- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Proximity Ligation Assay (PLA) Protocol Using Duolink® for T Cells
Published: Vol 6, Iss 10, May 20, 2016 DOI: 10.21769/BioProtoc.1811 Views: 20588
Reviewed by: Jia LiGriselda Zuccarino-CataniaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Flow-cytometric Detection of Low-level Reactive Oxygen Species in Cell Lines and Primary Immune Cells
Kevin Bode [...] Heiko Weyd
Sep 5, 2020 7243 Views

Sample Preparation and Integrative Data Analysis of a Droplet-based Single-Cell ATAC-sequencing Using Murine Thymic Epithelial Cells
Tatsuya Ishikawa [...] Taishin Akiyama
Jan 5, 2023 2416 Views

Proliferation Assay Using Cryopreserved Porcine Peripheral Mononuclear Cells Stimulated With Concanavalin A and Analyzed With FCS ExpressTM 7.18 Software
Marlene Bravo-Parra [...] Luis G. Giménez-Lirola
Jun 5, 2025 2620 Views
Abstract
Protein-protein interaction experiments, such as co-immunoprecipitation (IP) assays, classically require tremendous amount of cells. This becomes a problem when your work focuses on rare cell populations (e.g., lymphocyte subtypes). O-link Bioscience has developed Proximity Ligation Assay (PLA) reagents and procedures to alleviate and solve this kind of issue. Moreover PLA experiments are read out using fluorescence or bright field microscopy, providing additional information on intracellular interactions localization significantly bettering classical IP procedures.
PLA reagents are made of complementary small oligonucleotides “minus” and “plus” probes which specifically recognize host species from the primary antibodies (Abs) targeting the two proteins you are interested in. Experiments have to be designed with primary Abs from different species (rabbit, mouse or goat) as PLA probes “minus” or “plus” react against a specific host species of the primary antibody (e.g. “plus” anti-rabbit with “minus” anti-mouse or “plus” anti-mouse with “minus” anti-rabbit combos are allowed if mouse and rabbit primary Abs are used). When the two PLA probes are close enough (40 nm) a ligation occurs upon ligase incubation generating a circle DNA. These circle-forming DNAs are next amplified thanks to a polymerase and complementary fluorescent nucleotides, being incorporated at this step. Each luminescent spot is thereafter considered being an interaction site between the two proteins (Figure 1).
Figure 1. Schematic representation of Duolink® experiment. Primary Abs from different host species were used [i.e., Mouse (Ms) anti-Nlrp3 and Rabbit (Rb) anti-IRF4]. When protein-protein interaction occurs PLA probes allow incorporation of fluorescent oligonucleotides which are analyzed by microscopy.
PLA experiments have been performed on differentiated T cells from mice. T cells were grown on cover slip coated with poly-L-Lysine. Please be aware that for T cells and other non-adherent cells, experiments and staining are also possible in 500 µl microtubes until the last washing step before mounting cover slip on slide. This microtube method saves cytokines reagents and allows T cells to grow with usual methods but centrifugation repetition for washing steps is hazardous. Primary Abs incubation also requires also a specific setup if microtube method is chosen.
Materials and Reagents
- Glass cover slip (Ø13 mm thickness, No. 1, 5) (VWR International, catalog number: 631-0150 )
- Primary antibodies (Abs)
- Mouse anti-Nlrp3 (1/100) (Adipogen International, catalog number: AG-20B-0014 )
Note: Starting concentration of mouse anti-Nlrp3 is1 µg/µl. - Rabbit anri-IRF4 (1/100) (Novus Biologicals, catalog number: NBP1-00893 )
Note: Starting concentration of rabbit anri-IRF4 is 0.08 µg/µl.
- Mouse anti-Nlrp3 (1/100) (Adipogen International, catalog number: AG-20B-0014 )
- Duolink® reagents
- Anti-rabbit PLUS probe (1/5) (Sigma-Aldrich, catalog number: Duo92002 )
- Anti-mouse MINUS probe (1/5) (Sigma-Aldrich, catalog number: Duo92004 )
- Ligation reagents (5x) (Sigma-Aldrich, catalog number: Duo92007 )
Note: Ligation reagents, ligase and amplification reagents are all included in the pack # DUO92007 . - Ligase (1 unit/μl) (Sigma-Aldrich, catalog number: Duo82029 )
- Amplification reagents (containing orange labeled oligonucleotides) (5x) (Sigma-Aldrich, catalog number: Duo92007 )
- Polymerase (10 unit/μl) (Sigma-Aldrich, catalog number: Duo82030 )
- Wash buffer A (Sigma-Aldrich, catalog number: Duo82049 )
- Wash buffer B (Sigma-Aldrich, catalog number: Duo82049 )
- Anti-rabbit PLUS probe (1/5) (Sigma-Aldrich, catalog number: Duo92002 )
- Poly-L-Lysine (Sigma-Aldrich, catalog number: P4707 )
- Triton X-100 (0.1% in PBS) (Sigma-Aldrich, catalog number: T8787 )
- Blocking & Abs dilution buffer (0.5% BSA in PBS)
- Mounting medium containing DAPI (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: P36931 )
- 1x PBS (Lonza, catalog number: 17-516F )
- RNase free water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977035 )
- 4% formaldehyde (PFA) (VWR International, catalog number: 9713.1000 )
Equipment
- Pipets (from 1 to 1,000 µl)
- Fluorescence microscope (with DAPI and Cy2 or Cy3 emission filter) (ZEISS, model: Imager M2)
Note: Objective 63x is recommended for T cells imaging. - 37 °C incubator
- Freeze block for enzymes
- Orbital shaker
Software
- ImageJ
Procedure
- Day 0: Th2 and naïve T cells were isolated and differentiated as previously described (Bruchard et al., 2015). Approximately 107 cells were then transferred to 12-wells dishes containing a cover slip coated with a room temperature 10 min treatment with a 100-150 µl drop of poly-L-Lysine. Lysine was then removed by pipetting and wells were washed with 1 ml PBS. Cells were then seeded and allowed to attach to cover slip for 24 h at 37 °C.
- Day 1: Please note that all procedures from 1 to 5 are performed in the same 12-wells dishes for cell culture. T cells were briefly washed 2 times in 1 ml PBS by removing medium with a pipette and replacing with fresh PBS. Please be aware that cover slip shall never dry.
- T cells’ fixation was performed using 200 µl 4% PFA during 10 min at 4 °C followed by 2 washes of 1 ml PBS 1x in the well containing the cover slip.
- Permeabilization was obtained using 200 µl 0.1% Triton X-100 during 2 min at RT followed by 2 washes of 1 ml 1x PBS.
- Cover slips were blocked using 1 ml 0.5% BSA in PBS during 20 min at RT.
- Cover slips were incubated in a 30 to 40 µl drop of diluted primary antibodies (Abs) overnight at 4 °C (Figure 2).
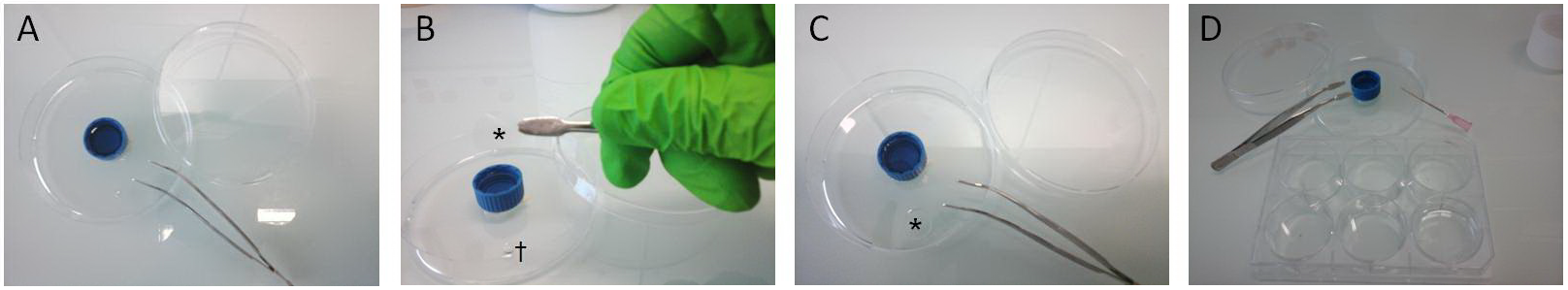
Figure 2. Cover slip handling for PLA procedures. A. A 60 mm diameter plastic petri dish was used as humidity chamber for Abs incubation and PLA experiments. Humidity was obtained with a 15 ml cap filled with water. Flat tip forceps were used to handle cover slips all along procedure. B. After being removed from culture dish, cover slips (*) were incubated face down on a 30 µl drop of Abs (†). C. Cover slip (*) placed upon the drop. From this step, the petri dish was closed and sealed with parafilm and placed overnight at 4 °C. D. The following day, cover slips were carefully picked up from the humidity chamber using forceps and a needle. Cover slips were then individually placed face up for washing steps in a well of a 6-wells plate filled with 3 ml of 1x PBS.
Note: This procedure was also repeated for incubation of Duolink® products. It enabled to save Duolink® reagents compared to manufacturer’s instructions. All other manufacturer’s instructions (incubation times, washing steps and buffers...) were carefully followed. - Day 2: Cover slips were washed 3 times 5 min in 1x PBS at RT.
- PLA experiment was performed using manufacturer’s instructions. The procedure is also available on bio-protocol website (Lin et al., 2015) until step 9.
- All samples were allowed to dry in the dark at RT.
- A drop of mounting medium containing DAPI was placed between each samples and slide. Slides were kept at 4 °C for short term storage.
- Day 3: Imaging procedures were performed using a fluorescence microscope with 63x objective equipped with appropriate filters (DAPI: Ex 365, Em: BP 445/50; Duolink spots: Ex: BP 550/25, Em: 605/70; Figure 3). Spots inside nuclei (as DAPI as the reference) were quantified using Image J and its Analyse particles tab. Slides were then kept at -20 °C for long term storage.
Representative data
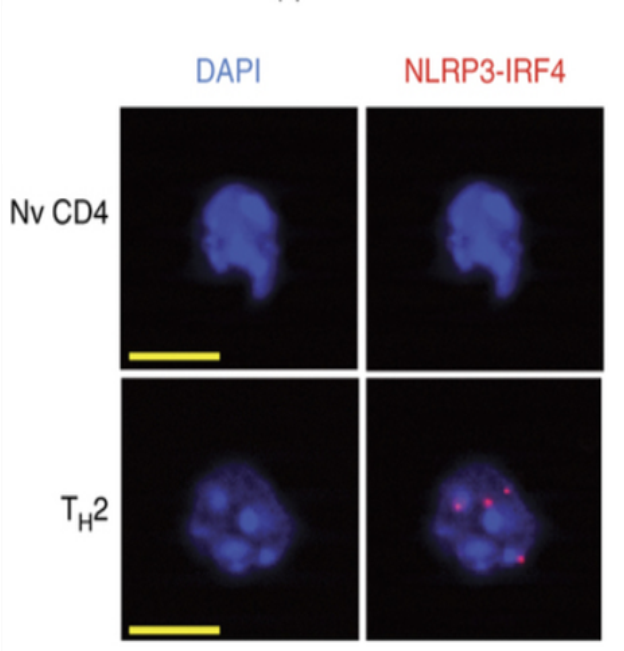
Figure 3. Reprensative images for Duolink spots in TH2 cells. T cells pictures obtained with our protocol (Nv: Naïve, Scale bar in yellow: 5 µm). Red dots in Th2 cells are Duolink spots.
Notes
- Use freshly prepared BSA in PBS anytime as possible or filter the preparation if using an old solution. If this step is not taken into account, sediments could appear and interfere with spots detection.
Acknowledgments
This work was supported by a French Government grant managed by the French National Research Agency under the program “Investissements d’Avenir” with reference ANR-11-LABX-0021 (Lipstic Labex). FG team is « Equipe labélisée Ligue Nationale Contre le Cancer ». V. D. is the recipient of a « poste d’accueil INSERM »; This protocol was adapted from previous work of Fredriksson et al. (2007) and Lin et al. (2015).
References
- Bruchard, M., Rebe, C., Derangere, V., Togbe, D., Ryffel, B., Boidot, R., Humblin, E., Hamman, A., Chalmin, F., Berger, H., Chevriaux, A., Limagne, E., Apetoh, L., Vegran, F. and Ghiringhelli, F. (2015). The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol 16(8): 859-870.
- Fredriksson, S., Dixon, W., Ji, H., Koong, A. C., Mindrinos, M. and Davis, R. W. (2007). Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods 4(4): 327-329.
- Lin, M. Z., Martin, J. L. and Baxter, R. C. (2015). Proximity ligation assay (PLA) to detect protein-protein interactions in breast cancer. Bio-protocol 5(10): e1479.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Derangère, V., Bruchard, M., Végran, F. and Ghiringhelli, F. (2016). Proximity Ligation Assay (PLA) Protocol Using Duolink® for T Cells. Bio-protocol 6(10): e1811. DOI: 10.21769/BioProtoc.1811.
Category
Immunology > Immune cell function > General
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link