- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
VIGS Assays
Published: Vol 4, Iss 5, Mar 5, 2014 DOI: 10.21769/BioProtoc.1057 Views: 30207
Reviewed by: Feng Li

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Real-time PCR Analysis of PAMP-induced Marker Gene Expression in Nicotiana benthamiana
Fan Liu [...] Yuanchao Wang
Oct 5, 2018 9308 Views

RNA Stability Measurements Using RT-qPCR in Arabidopsis Seedlings
Tianran Jia and Brandon H. Le
Jul 20, 2020 7007 Views

Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
Valentín Luna-García and Stefan de Folter
Sep 5, 2024 2085 Views
Abstract
Virus-induced gene silencing (VIGS) is a powerful method to study gene function in plants. Tobacco rattle virus (TRV)-based VIGS vector is the most efficient VIGS vector so far. This method was originally developed by the Dinesh-Kumar's group (Liu et al., 2002) . Here, we describe a rapid and high efficient TRV-based VIGS method for knocking down genes in Nicotiana benthamiana. For TRV-based VIGS, Agrobacterium culture containing pTRV1 and Agrobacterium culture containing pTRV2 with plant target gene fragment are mixed and infiltrated into the lower leaves of plant. After 2-3 weeks post infiltration, plant target gene will be silenced.
Materials and Reagents
- 6-leaf-stage Nicotiana benthamiana plants
Note: Nicotiana benthamiana can be obtained from our lab (Figure 1).

Figure 1. 6-leaf-stage Nicotiana benthamiana plant
- Bacteria strains
a. Escherichia coli strains, such as DH5α
b. Agrobacterium strains, such as GV3101
Note: All strains were obtained from our lab. - pTRV1 and pTRV2-LIC based expression vectors (Dong et al., 2007)
- pTRV1: a T-DNA vector containing duplicated CaMV 35S promoter, NOS terminator and cDNA clone of TRV RNA1 of Ppk20 strain.
- pTRV2-LIC: a T-DNA vector containing duplicated CaMV 35S promoter, NOS terminator and cDNA clone of TRV RNA2, of which non-structural genes were replaced by a multiple cloning site (MCS).
- pTRV2-NbPDS: gene fragement of NbPDS was inserted at MCS into pTRV2-LIC. This construct was usually used as a control to show the successful gene silencing.
- pTRV1 (ABRC, catalog number: CD3-1039 ) and pTRV2-LIC (ABRC, catalog number: CD3-1042 ) could be ordered at http://www.arabidopsis.org/abrc/catalog/vector_3.html.
- pTRV1: a T-DNA vector containing duplicated CaMV 35S promoter, NOS terminator and cDNA clone of TRV RNA1 of Ppk20 strain.
- Media for Agrobacteria
- Liquid Luria-Bertani (LB) medium
- Solid LB plates with 0.12% agar
Note: LB medium is autoclaved under 120 °C for 20 min.
- Liquid Luria-Bertani (LB) medium
- Antibiotics
a. Kanamycin
b. Rifampicin
c. Gentamicin
- Easy Taq DNA polymerase (Beijing TransGen Biotech)
- dNTP (Roche)
- TIANprep Mini Plasmid Kit (Beijing TransGen Biotech)
- MgCl2 (Sigma-Aldrich)
- MES (AMRESCO)
- Acetosyringone (Sigma-Aldrich)
- DMSO (AMRESCO)
- Infiltration buffer (see Recipes)
Equipment
- Centrifuge tubes
- Plant growth chamber (24 °C, 16 h photoperiod conditions, 50% huminity)
- Sterile 1 ml syringe without needle
- Sterile bacterial culture tubes
- Centrifuge
- PCR instrument
- 37 °C and 28 °C incubators with shaking
Procedure
- Clone plant target gene fragment into pTRV2-LIC as described previously (Liu et al., 2002), and transform it into Escherichia coli DH5α. Positive clones were picked up and the plasmid DNA was amplified by PCR, using specific primer pairs, and then confirmed for correct insertion by DNA sequencing. Grow a positive clone in 5 ml LB liquid medium (containing 50 µg/ml Kanamycin) in 37 °C incubator at 200 rpm shaking overnight. Collect the bacteria by 14,000 x g centrifuging for 1 min at room temperature and then extract plasmids using mini plasmid kit.
- Transform pTRV1, pTRV2 or its derivatives into Agrobacterium strain GV3101 respectively. Transformed Agrobacteria were grown for 2 days on LB plates containing 50 µg/ml Kanamycin, 30 µg/ml Rifampicin and 50 µg/ml gentamicin.
- Pick several clones and confirm that the grown Agrobacteria contain correct plasmid using PCR with specific primers.
- Grow one positive clone from each transformant containing pTRV1, pTRV2 or pTRV2 derivatives in 5 ml liquid LB media (containing 50 µg/ml Kanamycin, 50 µg/ml Rifampicin and 50 µg/ml Gentamicin) in 28 °C incubator shaking at 200 rpm overnight.
Note: Inoculating Agrobacteria into media for culturing should be done on super-clean bench, all equipments used needs to be sterile.
- Take the culture tubes out of the incubators. Adjust all the Agrobacterium culture to OD600=1.0. Taking equal volumes of agrobacterium culture (OD600 = 1.0) with pTRV1 and that with pTRV2 or pTRV2 derivatives. Mix them together and pellet by centrifuging at 3,000 x g for 5 min, at room temperature.
- Pour off the supernatant, re-suspend the agrobacterium pellet in infiltration buffer of equal volume to that of Agrobacterium culture (as to keep the OD600 at around 1.0). Keep the re-suspended culture at room temperature for 2-4 h.
- Select 6-leaf-stage plants and inflitrate the re-suspended Agrobacterium culture into abaxial side of expanded leaves, using 1 ml-syringe (without needles). 2 or 3 leaves of each plant need to be injected. Plants were grown in a growth room with a 16-h/8-h photoperiod at a light intensity of 10,000 lux at 24 °C. Figure 2 shows the schematic diagram of infiltration. Figure 3 showed the leaf state right afer inoculation.
Note: Each leaf for inoculation is often injected 2 circles with 1 cm diameters.
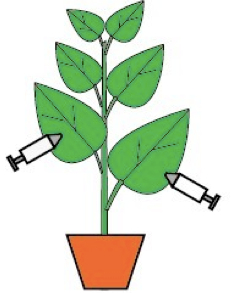
Figure 2. Schematic diagram of infiltration
Figure 3. The right picture showed the state right after inoculation and the left one showed a non-infected leaf
- 2 weeks post inoculation, target gene will be silenced at whole plant level. The new leaves always show the strong silencing phenotype, and are appropriate for following study. Figure 4 shows the upper leaves of plants that PDS gene was silenced by VIGS.

Figure 4. Successful silencing of NbPDS. Photograph was taken 2 weeks post inoculation.
Recipes
- Infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 µM acetosyringone) (100 ml)
1 M MgCl2: 20.33 g MgCl2 dissolved in 100 ml dH2O, autoclaved by 120 °C, 20 min. 1 M MgCl2 stock was stored at 4 °C.
1 M MES: 21.325 g MES dissolved in 100 ml dH2O, filter sterilized with 0.22 µm filter membrane
200 mM acetosyringone: 0.3924 g acetosyringone dissolved in 10 ml DMSO. 1 M MES stock was stored at room temperature. 200 mM acetosyringone was stored at -20 °C.
100 ml infiltration buffer
1 ml 1 M MgCl2
1 ml 1 M MES
100 μl 200 mM acetosyringone
Add dH2O to 100 ml
Acknowledgments
This protocol was adapted from the research article: Wang et al. (2013).
References
- Dong, Y., Burch-Smith, T. M., Liu, Y., Mamillapalli, P. and Dinesh-Kumar, S. P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145(4): 1161-1170.
- Liu, Y., Schiff, M., Marathe, R. and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30(4): 415-429.
- Wang, Y., Yu, B., Zhao, J., Guo, J., Li, Y., Han, S., Huang, L., Du, Y., Hong, Y., Tang, D. and Liu, Y. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25(4): 1383-1399.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhang, H. and Liu, Y. (2014). VIGS Assays. Bio-protocol 4(5): e1057. DOI: 10.21769/BioProtoc.1057.
Category
Plant Science > Plant molecular biology > RNA > Transcription
Molecular Biology > RNA > RNA interference
Plant Science > Plant immunity > Disease bioassay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










