- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Poly (A) RNA in Mesophyll Cells of Nicotiana benthamiana Using in situ Hybridization
Published: Vol 5, Iss 17, Sep 5, 2015 DOI: 10.21769/BioProtoc.1576 Views: 9393
Reviewed by: Zhaohui LiuPablo Bolanos-VillegasNing Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Method to Map Small RNAs with High Resolution
Kun Huang [...] Jeffrey L. Caplan
Aug 20, 2021 4681 Views

Quantitative Analysis of RNA Editing at Specific Sites in Plant Mitochondria or Chloroplasts Using DNA Sequencing
Yang Yang and Weixing Shan
Sep 20, 2021 3292 Views

Profiling of Single-cell-type-specific MicroRNAs in Arabidopsis Roots by Immunoprecipitation of Root Cell-layer-specific GFP-AGO1
Lusheng Fan [...] Xuemei Chen
Dec 20, 2022 2231 Views
Abstract
Export of transcribed mRNAs from nucleoplasm to cytosol is an essential process for the translation of genes into proteins. This process is tightly regulated by nuclear pores, composed of about 30 nucleoporin proteins (Nups). Whether or not the mRNAs are able to be appropriately exported to cytoplasm is of an importance for understanding the role of Nups. Here, we describe a practical protocol to detect the intracellular localization of mRNAs in mesophyll cells of Nicotiana benthamiana (N. benthamiana). This protocol is based on poly (A) in situ hybridization method using an oligo d(T) probe conjugated with Alexa Fluor-488.
Keywords: poly (A) in situ hybridizationMaterials and Reagents
- Leaves of 3-4 weeks old wild type or Nup gene silenced N. benthamiana
Notes:
a.This method can be applied for other dicot plants with relatively soft leaves.
b.For Virus-induced gene silencing (VIGS) of N. benthamiana. See Zhang and Liu (2014). - 10 µM 48-mer oligo d(T) 5’-labeled with Alexa Fluor-488, HPLC-purified [purchased from custom oligo services (e.g. Eurofins Genomics)]
Note: Dissolved in TE (Tris-EDTA) buffer and store at -80 °C, shaded. - 99.8% Methanol (4 ml per sample) (Wako pure chemical, special glade, catalog number: 131-01826 )
- 99.5% Ethanol (5 ml per sampl) (Wako pure chemical, special glade, catalog number: 057-00456 )
- 99.5% Ethanol/Xylene (1:1 v/v, 1 ml per sample) (Wako pure chemical, special glade, catalog number: 244-00086 )
- 99.8%Methanol/Fixation solutionB (1:1 v/v, 1 ml per sample)
- PerfectHyb Plus Hybridization Buffer (2 ml per sample) (Sigma-Aldrich, catalog number: H7033 )
- Fixation cocktail (see Recipes)
- Fixation solution A (see Recipes)
- Fixation solution B (see Recipes)
Equipment
- 3 ml vial containers
- Rotary shaker (e.g. BioCraft, model: BC-730 )
- Hybridization oven or incubator (50 °C) with rotary shaker (e.g. TAITEC, model: BR-23FP MR )
- Confocal laser-scanning microscope (preferred) or ordinary fluorescent microscope with the filter set for GFP or Alexa Fluor-488 (e.g. Olympus, model: FV1000-D )
Procedure
- Poly (A) RNA in situ hybridization
- Cut leaves of N. benthamiana into small pieces (e.g. 3 mm x 3 mm) by knife and put ~10 leaf pieces/sample in 3 ml vial container.
Note that the center of larger pieces won’t be stained with AF488 probe. - Add 1 ml Fixation solution A in the vial container and shake (Approx. 80 rpm) for 30 min at room temperature (RT). Leaf pieces will float on the surface of the solution at the start, but will sink later (Figure 1).


Figure 1. Leaf discs of N. benthamiana in Fixation solution A before (left) and after (right) shaking for 30 min (step A2) - Remove Fixation solution A by pipeting.
- Add 1 ml methanol. Shake (80 rpm) for 5 min at RT and remove methanol (twice). Note that the inner side of the vial should be washed well with methanol.
- Add 1 ml ethanol, shake (80 rpm) for 5 min at RT and remove ethanol (three times).
- Add 1 ml ethanol/xylene (1:1). Shake (80 rpm) for 30 min at RT and remove the solution. Note that leaf tissues will become white during steps A4-6.
- Add 1 ml 100% ethanol. Shake (80 rpm) for 5 min at RT and remove ethanol (twice).
- Add 1 ml 100% methanol. Shake (80 rpm) for 5 min at RT and remove ethanol (twice).
- Add methanol/Fixation solution B (1:1). Shake (80 rpm) for 5 min at RT and remove the solution.
- Add 1 ml Fixation solution A and shake (80 rpm) for 30 min at RT.
- Remove Fixation solution A.
- Add 1 ml Fixation solution B. Shake (80 rpm) for 5 min at RT and remove the solution (twice).
- Add 1 ml PerfectHyb Plus, and shake (80 rpm) for 5 min at RT.
- Remove the solution and add 1 ml of fresh PerfectHyb Plus. Shake (80 rpm) at 50 °C in hybridization oven (incubator with rotary shaker can be used) for more than 1 h.
- Add 1 µl of 10 µM Alexa Fluor-488-labeled oligo d(T). Shake (80 rpm) at 50 °C overnight. It is preferred to perform fluorescent microscopy immediately after the preparation of samples, but the samples can be kept at least for a couple of days at RT, shaded.
- Cut leaves of N. benthamiana into small pieces (e.g. 3 mm x 3 mm) by knife and put ~10 leaf pieces/sample in 3 ml vial container.
- Microscopic observation
- Mount the sample in PerfectHyb Plus on slide glass with cover slip.
- Observe the localization of mRNA with confocal laser-scanning microscopy. Use appropriate setting for the detection of Alexa Fluor-488 (excitation peak 490 nm, emission peak 525 nm). We generally use 488-nm excitation source, and Alexa Fluor-488 fluorescence is recorded between 500 and 600 nm.
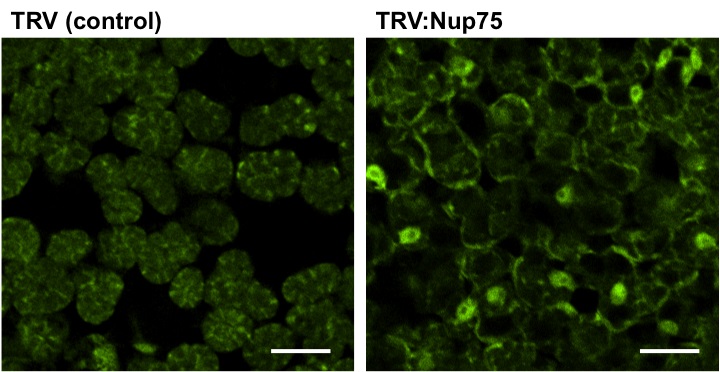
Figure 2. Distribution of Poly (A) RNA in mesophyll cells of control (TRV-infected) or NbNup75-silenced Nicotiana benthamiana. In NbNup75-silenced plant, abnormal accumulation of mRNA in nuclei is observed as a result of impaired export of mRNA. Bars=30 µm. (Ohtsu et al. 2014)
- Mount the sample in PerfectHyb Plus on slide glass with cover slip.
Recipes
- Fixation cocktail (x 4)
240 mM NaCl
14 mM Na2HPO4
6 mM NaH2PO4
5.4 mM KCl
160 mM EGTA
Autoclaved and stored at RT - Fixation solution A (for 10 ml)
Fixation cocktail (x 4) 2.5 ml
Formaldehyde 250 µl
DMSO 500 µl
Tween20 10 µl
Heptan 5 ml
H2O 1.74 ml
Prepare before use - Fixation solution B (for 10 ml)
Fixation cocktail (x 4) 2.5 ml
DMSO 500 µl
Tween 20 10 µl
Heptan 5 ml
H2O 1.99 ml
Prepare before use
Acknowledgments
This protocol was adapted from Parry et al. (2006) and Germain et al. (2010). The work was supported by a Grant-in-Aid for Scientific Research (B) (26292024) from the Japan Society for the Promotion of Science and by Grant for Basic Science Research Projects from the Sumitomo Foundation.
References
- Germain, H., Qu, N., Cheng, Y. T., Lee, E., Huang, Y., Dong, O. X., Gannon, P., Huang, S., Ding, P., Li, Y., Sack, F., Zhang, Y. and Li, X. (2010). MOS11: a new component in the mRNA export pathway. PLoS Genet 6(12): e1001250.
- Ohtsu, M., Shibata, Y., Ojika, M., Tamura, K., Hara-Nishimura, I., Mori, H., Kawakita, K. and Takemoto, D. (2014). Nucleoporin 75 is involved in the ethylene-mediated production of phytoalexin for the resistance of Nicotiana benthamiana to Phytophthora infestans. Mol Plant Microbe Interact 27(12): 1318-1330.
- Parry, G., Ward, S., Cernac, A., Dharmasiri, S. and Estelle, M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18(7): 1590-1603.
- Zhang, H. and Liu, Y. (2014). VIGS assays. Bio-protocol 4(5): e1057.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mizuno, Y. and Takemoto, D. (2015). Detection of Poly (A) RNA in Mesophyll Cells of Nicotiana benthamiana Using in situ Hybridization. Bio-protocol 5(17): e1576. DOI: 10.21769/BioProtoc.1576.
Category
Plant Science > Plant cell biology > Cell staining
Plant Science > Plant molecular biology > RNA > RNA detection
Cell Biology > Cell staining > Nucleic acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link