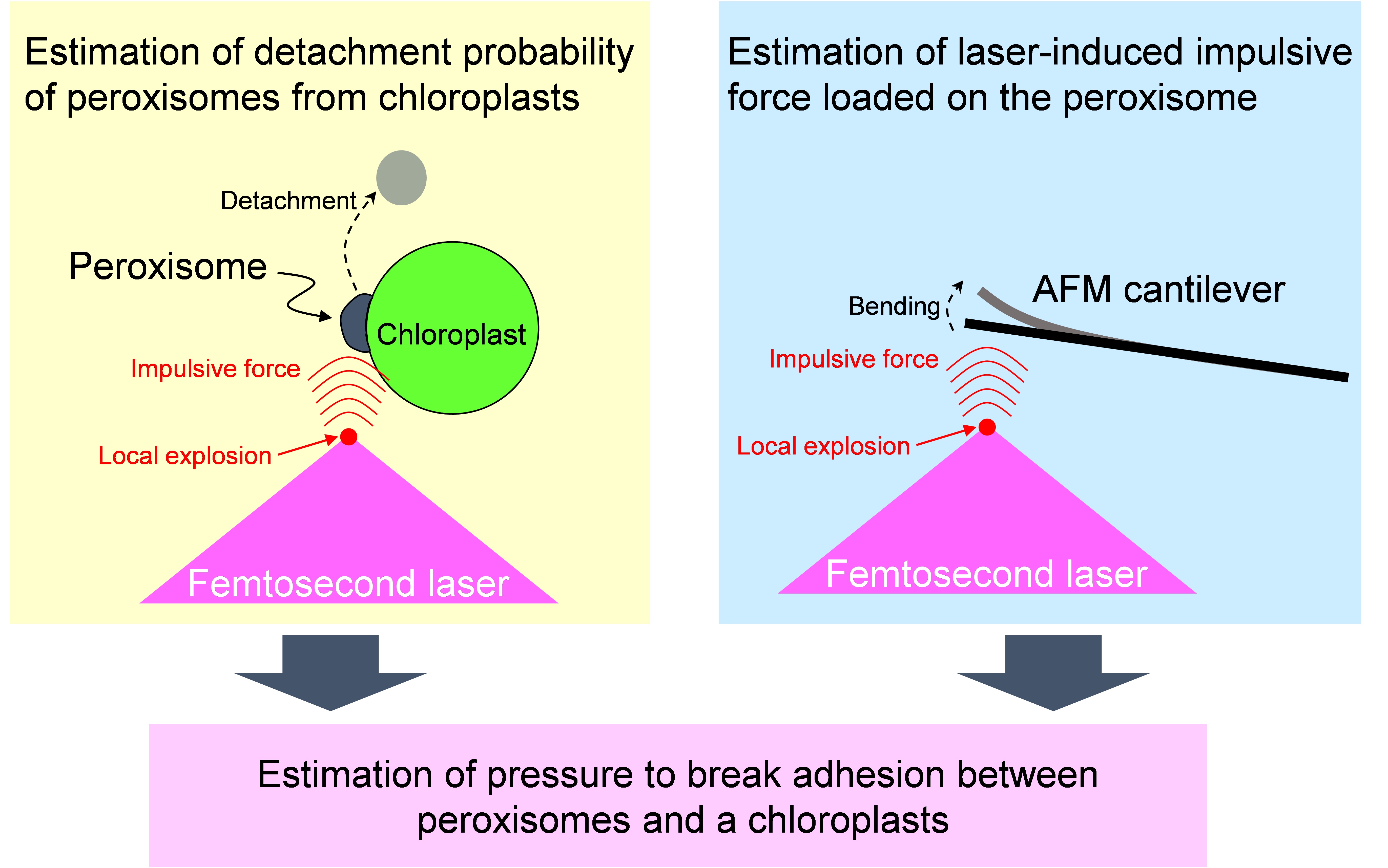
Figure 1. Flow chart estimating adhesion strength between a peroxisome and a chloroplast by utilizing femtosecond laser and atomic force microscope
Materials and Reagents
- Glass slide (26 x 76 No.1, thickness 0.8-1.0 mm) (Matsunami Glass Ind.)
Note: Any types of glass slide suitable for fluorescence observation can be used. We attached black tape on the glass slide to envelop the sample with the cover slip (see Figure 4D). - Cover slip (24 x 60 No.1, thickness 0.12-0.17 mm) (Matsunami Glass Ind.)
- 10 ml disposable syringe (Terumo Medical Corporation)
- Razor blade (43 x 23)
- Arabidopsis thaliana (ecotype Columbia) expressing peroxisome-targeted GFP (GFP-PTS1) (Mano et al., 2002)
- Agar powder (Funakoshi, catalog number: BA-10 )
- Distilled water
- 1/3x Murashige and Skoog salts (MS) medium (Wako Pure Chemical Industries, catalog number: 392-00591 )
Equipment
- Confocal microscope combined with amplified Ti:Sapphire femtosecond laser system (Figure 2)
- Ti:Sapphire femtosecond laser system (Cyber Laser Inc., model: IFRIT-SP-01 )
- Confocal microscope (Olympus Corporation of the America, model: FV300-IX71 )
- Objective (OLYMPUS CORPORATION, model: PlanN100x )
- Mechanical shutter (Gate time: 1/125 s) [SIGMAKOKI, model: 65GR (discontinued product); compatible product: SIGMAKOKI, model: SSH-25RA ]
- Collimator lenses
- λ/2 plate
- Polarizer
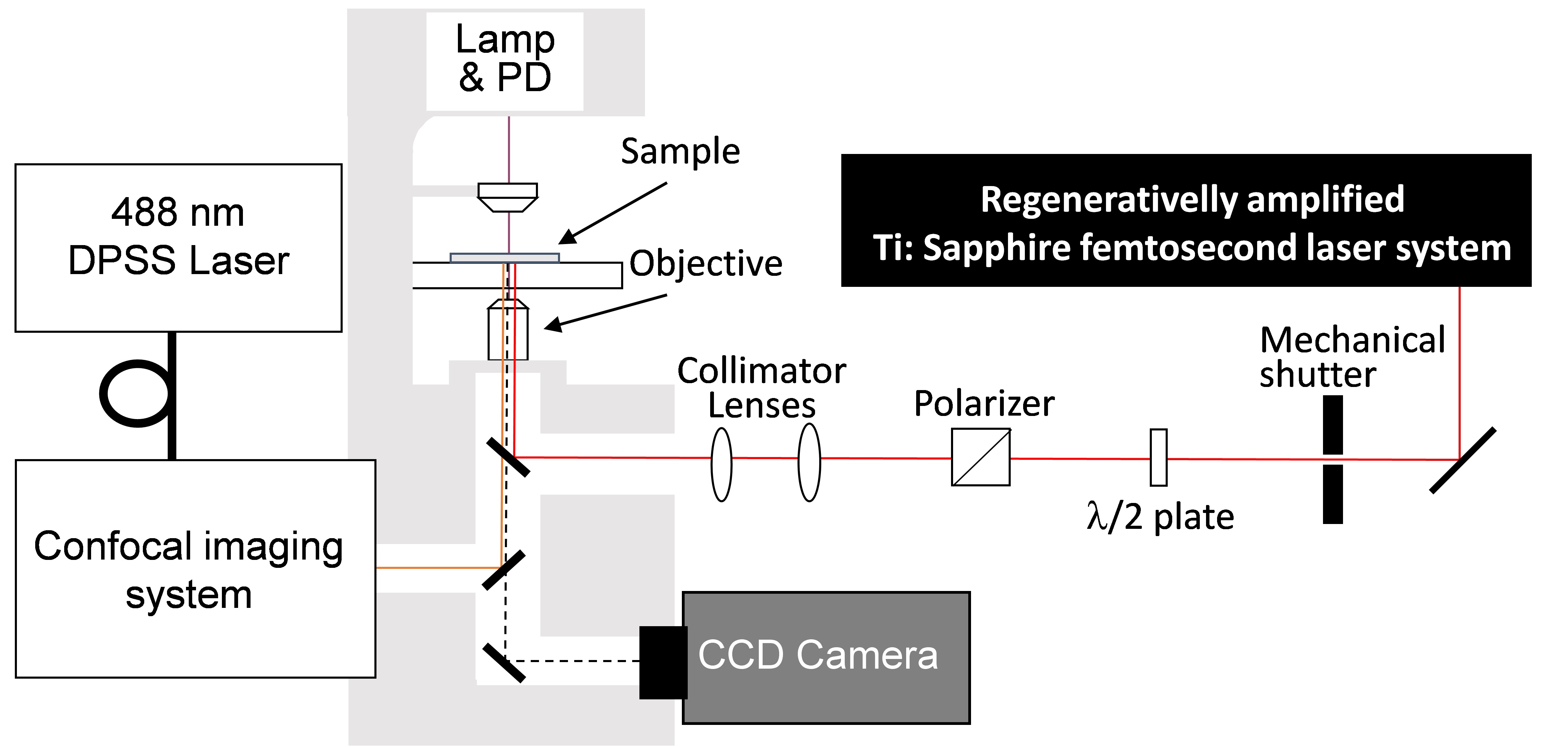
Figure 2. Experimental setup of confocal microscope and femtosecond laser system. Femtosecond laser pulses from an amplified Ti:Sapphire femtosecond laser system (Wavelength: 780 nm, Pulse duration: 230 fs, Pulse energy: <1 mJ/pulse, Repetition rate: 125 Hz) are introduced to a confocal microscope (Excitation wavelength: 488 nm, Detection wavelength: 515-550 nm). Mechanical shutter (Gate time: 1/125 s), collimator lenses, λ/2 plate and polarizer were placed in the beam line between the laser and the microscope. - Atomic force microscope (AFM) (Figure 3)
- AFM [Pacific Nanotechnology, model: Nano-R2 (discontinued product); compatible product: JPK Instruments, model: NanoWizard4 NanoScience ]
- Ti:Sapphire femtosecond laser system (see Equipment Section, 1a)
- Confocal microscope (see Equipment Section, 1b)
- Objective (see Equipment Section, 1c)
- A tipless AFM cantilever (NanoWorld AG, NANOSENSORSTM, model: TL-NCH )
- Motorized stage (SIGMAKOKI, model: BIOS-206T )
- Oscilloscope [TEKTRONIX, model: DP4104 (discontinued product); compatible product: TEKTRONIX, model: MDO4104c ]
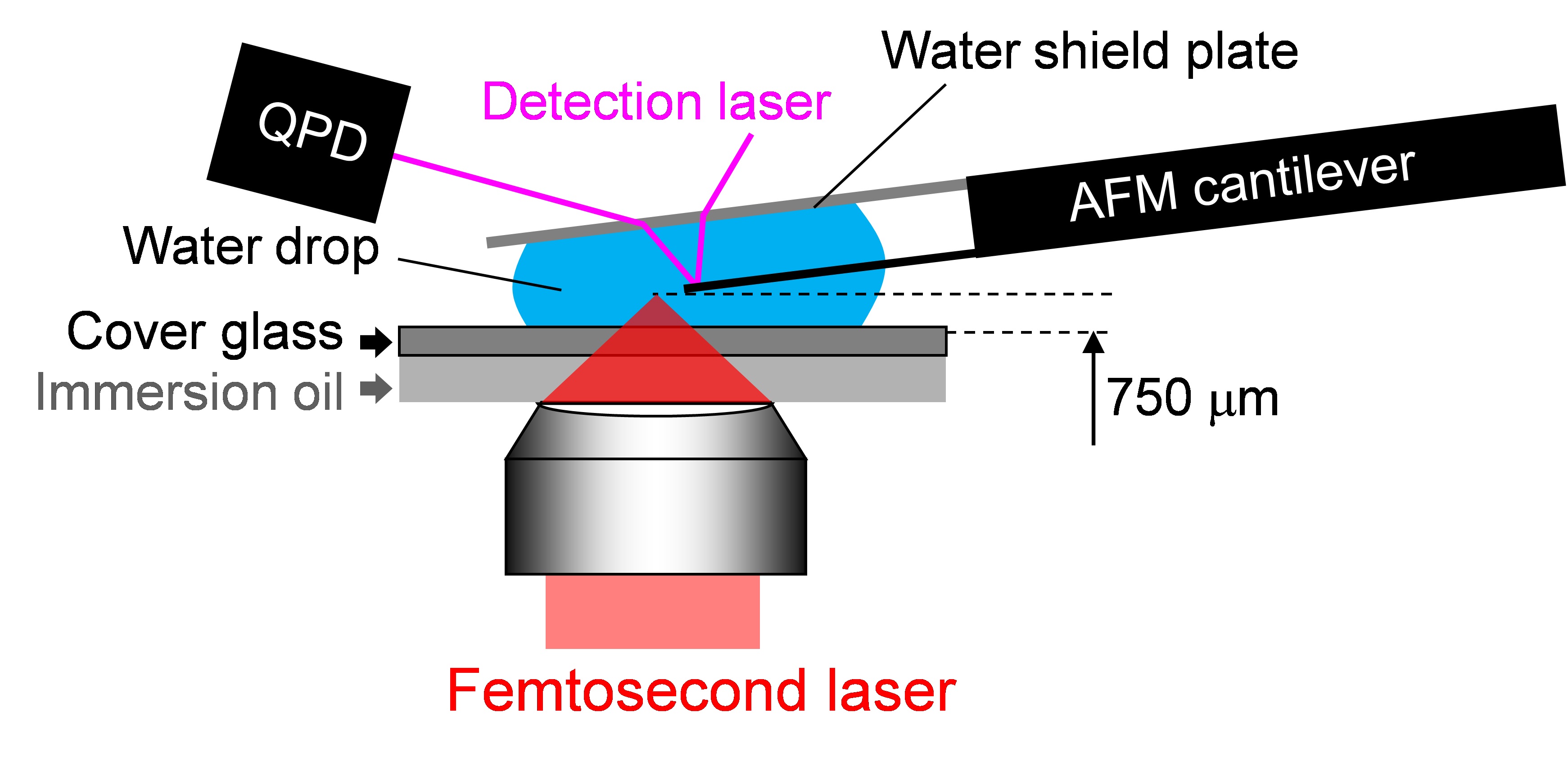
Figure 3. Experimental setup of AFM head on the microscope stage. The head of an AFM was mounted on the confocal microscope (Objective, PlanN100x) combined with the amplified Ti:Sapphire femtosecond laser system with the same setting as that mentioned in the former item. A tipless AFM cantilever is attached to the AFM head. The cantilever is placed between a water shield plate and cover glass, and immersed in distilled water or culture medium.
Note: The distance between the top of the cantilever and the laser focal point was tuned by a motorized stage equipped on the microscope. The normal function of the AFM system [feedback system between quadrant photo diode (QPD) and piezoelectric (PZT) motor] is deactivated. The signal of the QPD in the AFM head is directly monitored with an oscilloscope. The detailed settings are described in Hosokawa et al., 2011.
Software
- IGOR Pro 6.22J (WaveMetrics, https://www.wavemetrics.com)
- Visual Basic 6.0J (Microsoft, https://msdn.microsoft.com/en-us/vstudio/ms788229.aspx)
Note: Multivariate least-square fittings in procedures C and D were performed by a data analysis and graphing software, IGOR Pro 6.22J. For the least-square fitting in step C8, a user defined macro-program on the IGOR Pro was produced. The calculation program for step C9 was produced using a GUI programming language software, Visual Basic 6.0J.
Procedure
- Preparation of leaf section
- Pick a rosette leaf from a 3-week-old Arabidopsis plant and place in a 10 ml disposal syringe filled with distilled water (Figure 4B).
- Seal the top of the syringe by finger and pull plunger to deaerate the leaves (Figure 4B).
- Cut middle parts of the deaerated-leaf in quadrangle pieces with a side length of 1 cm and mount one piece of the deaerated-leaf section (Figure 4C) on a glass slide with distilled water, and cover with a cover slip (Figure 4D).
- Prepare two or three samples simultaneously.
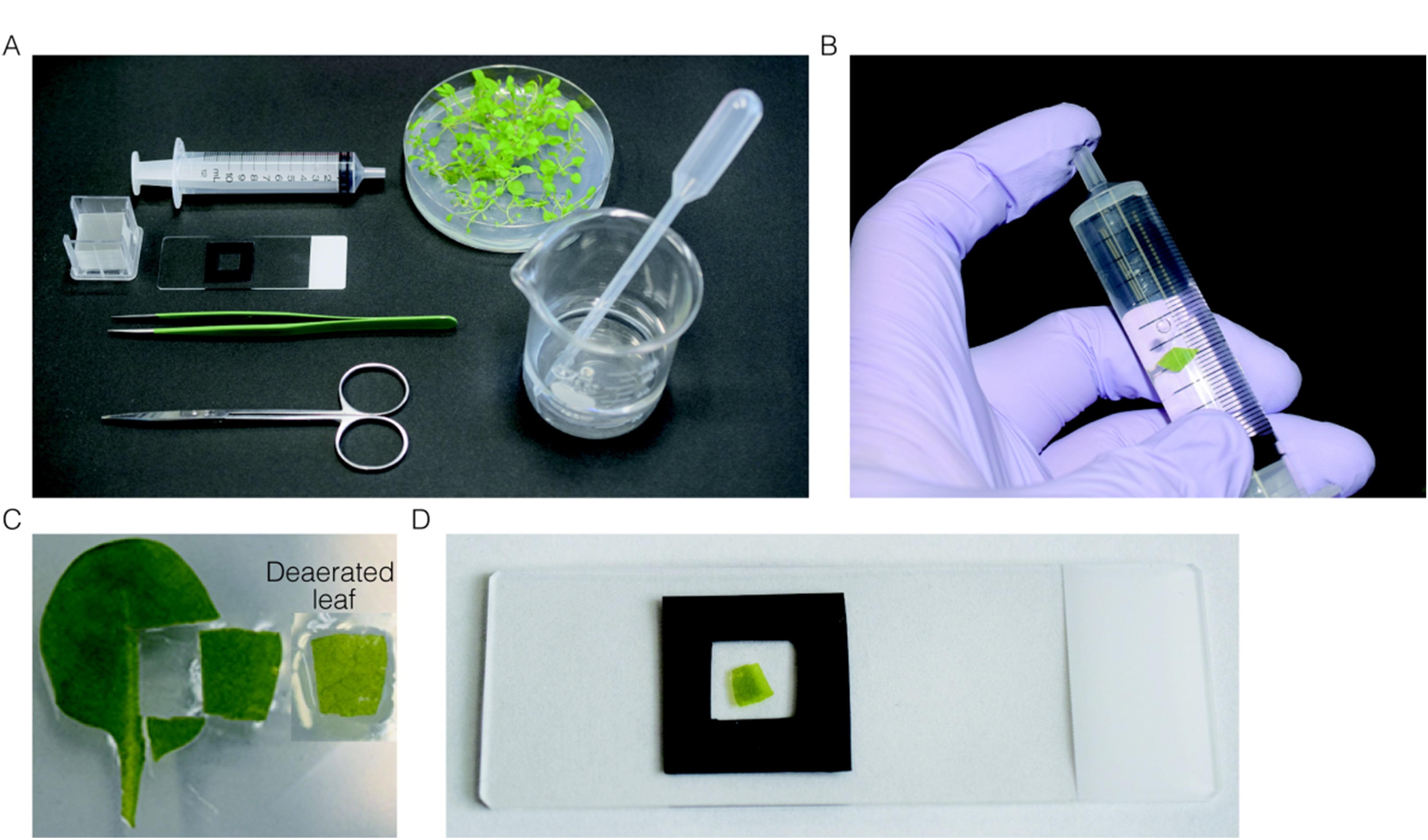
Figure 4. Preparation of leaf section for the estimation of the adhesion strength between peroxisomes and chloroplasts. A. All instruments used for preparation of leaf section. B. Deaeration of the leaves by pulling plunger while plugging the top of syringe. C. Leaf section used for observation. Before (left) and after (right) deaeration. D. Deaerated-leaf section mounted on the slide glass.
- Pick a rosette leaf from a 3-week-old Arabidopsis plant and place in a 10 ml disposal syringe filled with distilled water (Figure 4B).
- Estimation of detachment probability of peroxisomes from chloroplasts
- ncubate the leaf section sample (Figure 4D) in dark or light for 2 h in growth chamber at 22 °C to confirm the effect of light/darkness on the interaction. When investigating the contribution of actin filaments in the adhesion, add a drug, such as Latrunculin B and Cytochalasin D, to specifically depolymerase actin filaments.
- Mount the sample on the confocal microscope with facing adaxial side of the leaf section to the objective lens and focus on organelles in leaf palisade mesophyll cells (Figure 5). Adjust the focus into cells of the first inside layer, because the number of peroxisomes and chloroplast in the first inside layer are larger than those in the epidermis layer.
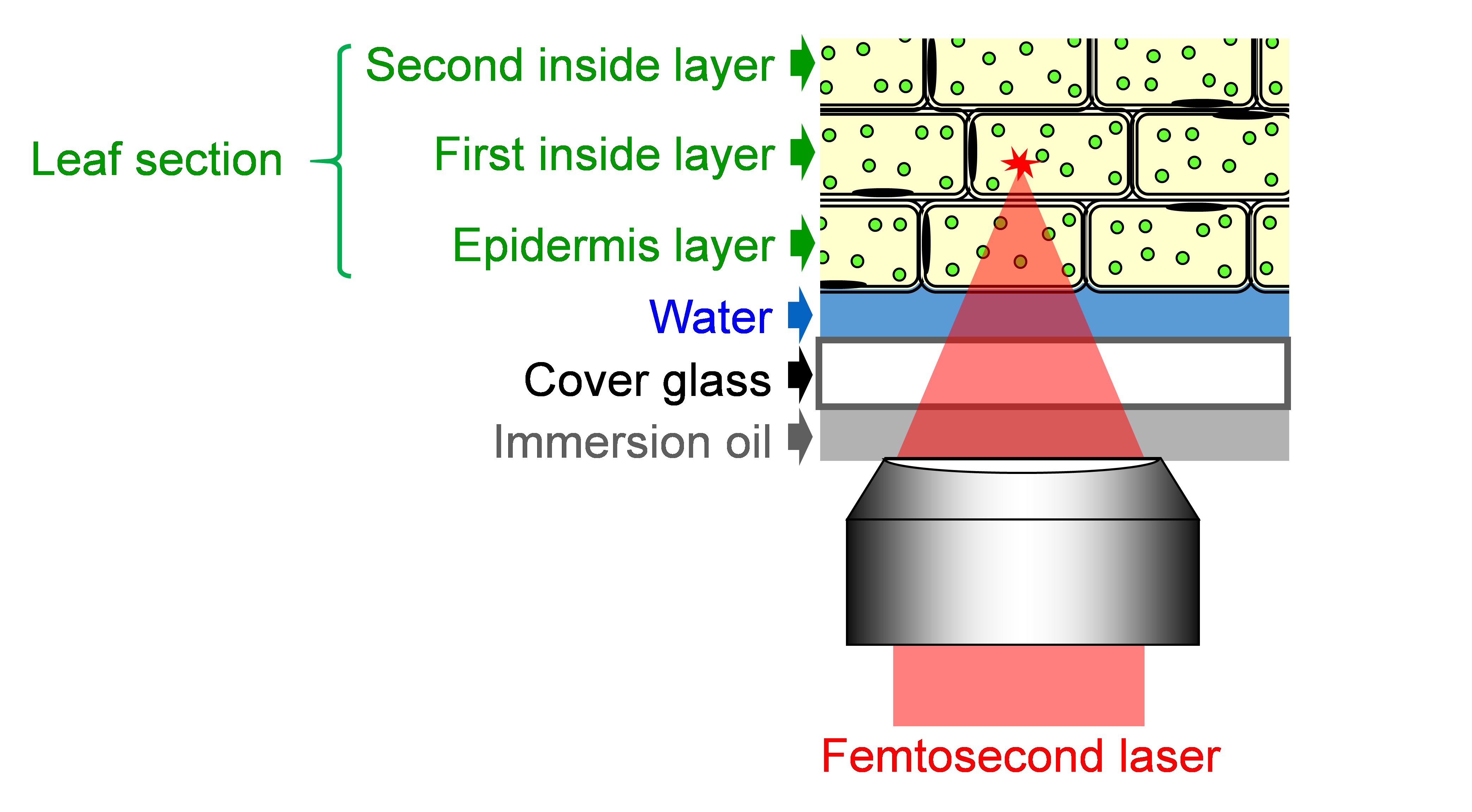
Figure 5. Schematic of focusing femtosecond laser into the leaf section - Tune the laser pulse energy by a λ/2 plate and polarizer shown in Figure 2.
- Tune the focal position of the femtosecond laser 5 μm away from the adhesion interface between a peroxisome and a chloroplast. Conduct the tuning by moving the motorized stage on the confocal microscope (precision: Less than 1 μm) with checking the distance between a peroxisome and a chloroplast by real time-sequential images of the CCD camera.
- Capture fluorescence image before laser shot (i.e., pictures on the left-hand side of Figure 6).
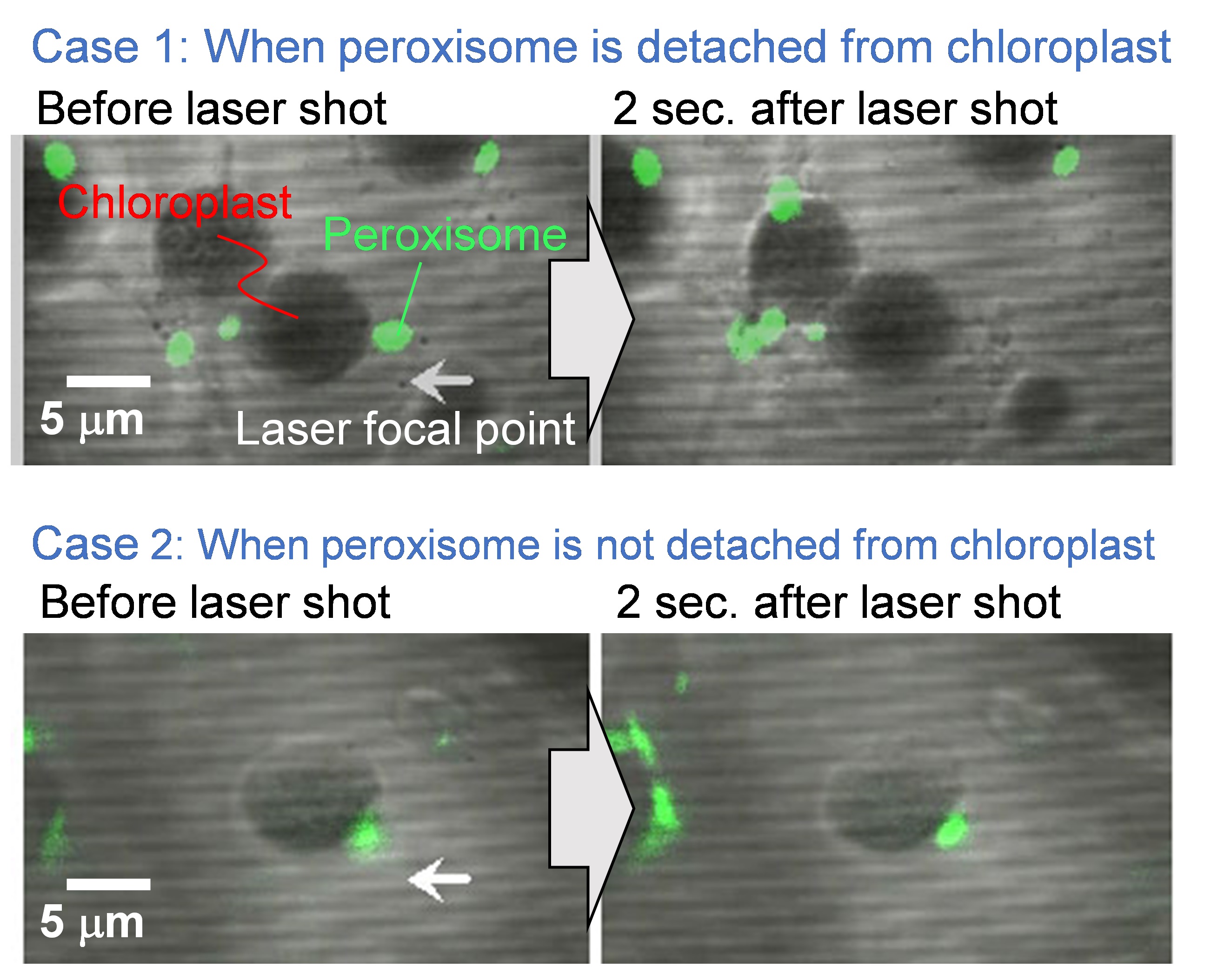
Figure 6. Representative experimental results before and after the laser exposure. Scale bar, 5 µm. - Shoot a single shot laser pulse by opening the mechanical shutter between the laser and the microscope, and load the laser-induced impulsive force to the targeted peroxisome.
- Capture fluorescence image after laser shot (i.e., pictures on the right-hand side of Figure 6).
- Check the existence of the targeted peroxisome. Cases 1 and 2 in Figure 6 indicate the representative examples that the peroxisome is detached and not detached from the chloroplast, respectively.
- Change the targeted peroxisome and repeat the steps B4-8.
- Calculate the detachment probability of peroxisomes as
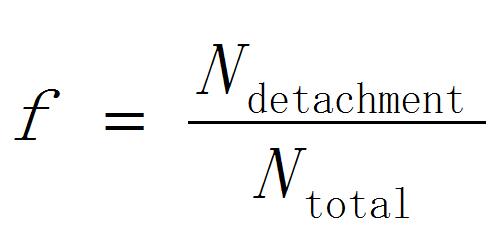 [1]
[1]
Where Ntotal is the total number of the accessed peroxisomes and Ndetachment is the number of peroxisomes detached from chloroplasts. - Repeat the steps B3-10 by changing the laser pulse energy.
Note: The experiment should be finished within 30 min after mounting the sample on the microscope, since light irradiation in the experiment affects the organelle motility and interaction. When the experiment is performed continuously, the sample should be replaced with a fresh sample after 30 min.
- ncubate the leaf section sample (Figure 4D) in dark or light for 2 h in growth chamber at 22 °C to confirm the effect of light/darkness on the interaction. When investigating the contribution of actin filaments in the adhesion, add a drug, such as Latrunculin B and Cytochalasin D, to specifically depolymerase actin filaments.
- Estimation of laser-induced impulsive force loaded on the peroxisome
- Mount the AFM head on the stage of the confocal microscope instead of the plant sample.
- Tune the laser pulse energy to that used in the estimation of the detachment probability of the peroxisome by the λ/2 plate and the polarizer shown in Figure 2.
- Tune the laser focal point of the femtosecond laser at 10 μm away from the tip of the AFM cantilever.
- Shoot a single shot laser pulse by opening the mechanical shutter between the laser and the microscope, and load the impulsive force on the AFM cantilever.
- Detect the oscillation of the AFM cantilever s(t), which is the difference between the top and bottom side voltages of the QPD, by the oscilloscope.
- Change optical axis (z axis) of the laser focal point by moving the microscope stage mechanically
- Repeat the steps C4-6.
- Perform the least-square fitting of oscillation data of the AFM cantilever by the equation:
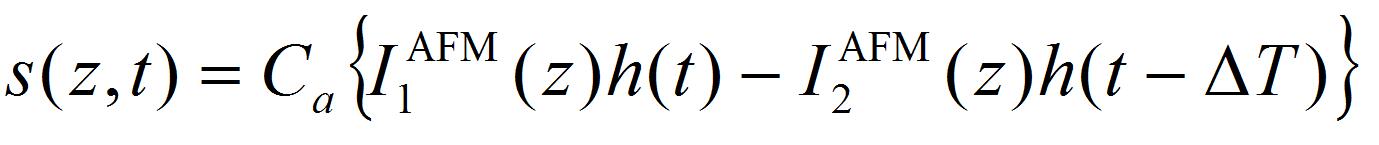 [2]
[2]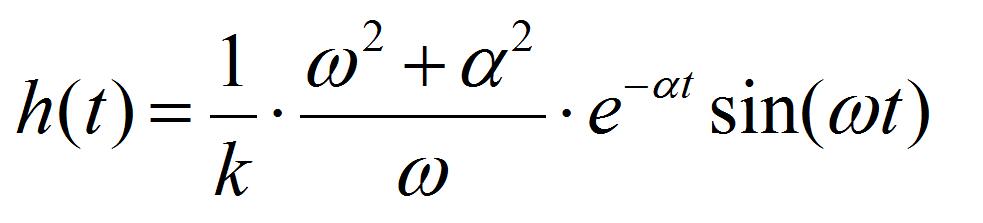 [3]
[3]
Where k is the spring constant of the AFM cantilever (44 N/m), ω the angular velocity of the oscillation, and a the damping constant. Ca is the coefficient to convert voltage difference of QPD to the position shift of the AFM cantilever, which is estimated from the force curve, on the condition that the cantilever is contacted to a hard material (Figure 7). I1AFM (z) and I2AFM (z) are the positive and negative impulses loaded on the AFM cantilever, respectively. ΔT is the time duration between positive and negative impulses. Variable parameters for the fitting are ω, α, I1AFM, I2AFM, and ΔT. The z position dependence of I1AFM, I2AFM is calculated by a macro program constructed with multivariate analysis software, IGOR Pro. The user defined functions of Eqs. [2] and [3] in the macro program and the analysis windows are shown in program 1 (supplementary file S1) and Figure 8. The z position dependence of I1AFM, I2AFM is estimated by fitting the calculation data (blue lines in Figure 8) to experimental data (red lines in Figure 8).
Note: Parameters, ω, α, and ΔT do not depend on the z axis. If they show a strong z axis dependency, there is the possibility of the presence of error(s) in the experimental system or the procedure.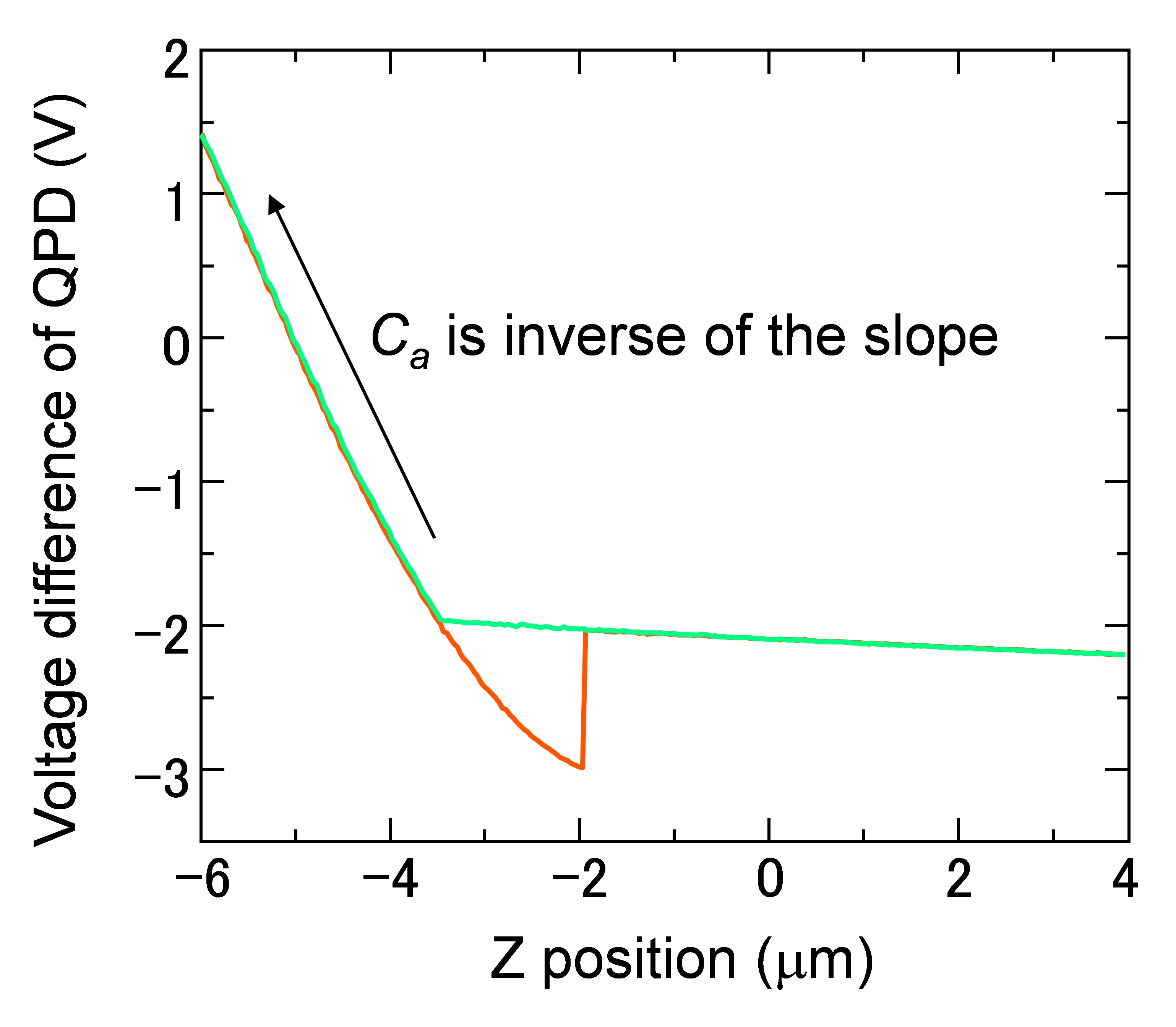
Figure 7. Force curve of the AFM cantilever. Ca was estimated from the slope in the region of Z position between -4 and -6 μm, where the cantilever contacts with the glass substrate.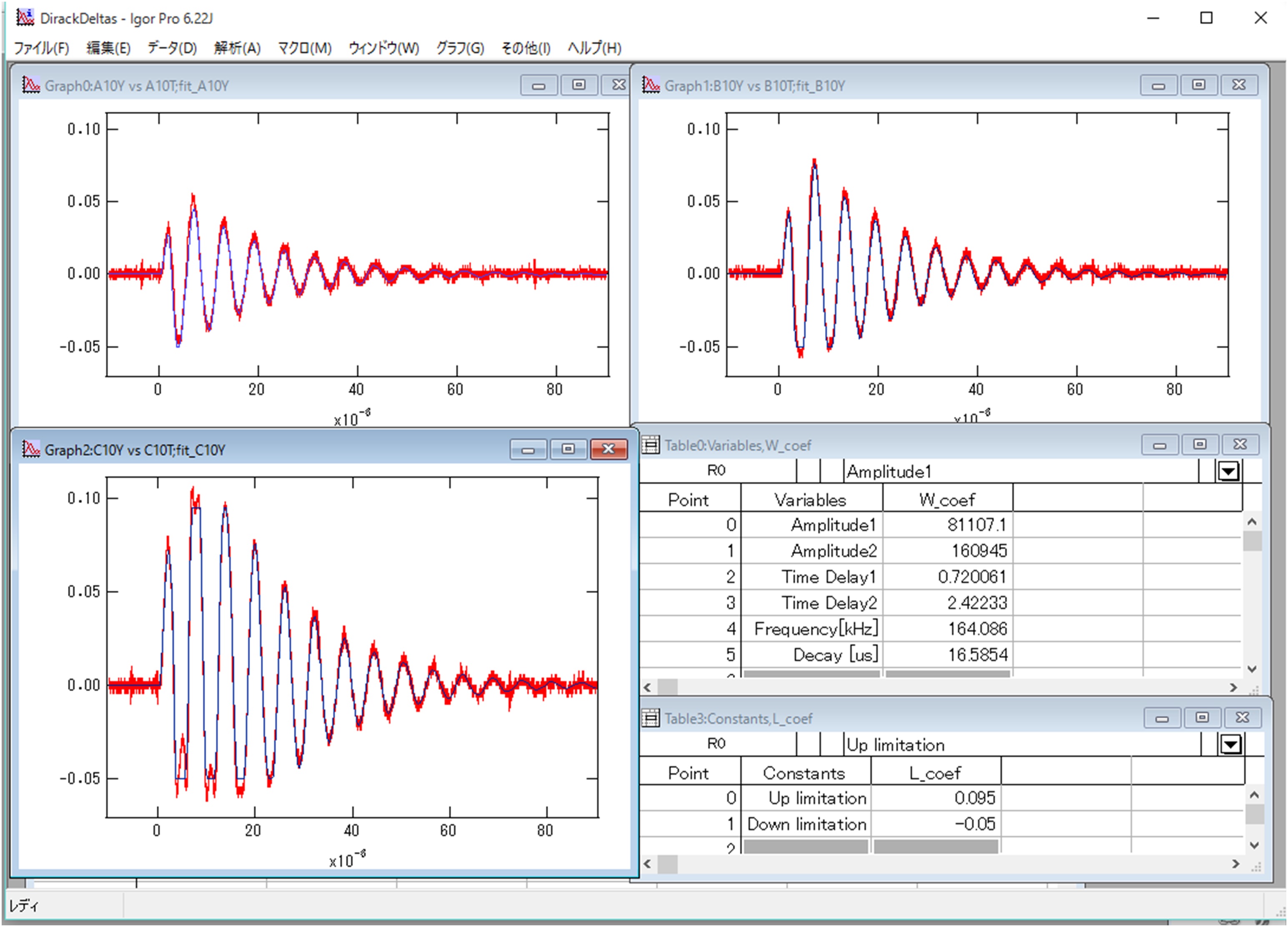
Figure 8. Analysis of oscillation data of the AFM cantilever by IGOR Pro. Graphs in the window are the Z position dependence of oscillations of the cantilever (Top-left graph: Z=-10 μm, Bottom-left: Z=-20 μm, Right-top: Z=-30 μm). Red lines are experimental data observed by the oscilloscope. Blue lines are results by the least-square fitting. The right-middle table shows variable parameters for the fitting (w_coef). The right-bottom table represents the constant values to define a range to conduct the analysis. They are used when the cantilever shifts over the detection limitation of QPD. - Perform least-square fitting of I1 AFM (z) and I2 AFM (z) by the equation:
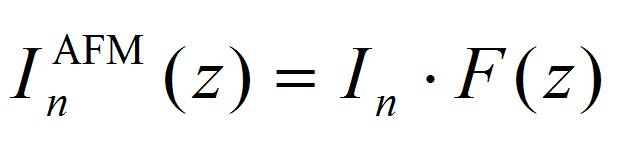 [4]
[4]
F(z) is the ratio of impulse loaded on the cantilever from the laser focal point, corresponding to integral of impulse at the small fraction Δs on the AFM cantilever. Details for Δs are given in the supporting information of the literature (Hosokawa et al., 2011). The calculation program and typical calculation results are shown in Program 2 (supplementary file S2) and Figure 9. The obtained fitting parameters of I1 and I2 are positive and negative impulses generated at the laser focal point.
Impulses of I1 AFM and I2 AFM estimated in the step C8 were plotted as a function of Z position, and compared with a suitable curve in the figure, corresponding to the experimental condition. Linear coefficients of I1 and I2 in Eq. [4] are determined by fitting the experimental data [I1 AFM (z) and I2 AFM (z)] to the calculation data [F(z)].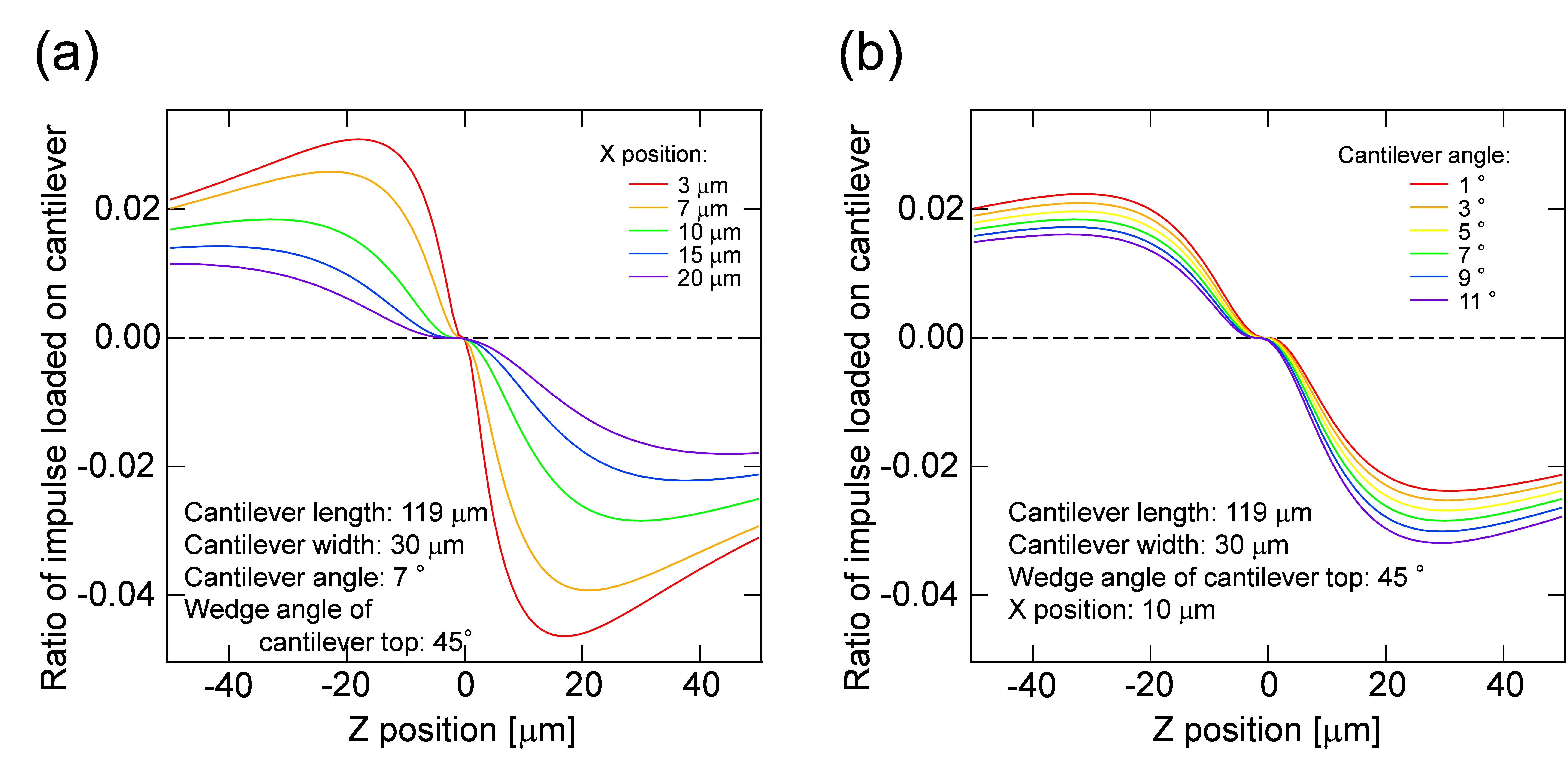
Figure 9. Ratio of impulses loaded on the cantilever and its dependency on the distance between the laser focal point and the cantilever top in the image plane (x position) (a) and the cantilever angle (b). Z position indicates the distance between the laser focal point and the cantilever top in the direction of optical axis. - Calculate the positive pressure loaded on the peroxisome by the equation:
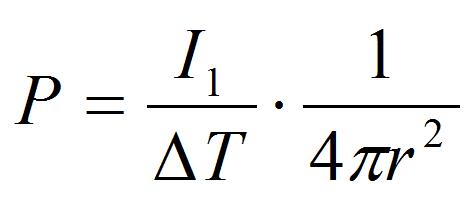 [5]
[5]
Where r is the distance between the laser focal point and the peroxisome (r = 5 μm in the procedure B). This pressure is an index of adhesion strength between peroxisome and chloroplast.
Note: It is better to make a calibration curve of P as a function of the laser pulse energy. The pulse energy dependency of P should be evaluated by repeating the steps B2-9 in the range used in the estimation of the detachment probability of the peroxisome. The P in procedure B will be precise when the estimation is performed from the calibration curve based on the pulse energy dependency.
- Mount the AFM head on the stage of the confocal microscope instead of the plant sample.
- Estimation of pressure to break adhesion between a peroxisome and a chloroplast
- Calculate Z-score, z, from the detachment probability of peroxisomes, f, estimated by procedure B. The relation between z and f is expressed as
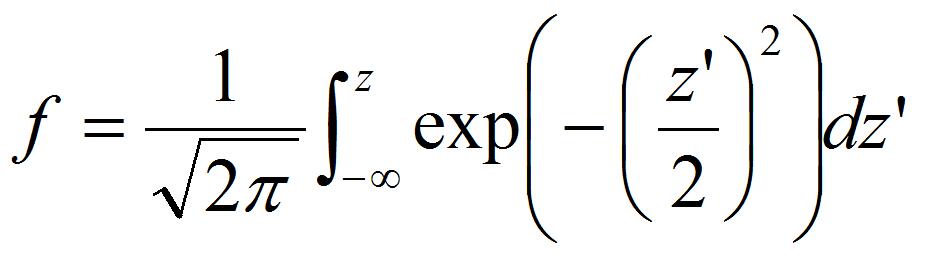 [6]
[6] - Plot the Z-score as a function of pressure loaded on the peroxisome P estimated by procedure C.
- Perform least-square fitting by equation
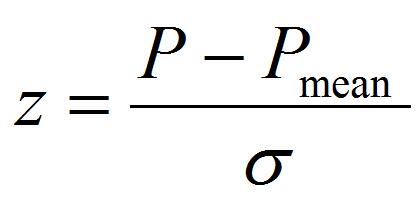 [7]
[7]
The fitting parameters of Pmean and σ are the mean value of pressure to detach the peroxisome and its standard deviation, which is an index of individual difference of adhesion strength.
- Calculate Z-score, z, from the detachment probability of peroxisomes, f, estimated by procedure B. The relation between z and f is expressed as
Acknowledgments
This work was supported by MEXT KAKENHI (Grant-in-Aid for Scientific Research on Innovative Areas) to M. N. (no. 22120007) and Y. H. (no. 22120010).
References
- Hosokawa, Y., Hagiyama, M., Iino, T., Murakami, Y. and Ito, A. (2011). Noncontact estimation of intercellular breaking force using a femtosecond laser impulse quantified by atomic force microscopy. Proc Natl Acad Sci U S A 108(5): 1777-1782.
- Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M. and Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol 43(3): 331-341.
- Oikawa, K., Mano, S., Yamada, K., Hosokawa, Y. and Nishimura, M. (2016). Measuring the interactions between peroxisomes and chloroplasts by in situ laser analysis. Bio-protocol 6(8): e1790.
- Oikawa, K., Matsunaga, S., Mano, S., Kondo, M., Yamada, K., Hayashi, M., Kagawa, T., Kadota, A., Sakamoto, W., Higashi, H., Watanabe, M., Mitsui, T., Shigemasa, A., Iino, T., Hosokawa, Y. and Nishimura M. (2015). Peroxisome - chloroplast physical interaction elucidated by in situ laser analysis. Nature Plants 15035.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hosokawa, Y., Iino, T., Oikawa, K., Mano, S., Yamada, K. and Nishimura, M. (2016). Quantification of the Adhesion Strength between Peroxisomes and Chloroplasts by Femtosecond Laser Technology. Bio-protocol 6(11): e1834. DOI: 10.21769/BioProtoc.1834.
Category
Plant Science > Plant cell biology > Cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











