- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
GUS Staining of Guard Cells to Identify Localised Guard Cell Gene Expression
Published: Vol 7, Iss 14, Jul 20, 2017 DOI: 10.21769/BioProtoc.2446 Views: 18078
Reviewed by: Scott A M McAdamHonghong WuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Inoculation of Maize with Sugarcane Mosaic Virus Constructs and Application for RNA Interference in Fall Armyworms
Iram Gull and Georg Jander
Jul 20, 2023 2206 Views

A Microplate-Based Expression Monitoring System for Arabidopsis NITRATE TRANSPORTER2.1 Using the Luciferase Reporter
Yoshiaki Ueda and Shuichi Yanagisawa
Dec 5, 2024 1784 Views

A Novel Gene Stacking Method in Plant Transformation Utilizing Split Selectable Markers
Guoliang Yuan [...] Xiaohan Yang
Feb 20, 2025 1979 Views
Abstract
Determination of a gene expression in guard cells is essential for studying stomatal movements. GUS staining is one means of detecting the localization of a gene expression in guard cells. If a gene is specially expressed in guard cells, the whole cotyledons or rosette leaf can be used for GUS staining. However, if a gene is expressed in both mesophyll and guard cells, it is hard to exhibit a clear expression of the gene in guard cells by a GUS staining image from leaf. To gain a clear guard cell GUS image of small G protein ROP7, a gene expressed in both mesophyll and guard cells, we peeled the epidermal strips from the leaf of 3-4 week-old plants. After removing the mesophyll cells, the epidermal strips were used for GUS staining. We compared the GUS staining images from epidermal strips or leaf of small G protein ROP7 and RopGEF4, a gene specifically expressed in guard cells, and found that GUS staining of epidermal strips provided a good method to show the guard cell expression of a gene expressed in both mesophyll and guard cells. This protocol is applicable for any genes that are expressed in guard cells of Arabidopsis, or other plants that epidermal strips can be easily peeled from the leaf.
Keywords: Guard cellsBackground
Stomatal movements regulate the gas exchange between plants and environment, therefore, it is important to reveal the mechanism of the opening or closure of stomata. Determination of the guard cell expression of a gene is essential for studying its role in stomatal movements. There are several ways to identify the expression of a gene in guard cells. One way is to check the RNA expression of a gene in guard cells by RT-PCR (Jeon et al., 2008; Takimiya et al., 2013). To do so, the protoplasts of mesophyll and guard cells need to be separated. Another way is to check the GUS signal in guard cells of the transgenic plant expressing GUS driven by a gene’s native promoter. In some reports, the evidence of both RNA expression and GUS signal in guard cells were provided (Zheng et al., 2002; Jeon et al., 2008). As for the GUS signal in guard cells, if a gene is specifically expressed in guard cells, like OST1, MYB60, ROP11 and RopGEF4, a distinguished GUS signal in guard cells can be obtained from a GUS staining image with whole leaf (Mustilli et al., 2002; Li et al., 2012; Li and Liu, 2012; Rusconi et al., 2013). However, if a gene is expressed in both mesophyll and guard cells, like ROP10 and RopGEF2, the GUS signal in guard cells is hard to be distinguished from the mesophyll background (Zheng et al., 2002; Li and Liu, 2012). After GUS staining procedure, the leaf will become soft, and it is very difficult to peel the epidermal strips. Therefore, just after the leaf was excised from the plants, we peeled the epidermal strips from the leaf, and the strips were used for GUS staining after the mesophyll cells were removed. By this method, we obtained a clear guard cell GUS image of ROP7, a gene expressed in both mesophyll and guard cells.
Materials and Reagents
- Pipette tips
- 1.5 ml Eppendorf tubes
- Sterilized filter paper
- Plastic Petri dishes for plant culture
- Slide
- Cover glass
- 0.45 micron filter
- Aluminum foil
- Arabidopsis thaliana seeds of ROP7pro:GUS and RopGEF4pro:GUS lines
- 100%, 75%, 40%, 20%, 10%, 5% ethanol in water
- 50% glycerol (Sangon Biotech, catalog number: A100854 )
- 100% methanol
- 37% hydrochloric acid (12 N)
- Sodium hydroxide (AMRESCO, catalog number: 0583 )
- Ethylenediaminetetraacetate acid (EDTA) (AMRESCO, catalog number: 0322 )
- Triton X-100 (AMRESCO, catalog number: 0694 )
- Potassium ferricyanide (AMRESCO, catalog number: 0713 )
- Potassium ferrocyanide (Sigma-Aldrich, catalog number: P9387 )
- X-Gluc (Sigma-Aldrich, catalog number: B5285 )
- Dimethylformamide (AMRESCO, catalog number: 0464 )
- Sodium dihydrogen phosphate (NaH2PO4·H2O) (Sigma-Aldrich, catalog number: S9638 )
- Disodium hydrogen phosphate (Na2HPO4·7H2O) (Sigma-Aldrich, catalog number: 431478 )
- 20% methanol in 0.24 N hydrochloric acid (see Recipes)
- 60% ethanol in 7% sodium hydroxide (see Recipes)
- GUS staining solution (see Recipes)
Equipment
- Pipetman 100 μl (Gilson, catalog number: F123615 )
- Pipetman 1,000 μl (Gilson, catalog number: F123602 )
- Tweezers
- Brush pen
- Plant growth chamber (Percival Scientific, model: CU-36L5 ) and greenhouse
- Pots
- Incubator at 37 °C (SANFA, model: DNP-9052 )
- Microscope (ZEISS, model: Axio Imager A1 )
- Water purification system (deionized water) (EMD Millipore, model: Elix® Essential , 5 L)
Procedure
- Arabidopsis plants were grown according to Arabidopsis growing guide http://www.bio-protocol.org/e126.
- Harvest the fully expanded rosette leaves from 3-4 week-old plants, and peel the epidermal strips from the abaxial surface of the leaf (Figure 1A), remove the mesophyll tissue from the strips with a brush pen (Figure 1B). Epidermal strips or leaf are then immersed in 1.5 ml Eppendorf tubes with GUS staining solution (see Recipes), and keep the tubes in an incubator at 37 °C under darkness for 16-20 h.
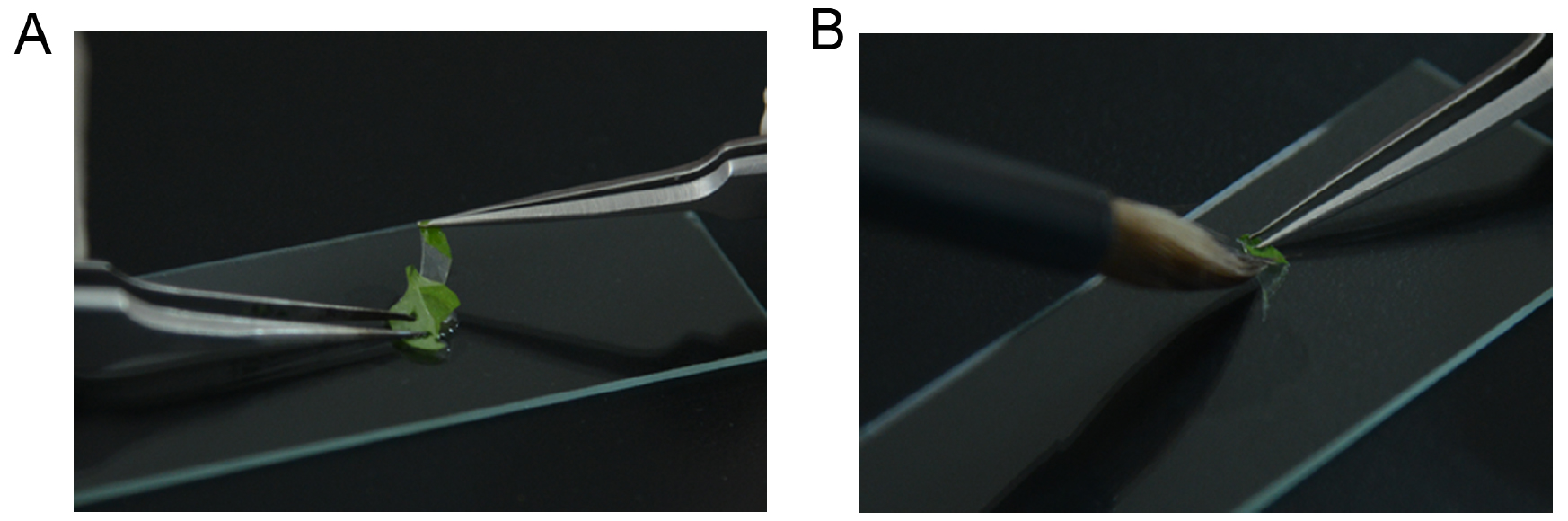
Figure 1. Images show the peeling of the epidermis from the abaxial surface of leaf (A) and removing the mesophyll tissue from epidermal strip by a brush pen (B) - Replace the supernatant with 1 ml 100% ethanol and incubate for 30 min, repeat this step for 2 times.
- Replace the supernatant with 1 ml 75% ethanol, and incubate for 5 min.
- Replace the supernatant with 1 ml 20% methanol in 0.24 N hydrochloric acid, and incubate at 37 °C for 15 min.
- Replace the supernatant with 1 ml 60% ethanol in 7% sodium hydroxide, and incubate for 15 min.
- Replace the supernatant with 1 ml 40% ethanol, and incubate for 5 min; replace the supernatant with 20%, 10% and 5% ethanol gradually each for 5 min. After all these steps, the leaf tissue will turn to white, and keep the samples in 5% ethanol.
- Put one drop of 50% glycerol on the slide, and place the samples in the glycerol solution; put the cover glass on the samples.
- Observe the samples under a microscope and take pictures:
- Switch on the light source of the microscope, and rotate the nosepiece to the lowest-power objective.
- Place the slide on the stage of the microscope, and move the slide to center the specimen under the lens.
- Adjust the large coarse focus knob until the specimen is in focus.
- Scan the slide at low power objective (4x or 10x objective), center the part of the specimen that one wants to view, and then rotate the nosepiece to the 20x or 40x objective.
- Adjust the small fine focus knob until the cells are in focus, and adjust the diaphragm until the cells have clear and sharp contrast.
- Take a picture with the CCD.
- Switch on the light source of the microscope, and rotate the nosepiece to the lowest-power objective.
Data analysis
As shown in Figure 2, the GUS signals in guard cells were very clear in the images of GUS staining from both the leaf (Figure 2C) and the epidermal strips (Figure 2D) of RopGEF4pro:GUS lines. The results were consistent with the specific expression of RopGEF4 in guard cells reported by Li and Liu (2012). However, the GUS signal was not clear when stained the whole leaf of ROP7pro:GUS line because of the strong background of mesophyll tissue (Figure 2A). When the epidermal strips were separated from mesophyll cells, we can see a clear guard cell GUS signal after GUS staining (Figure 2B). When this method is applied, it is better to use 3-4 week old plants of Arabidopsis, because the epidermal strips are easily peeled from the mesophyll tissue during this time. This method is applicable for any genes that are expressed in guard cells of Arabidopsis, or other plants that epidermal strips can be easily peeled from the leaf.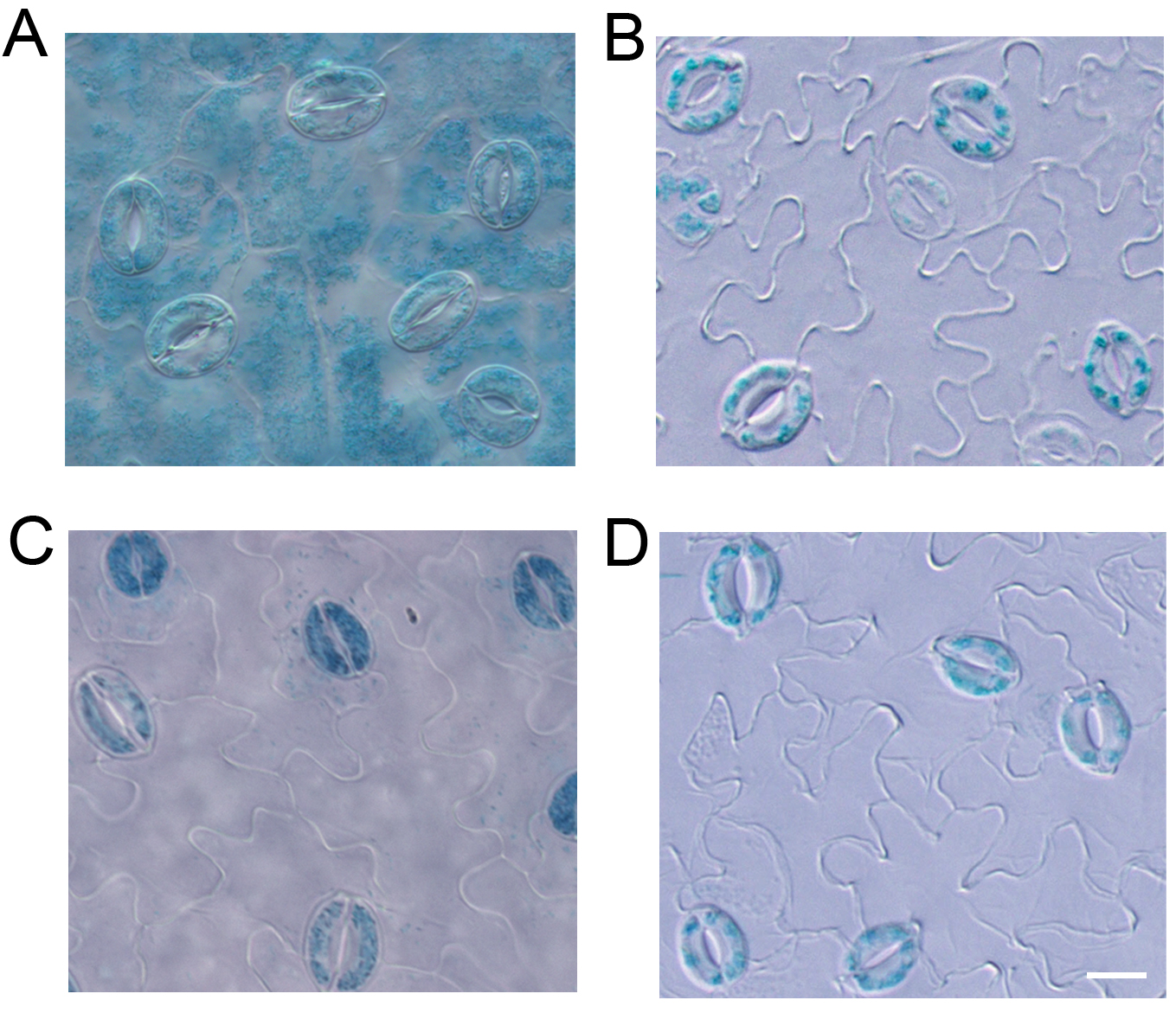
Figure 2. GUS staining with the leaf (A) and epidermal strips (B) from ROP7pro:GUS line, and leaf (C) and epidermal strips (D) from RopGEF4pro:GUS line. Scale bar = 10 μm.
Notes
- Please be very careful to keep the epidermal strips in the tubes in each step, try to avoid losing the epidermal strips when removing the supernatant.
- The epidermal strips became soft and easily broken after GUS staining procedure, so be very careful to flatten the epidermal strip on the slide.
Recipes
- 20% methanol in 0.24 N hydrochloric acid
Mix:
20 ml of 100% methanol
2 ml of 37% hydrochloric acid (12 N)
Deionized water 78 ml - 60% ethanol in 7% sodium hydroxide
Dissolve 28 g sodium hydroxide in deionized water to 100 ml to make 28% sodium hydroxide
Mix:
60 ml of 100% ethanol
25 ml of 28% sodium hydroxide
15 ml of deionized water - GUS staining solution
100 mM sodium phosphate buffer (pH = 7.0)
10 mM EDTA
0.1% Triton X-100
0.5 mM potassium ferricyanide
0.5 mM potassium ferrocyanide
1 mM X-Gluc- Dissolve 0.8892 g X-Gluc in 100 ml dimethylformamide to make a 20 mM stock and keep in the dark at -20 °C
- Make 200 mM sodium phosphate buffer (pH 7.0): dissolve 1.08 g sodium dihydrogen phosphate (NaH2PO4·H2O) and 3.27 g disodium hydrogen phosphate (Na2HPO4·7H2O) in deionized water up to 100 ml
- Mix:
50 ml of 200 mM sodium phosphate buffer (pH 7.0)
2 ml of 500 mM EDTA solution
0.5 ml of 20% (w/v) Triton X-100
1 ml of 50 mM potassium ferricyanide
1 ml of 50 mM potassium ferrocyanide
5 ml of 20 mM X-Gluc - Bring up to 100 ml with deionized water
- Filter-sterilize (0.45 micron filter) and keep at 4 °C wrapped with aluminum foil
- Dissolve 0.8892 g X-Gluc in 100 ml dimethylformamide to make a 20 mM stock and keep in the dark at -20 °C
Acknowledgments
This work was supported by the National Science Foundation of China (Grant No. 31571450 to Y.-L.C.). The protocol was modified according to Pecinka et al. (2009).
References
- Jeon, B. W., Hwang, J. U., Hwang, Y., Song, W. Y., Fu, Y., Gu, Y., Bao, F., Cho, D., Kwak, J. M., Yang, Z. and Lee, Y. (2008). The Arabidopsis small G protein ROP2 is activated by light in guard cells and inhibits light-induced stomatal opening. Plant Cell 20(1): 75-87.
- Li, Z. and Liu, D. (2012). ROPGEF1 and ROPGEF4 are functional regulators of ROP11 GTPase in ABA-mediated stomatal closure in Arabidopsis. FEBS Lett 586(9): 1253-1258.
- Li, Z., Kang, J., Sui, N. and Liu, D. (2012). ROP11 GTPase is a negative regulator of multiple ABA responses in Arabidopsis. J Integr Plant Biol 54(3): 169-179.
- Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F. and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14(12): 3089-3099.
- Pecinka, A., Rosa, M., Schikora, A., Berlinger, M., Hirt, H., Luschnig, C. and Mittelsten Scheid, O. (2009). Transgenerational stress memory is not a general response in Arabidopsis. PLoS One 4(4): e5202.
- Rusconi, F., Simeoni, F., Francia, P., Cominelli, E., Conti, L., Riboni, M., Simoni, L., Martin, C. R., Tonelli, C. and Galbiati, M. (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J Exp Bot 64(11): 3361-3371.
- Takemiya, A., Sugiyama, N., Fujimoto, H., Tsutsumi, T., Yamauchi, S., Hiyama, A., Tada, Y., Christie, J. M. and Shimazaki, K. (2013). Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat Commun 4: 2094.
- Zheng, Z. L., Nafisi, M., Tam, A., Li, H., Crowell, D. N., Chary, S. N., Schroeder, J. I., Shen, J. and Yang, Z. (2002). Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14(11): 2787-2797.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Liu, Z., Wang, W., Zhang, C., Zhao, J. and Chen, Y. (2017). GUS Staining of Guard Cells to Identify Localised Guard Cell Gene Expression. Bio-protocol 7(14): e2446. DOI: 10.21769/BioProtoc.2446.
- Wang, W., Liu, Z., Bao, L. J., Zhang, S. S., Zhang, C. G., Li, X., Li, H. X., Zhang, X. L., Bones, A. M., Yang, Z. B. and Chen, Y. L. (2017). The RopGEF2-ROP7/ROP2 Pathway Activated by phyB Suppresses Red Light-Induced Stomatal Opening. Plant Physiol 174(2): 717-731.
Category
Plant Science > Plant molecular biology > DNA > Gene expression
Molecular Biology > DNA > Gene expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link