- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction and Quantification of Polyphosphate in the Budding Yeast Saccharomyces cerevisiae
Published: Vol 6, Iss 14, Jul 20, 2016 DOI: 10.21769/BioProtoc.1874 Views: 12371
Reviewed by: Fanglian HeYanjie LiRosario Gomez-Garcia

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Glutamylation Inhibition of Ubiquitin Modification and Phosphoribosyl-Ubiquitin Ligation Mediated by Legionella pneumophila Effectors
Alan G. Sulpizio [...] Yuxin Mao
Nov 5, 2020 3931 Views

Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
Dina A. Abdulrahman and Michael Veit
Apr 5, 2025 1854 Views
Abstract
Inorganic polyphosphate (polyP) is a linear polymer present in both prokaryotic and eukaryotic organisms and made from three to hundreds of orthophosphate residues linked by phosphoanhydride bonds. The biological role of this molecule goes beyond serving as Pi store or energy source to replace ATP. For instance, in yeast polyP levels have been related to stress adaptation and this molecule has been shown to be the substrate for polyphosphorylation of proteins. Here we describe two different methods to purify polyP from the yeast Saccharomyces cerevisiae and the subsequent protocol to quantify polyP levels by spectrophotometrically measuring the Pi generated upon enzymatic hydrolysis of purified polyP. It must be noted that the purification protocol used greatly influences the polyP values obtained.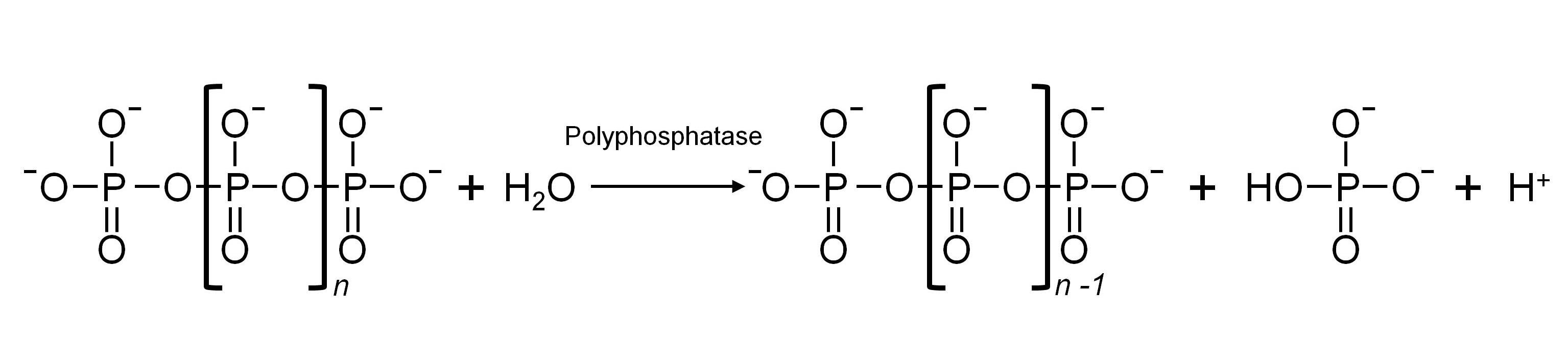
Figure 1. Enzymatic hydrolysis of polyP
Materials and Reagents
- 1.5 ml screw cap tubes
- Silica-gel columns from QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28706 )
- Inoculation loop
- Magnetic stirrer
- 96-Well assay microplate non-treated clear polystyrene (Thermo Fisher Scientific, NuncTM MicroWellTM, catalog number: 269620 )
- Yeast Saccharomyces cerevisiae
- Yeast extract, peptone (YP) base medium (Conda, catalog number: 1511 )
- AE buffer
- Phenol solution (Sigma-Aldrich, catalog number: P4557 )
- Sodium dodecylsulfate (SDS) (Panreac AppliChem, catalog number: A7249 )
- Chloroform (CHCl3) (Merck Millipore, catalog number: 102445 )
- RNase A (100 mg/ml) (QIAGEN, catalog number: 19101 )
- DNase I (Roche Diagnostics, catalog number: 10104159001 )
- Sodium acetate trihydrate (NaC2H3O2·3H2O) (Merck Millipore, catalog number: 106267 )
- Ethanol absolute (CH3CH2OH) (Panreac Applichem, catalog number: 131086 )
- Milli-Q water
- 98% sulfuric acid (H2SO4) (Merck Millipore, catalog number: 112080 )
- Sodium hydroxide (NaOH) (Panreac Applichem, catalog number: 131687 )
- Tris (2-amino-2-hydroxymethyl-propane-1,3-diol) buffer (Panreac Applichem, catalog number: A1379 )
- Neutral red (Sigma-Aldrich, catalog number: N4638 )
- Sodium iodide (NaI) (Sigma-Aldrich, catalog number: 383112 )
- Acetic acid glacial (CH3COOH) (Panreac Applichem, catalog number: 131088 )
- Disodium ethylenediaminetetraacetate 2-hydrate (EDTA) (Panreac Applichem, catalog number: 131669 )
- Recombinant Ppx1 (rPpx1)
Note: Ppx1 is a S. cerevisiae exopolyphosphatase that hydrolyses polyphosphate into Pi residues. It was obtained from E. coli containing a plasmid-borne, His-tagged version of the PPX1 gene, as described in (Ruiz et al., 2001), after one-step affinity purification with HisTrapTM HP columns (GE Healthcare Life Sciences ). - Potassium dihydrogen phosphate (KH2PO4) (Merck Millipore, catalog number: 104877 )
- Sodium phosphate glass Type 45 (polyP45) (Sigma-Aldrich, catalog number: S4379 )
- Sodium tripolyphosphate (polyP3) (Sigma-Aldrich, catalog number: 238503 )
- Malachite green oxalate salt (Sigma-Aldrich, catalog number: M9015 )
- Polyvinyl alcohol (Sigma-Aldrich, catalog number: P1763 )
- D-Glucose monohydrate (Panreac Applichem, catalog number: A1349 )
- Ammonium acetate (NH4C2H3O2) (Panreac Applichem, catalog number: 131114 )
- Magnesium acetate tetrahydrate [Mg(CH3COO)2·4H2O] (Merck Millipore, catalog number: 105819 )
- Sodium chloride (NaCl) (Panreac Applichem, catalog number: 131659 )
- Ammonium heptamolybdate tetrahydrate [(NH4)6Mo7O24·4H2O] (Sigma-Aldrich, catalog number: 09878 )
- YPD (see Recipes)
- 3 M sodium acetate (see Recipes)
- 0.5 M EDTA (see Recipes)
- Buffer AE (see Recipes)
- 10% SDS (see Recipes)
- RNase A (10 mg/ml) (see Recipes)
- DNase I (10 mg/ml) (see Recipes)
- 70% ethanol (see Recipes)
- 0.1% neutral red solution (see Recipes)
- 1 M Tris-HCl (pH 7.5) supplemented to 6% (v/v) with 0.1% neutral red solution (w/v) (see Recipes)
- 2 M NaOH (see Recipes)
- 1 M sulfuric acid (see Recipes)
- 6 M NaI (see Recipes)
- 1 M ammonium acetate (see Recipes)
- 1 M magnesium acetate (see Recipes)
- 5 M NaCl (see Recipes)
- Wash buffer (see Recipes)
- Polyphosphate assay buffer (see Recipes)
- 50 mM potassium dihydrogen phosphate (see Recipes)
- Phosphate calibration curve (see Recipes)
- 222 µM polyP45 (10 mM Pi) (see Recipes)
- 3.3 mM polyP3 (10 mM Pi) (see Recipes)
- 6 µM polyP45 (see Recipes)
- 200 µM polyP3 (see Recipes)
- 28 mM ammonium heptamolybdate in 2.1 M H2SO4 (see Recipes)
- 0.76 mM malachite green in 0.35% polyvinyl alcohol (see Recipes)
Equipment
- Novaspec Plus spectrophotometer GE
- Thermo LabSystems Multiskan Ascents 354 microplate reader
- Eppendorf Thermomixer® compact
- Vortex mixer (Heidolph)
- Centrifuge (Eppendorf, MiniSpin®)
Procedure
- Polyphosphate purification by precipitation
Protocol adapted from (Kumble and Kornberg, 1995)- Inoculate a single colony of the yeast strain with a sterile inoculation loop from a fresh plate into 5 ml of liquid medium (YPD or appropriate selection medium) and incubate overnight on a rotary shaker at 200-220 rpm and 28 °C.
- After 16-24 h of growth, determine the OD600 of the yeast culture by using a spectrophotometer. Inoculate cells in 5 ml YPD (final OD600 0.2) and incubate in the shaker at 200-220 rpm and 28 °C until the OD600 reaches 0.6-0.8 (This will take 3 to 5 h.).
- Collect from 0.5 to 1 OD600 units of yeast exponential culture (1-2 x 107 cells) by centrifugation at 12,000 x g for 1 min at room temperature and discard the supernatant.
- Resuspend the cell pellet with 400 μl of AE buffer prechilled at 4 °C.
- Transfer the cell resuspension to previously prepared 1.5 ml screw cap tubes containing 300 µl phenol and 40 μl 10% SDS and securely fasten the lid.
- Mix by inversion 4 times and vortex 5 sec to homogenize.
- Incubate at 65 °C for 5 min and chill the tubes for 1 min on ice.
- Add 300 µl of chloroform, securely fasten the lid, mix by inversion 4 times, and vortex 5 sec to homogenize.
- Centrifuge at room temperature for 2 min at 13,000 x g to separate the aqueous phase containing the polyP from the organic phase.
- Carefully transfer the aqueous phase (top) to prepared 1.5 ml screw cap tubes containing 350 µl chloroform. Typically, the recovered volume will be approximately 450 µl. Be sure not to carry over any phenol during pipetting by avoid touching the bottom phase or the white protein containing interphase.
- Securely fasten the lid, mix by inversion 4 times, and vortex 5 sec to homogenize.
- Centrifuge at room temperature for 2 min at 13,000 x g.
- Recover the aqueous phase, approximately 400 µl, and transfer it to a new 1.5 ml microcentrifuge tube. As before, be sure not to carry over any phenol during pipetting by avoid touching the bottom phase.
- Add 2 µl of RNase A (10 mg/ml) and 2 µl of DNase I (10 mg/ml) to each tube and incubate 1 h at 37 °C. RNase A and DNase I are added before precipitation to degrade RNA and DNA, respectively, thus avoiding RNA and DNA precipitation with polyP.
- Transfer the aqueous phase to pre-cold at -20 °C, 1.5 ml microcentrifuge tube containing 1 ml of absolute ethanol and 40 µl of 3M sodium acetate (pH 5.3) and leave 3 h at -20 °C to precipitate polyphosphate.
- Centrifuge for 20 min at 13,000 x g at 4 °C.
- Discard the supernatant by decantation and add 500 µl of 70% ethanol.
- Centrifuge for 5 min at 13,000 x g at 4 °C.
- Discard the supernatant by decantation and centrifuge 1 min at 13,000 x g to remove traces of ethanol by pipetting.
- Open the tubes and dry the small translucent-white polyphosphate pellet that can be observed at the bottom of the tube at room temperature for 5 min or until the pellet is completely dry.
- Resuspend in 50 µl of Milli-Q water.
- The polyphosphate sample can be directly measured or stored at -20 °C.
- Inoculate a single colony of the yeast strain with a sterile inoculation loop from a fresh plate into 5 ml of liquid medium (YPD or appropriate selection medium) and incubate overnight on a rotary shaker at 200-220 rpm and 28 °C.
- Polyphosphate purification with silica-gel columns
Protocol adapted from (Werner et al., 2005)- Inoculate a single colony of the yeast strain with a sterile inoculation loop from a fresh plate into 5 ml of liquid medium (YPD or appropriate selection medium) and incubate overnight on a rotary shaker at 200-220 rpm and 28 °C.
- After 16-24 h of growth, determine the OD600 of the yeast culture by using a spectrophotometer. Inoculate cells in 5 ml YPD (final OD600 0.2) and incubate in the shaking incubator at 200-220 rpm and 28 °C until the OD600 reaches 0.6-0.8 (This will take 3 to 5 h.).
- Collect from 0.5 to 1 OD600 units of yeast exponential culture (1-2 x 107 cells) by centrifugation at 12,000 x g for 1 min at room temperature and discard the supernatant.
- Add 50 µl of 1 M sulfuric acid over the pellet of cells, mix and leave 5 min at room temperature.
- Neutralize the suspension with 50 µl of 2 M NaOH and add 100 µl of 1 M Tris-HCl (pH 7.5) supplemented to 6% (v/v) with 0.1% neutral red solution (w/v).
- Remove the cell debris by centrifugation for 10 min at 800 x g at 4 °C.
- Recover 200 µl of supernatant, add 600 µl of 6 M NaI and mix well prior adjusting the pH of the sample. The pH should be around 7 (The sample colour becomes orange-red.). Samples too acidic (pink) or too basic (yellow) have to be corrected by addition of 2 M NaOH or 1 M sulfuric acid, respectively.
Note: The correct adjustment of pH is a critical step for the reproducibility of polyP extraction. - Apply the 800 µl of polyphosphate extract to the QIAquick purification column and centrifuge the column for 1 min at 13,000 x g at room temperature.
- Discard the flow-through and wash the column twice with 400 µl of wash buffer by centrifugation for 1 min at 13,000 x g at room temperature.
- After the last wash, discard the flow-through and dry the column by centrifugation for an additional 1 min at 13,000 x g at room temperature.
- Transfer the column to new 1.5 ml microcentrifuge tube and add 50 µl Milli-Q water. Let the column stand for 1 min and elute the polyphosphate by centrifugation for 1 min at 13,000 x g at room temperature. It is highly recommended to use Milli-Q water with pH > 7 for a better polyP elution.
- The eluted polyphosphate can be directly measured or stored at -20 °C.
- Inoculate a single colony of the yeast strain with a sterile inoculation loop from a fresh plate into 5 ml of liquid medium (YPD or appropriate selection medium) and incubate overnight on a rotary shaker at 200-220 rpm and 28 °C.
- Colum regeneration
If necessary, the columns can be reused after regeneration with the following protocol:- Wash the column once with 750 µl of 0.2 M acetic acid and 50 mM EDTA (pH 8) by centrifugation 1 min at 13,000 x g at room temperature.
- Wash three times with 750 µl Milli-Q water by centrifugation 1 min at 13,000 x g each time at room temperature.
- Normally, a column can be used 3-5 times. New columns must be used if a decrease in the yield of polyP recovery is observed.
- Wash the column once with 750 µl of 0.2 M acetic acid and 50 mM EDTA (pH 8) by centrifugation 1 min at 13,000 x g at room temperature.
- Polyphosphate digestion and plate preparation
- Use 96-well plate for the reaction of polyphosphate degradation and the subsequent measure of the released phosphate. An excess of rPpx1 (500 ng of rPpx1 enzyme per reaction) is employed for enzymatic polyphosphate digestion.
- Add 90 µl of polyphosphate assay buffer to wells that will contain phosphate standard curve, sample background measurements, and blanks. Add 90 µl of the mix of Polyphosphate assay buffer and rPpx1 enzyme to the sample wells.
- Add 10 µl of the sample (duplicate), phosphate standard curve (0 to 500 µM KH2PO4), and positive control consisting in 200 µM polyP3 or 6 µM polyP45 to the 90 µl of Polyphosphate assay buffer. Mix by pipetting up and down several times.
- Incubate the plate at 37 °C for 60 min.
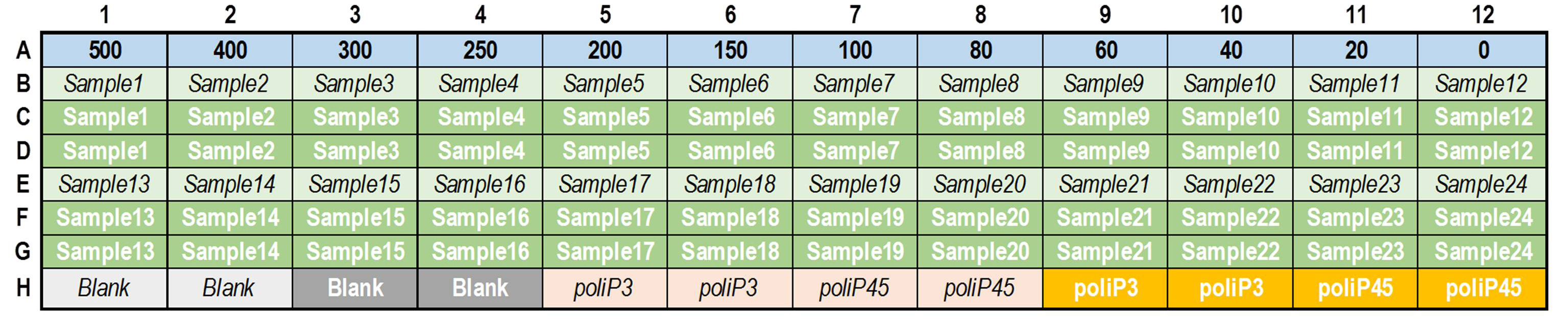
Figure 2. Example plate of polyphosphate measurement. Row A contains the phosphate calibration curve. Rows B and E contain the polyphosphate sample without enzyme (italics) to measure the background and rows C, D, F, & G contain duplicate samples with rPpx1 enzyme. Similarly, the last row contains blanks and positive controls of polyP3 and polyP45 that are incubated with and without enzyme (italics and bold respectively).
- Use 96-well plate for the reaction of polyphosphate degradation and the subsequent measure of the released phosphate. An excess of rPpx1 (500 ng of rPpx1 enzyme per reaction) is employed for enzymatic polyphosphate digestion.
- Phosphate quantification
Protocol adapted from (Cogan et al., 1999). Quantification of phosphate is made directly on the PolyP digestion plate- To quantify the released phosphate, add to each well 86 µl of 28 mM ammonium heptamolybdate in 2.1 M H2SO4 and 64 µl of 0.76 mM malachite green in 0.35% polyvinyl alcohol. Mix well by using the shaking option of the microplate reader or by pipetting up and down the samples to ensure a complete mixture of all reagents.
- After 20 min at room temperature measure the A595 in a multiplate reader. Do not let the reaction proceed more than 1 h as it can cause appearance of small precipitates in highly concentrated phosphate samples and polyP degradation in background samples, thus interfering with the correct phosphate measure.
- The amount of phosphate is obtained by comparing the absorbance value of each sample with the phosphate standard curve after subtracting the background value for each sample. The polyP3 and polyP45 are used as controls for the rPpx1 reaction over polyphosphates.
Figure 3. Example plate of phosphate quantification
- To quantify the released phosphate, add to each well 86 µl of 28 mM ammonium heptamolybdate in 2.1 M H2SO4 and 64 µl of 0.76 mM malachite green in 0.35% polyvinyl alcohol. Mix well by using the shaking option of the microplate reader or by pipetting up and down the samples to ensure a complete mixture of all reagents.
Notes
- The main advantage of silica column purification versus the precipitation protocol is that the former allows processing many samples at once. Even more, it is possible to use Qiagen 96 PCR purification plates instated of columns to process a high number of samples. However, the main drawback is that likely short chain polyPs are lost and the amount of PolyP is underestimated in comparison with the precipitation protocol. As a reference, when we used commercial polyP45, that is a mix of different polyP with enrichment in polyP chains with 45 phosphate residues, the polyP recovery using silica columns process was estimated to be 20-22%. Thus, it should be possible to approximately compare PolyP levels obtained by the column method with those generated by the precipitation method by multiplying by a factor of 4. As an example with S. cerevisiae samples, phenol-extracted material (that is, processed until step A14 of the "Polyphosphate purification by precipitation" protocol) was split into 2 halves. One half was processed following the same protocol and the other subjected to purification using the QIAquick columns. As shown in Figure 4, purification through the silica columns results in 3 to 4-fold decrease in the detected polyP levels. Consequently, if the objective of the extraction is to obtain high amounts of polyP of all sizes, the precipitation method is most suitable. A detailed method for assessing polyP size by gel electrophoresis can be found as a BioProtocol (Garcia, 2014).
In contrast, the precipitation method is more economic, as it does not require purchasing purification columns.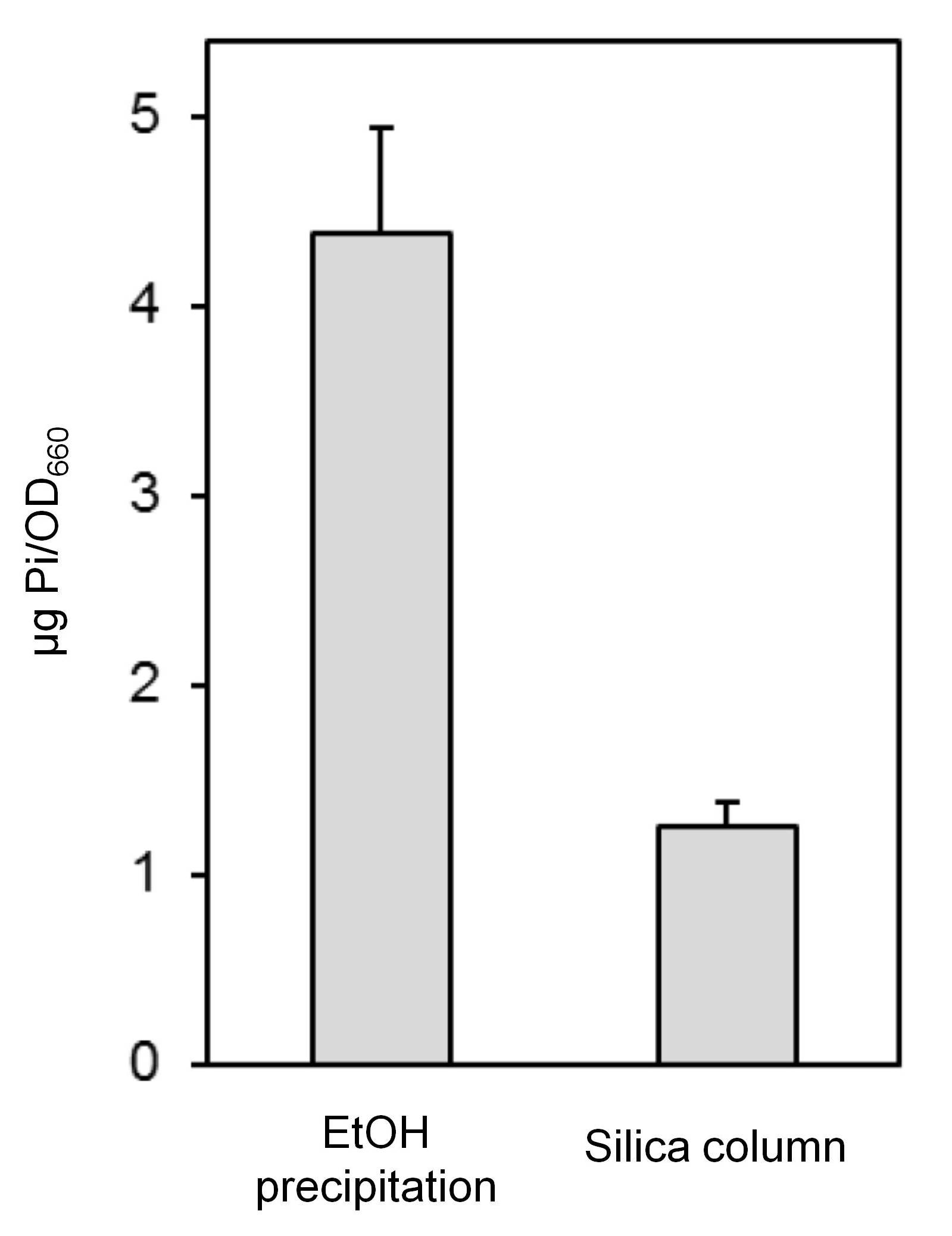
Figure 4. Comparison of ethanol precipitation and silica column purification methods. The figure shows the amount of Pi per unit of optical density of the culture. Data are presented as the mean ± SD from 5 experiments.
Recipes
- YPD
Mix 30 g YP base medium and 20 g glucose in 500 ml dH2O
Add dH2O to 1,000 ml and autoclave - 3 M sodium acetate (pH 5.3)
Mix 40.82 g sodium acetate trihydrate with 70 ml dH2O
Adjust pH to 5.3 with acetic acid glacial
Add dH2O to 100 ml and stored at room temperature - 0.5 M EDTA (pH 8)
Dissolve 18.61 g disodium ethylenediaminetetraacetate 2-hydrate in 80 ml dH2O
Adjust pH to 8 by adding ~2 g NaOH pellets and stir vigorously on a magnetic stirrer
Add dH2O to 100 ml and stored at room temperature - Buffer AE [50 mM sodium acetate (pH 5.3), 10 mM EDTA]
Mix 1.67 ml 3 M sodium acetate (pH 5.3) with 70 ml dH2O
Add 2 ml 0.5 M EDTA
Add dH2O to 100 ml and stored at room temperature - 10% SDS
Mix 10 g sodium dodecylsulfate (SDS) with 80 ml dH2O
Add dH2O to 100 ml and stored at room temperature - RNase A (10 mg/ml)
Mix 100 µl RNase A (100 mg/ml) with 900 µl dH2O
Stored at 4 °C - DNase I (10 mg/ml)
Mix 50 mg DNase I with 5 ml dH2O
Stored at 4 °C - 70% ethanol
Mix 70 ml of ethanol absolute with 30 ml dH2O
Stored at -20 °C - 0.1% neutral red solution (w/v)
Mix 0.1 g neutral red with 100 ml 70% ethanol
Stored at room temperature - 1 M Tris-HCl (pH 7.5) supplemented to 6% (v/v) with 0.1% neutral red solution (w/v)
Mix 12.1 g Tris with 70 ml dH2O
Add 6 ml 0.1% neutral red solution (w/v)
Adjust pH to 7.5 with HCl
Add dH2O to 100 ml and stored at room temperature - 2 M NaOH
Mix 8 g NaOH with 80 ml dH2O
Add dH2O to 100 ml and stored at room temperature - 1 M sulfuric acid (H2SO4)
Mix 5.45 ml 98% sulfuric acid with 94.55 ml dH2O
Stored at room temperature - 6 M NaI
Mix 13.5 g NaI with dH2O up to 15 ml
Solution has to be freshly prepared each time and protected from light - 1 M ammonium acetate
Mix 7.71 g ammonium acetate with 80 ml dH2O
Add dH2O to 100 ml and stored at room temperature - 1 M magnesium acetate
Mix 21.4 g magnesium acetate tetrahydrate with 70 ml dH2O
Add dH2O to 100 ml and stored at room temperature - 5 M NaCl
Mix 29.22 g sodium chloride with 70 ml dH2O
Add dH2O to 100 ml and stored at room temperature - Wash buffer [10 mM Tris-HCl buffer (pH 7.5), 50% ethanol, 1 mM EDTA and 100 mM NaCl]
Mix 500 µl 1M Tris-HCl (pH 7.5), 25 ml 100% ethanol, 100 µl 0.5 M EDTA, 1 ml 5 M NaCl, and 20 ml dH2O
Adjust pH to 7.5 with HCl
Add dH2O to 50 ml and stored at room temperature
0.2 M acetic acid and 50 mM EDTA
Mix 571 µl acetic acid glacial and 5 ml of 0.5 M EDTA
Add dH2O to 50 ml and stored at room temperature - Polyphosphate assay buffer [20 mM Tris-HCl (pH 7.5), 5 mM magnesium acetate, 100 mM ammonium acetate]
Mix 2 ml 1 M Tris-HCl (pH 7.5), 500 µl 1 M magnesium acetate, and 10 ml 1 M ammonium acetate
Adjust pH to 7.5 with HCl
Add dH2O to 100 ml and stored at 4 °C - 50 mM potassium dihydrogen phosphate
Mix 340 mg potassium dihydrogen phosphate with 50 ml dH2O
Stored room temperature - Phosphate calibration curve (0 to 500 µM KH2PO4)
Mix 100 µl 50 mM potassium dihydrogen phosphate with 9.9 ml dH2O
From the 500 µM KH2PO4 make serial dilutions of 400, 300, 250, 200, 150, 100, 80, 60, 40, and 20 µM KH2PO4
Stored room temperature or at 4 °C - 222 µM polyP45 (10 mM Pi)
Mix 10.4 mg sodium phosphate glass Type 45 (polyP45) with 10 ml dH2O
Stored at 4 °C - 3.3 mM polyP3 (10 mM Pi)
Mix 12.3 mg sodium tripolyphosphate (polyP3) with 10 ml dH2O
Stored at 4 °C - 6 µM polyP45
Mix 135.1 µl 222 µM polyP45 with 4.86 ml dH2O
Stored at 4 °C - 200 µM polyP3
Mix 300 µl 3.3 mM polyP3 with 4.7 ml dH2O
Stored at 4 °C - 28 mM ammonium heptamolybdate in 2.1 M H2SO4
Mix 3.46 g ammonium heptamolybdate tetrahydrate with 80 ml dH2O
Add 11.2 ml 98% sulfuric acid
Add dH2O to 100 ml and stored at room temperature - 0.76 mM malachite green in 0.35% polyvinyl alcohol
Mix 350 mg polyvinyl alcohol in 100 ml dH2O at 80 °C and stir vigorously on a magnetic stirrer until all polyvinyl alcohol dissolves completely
Add 35 mg malachite green oxalate salt
Add dH2O to 100 ml and stored at room temperature
Acknowledgments
We thank Dr. Roberto Docampo (University of Georgia, Athens, USA.) for the E. coli clone expressing rPpx1. JA is recipient of grants BFU2011-30197-C03-01 and BFU2014-54591-C2-1-P from the Ministerio de Economía y Competitividad (MINECO), Spain, and 2014SGR-4 from the Generalitat de Catalunya. JC is recipient of grant BFU 2013-44189-P from Ministerio de Economia y Competitividad (MINECO), Spain, and 2014 SGR-1014 from the Generalitat de Catalunya.
References
- Cogan, E. B., Birrell, G. B. and Griffith, O. H. (1999). A robotics-based automated assay for inorganic and organic phosphates. Anal Biochem 271(1): 29-35.
- Freimoser, F. M., Hurlimann, H. C., Jakob, C. A., Werner, T. P. and Amrhein, N. (2006). Systematic screening of polyphosphate (poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism. Genome Biol 7(11): R109.
- Garcia, M. R. (2014). Extraction and quantification of Poly P, Poly P analysis by Urea-PAGE. Bio-protocol 4(9): e1113.
- Kumble, K. D. and Kornberg, A. (1995). Inorganic polyphosphate in mammalian cells and tissues. J Biol Chem 270(11): 5818-5822.
- Rosenfeld, L., Reddi, A. R., Leung, E., Aranda, K., Jensen, L. T. and Culotta, V. C. (2010). The effect of phosphate accumulation on metal ion homeostasis in Saccharomyces cerevisiae. J Biol Inorg Chem 15(7): 1051-1062.
- Ruiz, F. A., Rodrigues, C. O. and Docampo, R. (2001). Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J Biol Chem 276(28): 26114-26121.
- Werner, T. P., Amrhein, N. and Freimoser, F. M. (2005). Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch Microbiol 184(2): 129-136.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Canadell, D., Bru, S., Clotet, J. and Ariño, J. (2016). Extraction and Quantification of Polyphosphate in the Budding Yeast Saccharomyces cerevisiae. Bio-protocol 6(14): e1874. DOI: 10.21769/BioProtoc.1874.
Category
Microbiology > Microbial biochemistry > Other compound
Microbiology > Microbial biochemistry > Protein > Modification
Biochemistry > Other compound > Ion > Phosphate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link















