- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of the Number of Peroxisomes
Published: Vol 4, Iss 21, Nov 5, 2014 DOI: 10.21769/BioProtoc.1284 Views: 10393
Reviewed by: Ru ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views

A Simple Protocol for Periodic Live Cell Observation of Flagellate Stages in the Lichen Alga Trebouxia
Enrico Boccato [...] Mauro Tretiach
Jan 20, 2026 187 Views
Abstract
This is the detailed protocol for the measurement of the number of peroxisomes described by Shibata et al. (2013). It is difficult to count the number of organelles in a cell because of the thickness of plant leaves. To overcome this challenge, protoplasts were isolated from leaves, and the number of peroxisomes per protoplast was counted. This method can be applied to other organelles such as mitochondria that are labeled with GFP or its derivatives.
Keywords: PeroxisomeMaterials and Reagents
- Arabidopsis plant expressing peroxisome-targeted GFP (Mano et al., 2002)
- Cellulase Onozuka R-10 (Yakult Honsha)
- Macerozyme R-10 (Yakult Honsha)
- MES
- Mannitol
- Enzyme solution (see Recipes)
- Wash solution (see Recipes)
Equipment
- Time tape (Time Med)
- Mending tape (3M)
- Aluminum foil
- 20 ml beaker
- Shaker
- Pasteur pipette
- Test tube
- Swing-out rotor centrifuge machine
- Confocal laser scanning microscope (Zeiss, model: LSM510 META )
- MAS-coated glass slide (super frost) (Matsunami Glass, catalog number: S9441 )
- Cover slip (24 x 60 No.1, 0.12-0.17 mm) (Matsunami Glass, catalog number: C024601 )
Software
- NIH ImageJ software 1.46 for Windows (http://imagej.nih.gov/ij)
Note: ImageJ is also available on Mac OS X and Linux.
Procedure
- Protoplast isolation
There are several methods for protoplast isolation. The following method is based on the Tape-Sandwich method (Wu et al., 2009) with slight modifications.- Collect the sixth and seventh leaves of 3-week-old Arabidopsis plants. Four leaves are enough for this experiment.
- Remove the leaf epidermis by peeling using two types of tape, Time tape and mending tape. Put a Time tape and a Mending tape on the upper side and the underside of a leaf, respectively. Gently remove the Mending tape to peel the epidermis of a leaf. Please see Wu et al. (2009) for more detail.
Note: Once you find a compatible tape-set, you can peel the epidermis easily and cleanly. However, there are inconvenient lots even in same products. - Put the Time tape with the leaf into a beaker with 5 ml of enzyme solution (see Recipes below).
- Wrap the beaker in aluminum foil to avoid light damage and gently shake (~40 rpm) for 30 min at room temperature. During this step, cell walls are digested with enzyme solution, and protoplasts are isolated from the leaves.
- Gently transfer the protoplast-containing solution into a test tube by decantation.
- Spin down at 100 x g for 1 min at room temperature using a swing-out rotor.
- Discard the supernatant and gently add 2 ml of wash solution (see Recipes below).
- Spin down at 100 x g for 1 min at room temperature using a swing-out rotor.
- Repeat the steps A7-8 once again.
- Discard the supernatant and gently add 1 ml of wash solution.
- Collect the sixth and seventh leaves of 3-week-old Arabidopsis plants. Four leaves are enough for this experiment.
- Acquisition of protoplast images by microscope
- Carefully put the protoplast solution on a MAS-coated glass slide using Pasteur pipette.
- Gently put a cover slip on the slide.
- Mount prepared specimen on microscope and focus. Look for protoplasts which are 40- 55 µm in diameter.
- Acquire the Z-series images of a protoplast from the upper to the lower (or middle) position.
Our condition using LSM510 is as follows: Argon laser 488 nm, Emission filter BP505-550, 40x dry objective (Plan-Neofluar 40x/0.75), 1 x zoom, pixel size 0.22 µm. The step size between each focal plane is 0.887 µm. To obtain images quickly, minimize the scanning area using “ROI”. - Execute Z-projection to compile the acquired sequential images in one image by operating software of LSM510 META.
Note: The observation of protoplasts should be finished within one hour after the isolation, since isolated protoplasts become quickly unhealthy and easily burst. Z projection can also be done in ImageJ: Image>Stacks>Z project>Maximum intensity projection.
- Carefully put the protoplast solution on a MAS-coated glass slide using Pasteur pipette.
- Peroxisome counting
- Launch an ImageJ software and open the Z-projection image.
- Calculate the diameter of each protoplast using the menu of Analyze>Measure.
Note: You need to carry out “Set scale” before calculation. Click “Straight” tool and trace the scale bar of opened image. Go Analyze>Set Scale, input the distance in the box “Known distance”, check Global, and click OK to close the window. - Go Plugins>Analyze>Cell Counter and count up peroxisomes manually (Figure 1).
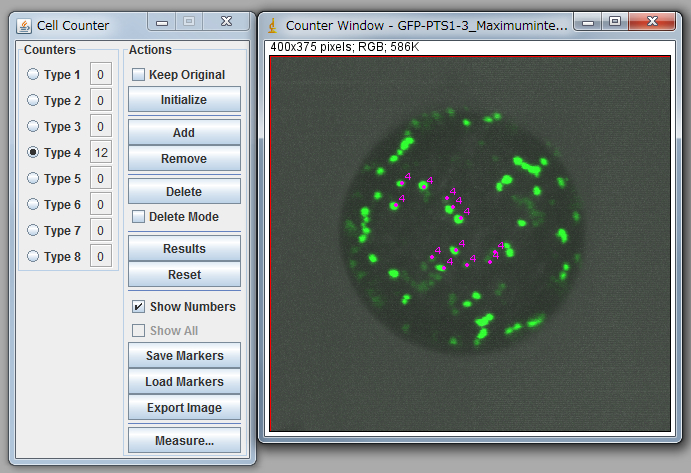
Figure 1. Peroxisome counting using ImageJ.
Note: For statistical analysis, we counted 15-20 protoplasts. See Table 1 and 2. It is necessary to confirm that the average of diameters of protoplasts between the control and your targets are nearly equal. In the case of table 1, a protoplast with small diameter (sample 1) was eliminated from the analysis due to too small diameter. Carefully discriminate between organelles and noises because signals from the bottom side are low, and some of them are observed as a connected signal. To minimize the subjectivity of people, it is recommended that multiple people independently count peroxisomes against same data as a blind-test.
Table 1. Diameters and numbers of peroxisome in a protoplast*1A protoplast with small diameter, such as sample 1, was eliminated from the analysis due to too small diameter.Diameter Number sample 1*1 29.8 31 sample 2 36.1 29 sample 3 36.6 25 sample 4 37.7 47 sample 5 40.4 42 sample 6 42.0 40 sample 7 43.5 59 sample 8 44.0 42 sample 9 45.0 58 sample 10 45.5 49 sample 11 45.9 54 sample 12 45.9 81 sample 13 46.0 45 sample 14 47.0 52 sample 15 47.3 33 sample 16 47.6 66 sample 17 47.9 52
Table 2. The average of diameters and numbers of peroxisome in a protoplastSD and SE represent standard deviation and standard error, respectively.Diameter (D) Number n 35<D<50 43.6 48.4 16 SD 5.1 14.2 SE 1.3 3.6
- Launch an ImageJ software and open the Z-projection image.
Recipes
- Enzyme solution
1% (v/w) cellulase Onozuka R-10
0.25% (v/w) macerozyme R-10
0.4 M mannitol
20 mM MES (pH 5.7) - Wash solution
0.4 M mannitol
20 mM MES (pH 5.7)
Acknowledgments
This is the detailed protocol for the measurement of the number of peroxisomes described by Shibata et al. (2013).
References
- Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M. and Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol 43(3): 331-341.
- Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., Hayashi, M., Sakamoto, W., Ohsumi, Y. and Nishimura, M. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25(12): 4967-4983.
- Wu, F. H., Shen, S. C., Lee, L. Y., Lee, S. H., Chan, M. T. and Lin, C. S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shibata, M., Oikawa, K., Mano, S. and Nishimura, M. (2014). Measurement of the Number of Peroxisomes. Bio-protocol 4(21): e1284. DOI: 10.21769/BioProtoc.1284.
Category
Plant Science > Plant cell biology > Cell structure
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










