- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Confocal Imaging of Myeloid Cells in the Corneal Stroma of Live Mice
Published: Vol 5, Iss 13, Jul 5, 2015 DOI: 10.21769/BioProtoc.1517 Views: 12369
Reviewed by: Jia LiAndrea IntroiniAlka Mehra

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2917 Views

Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient
Vikram Aditya [...] Wei Yue
Apr 5, 2025 2880 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
The accessibility and transparency of the cornea makes it a good tissue model for monitoring immunological responses using in vivo real time imaging analysis (Lee et al., 2010; Tan et al., 2013). These corneal qualities have also allowed for high-resolution in vivo imaging of non-ocular tissue transplanted into the anterior chamber of the mouse eye (Speier et al., 2008a; Speier et al., 2008b). This protocol was adapted from Speier (2008) to successfully assess real-time in vivo myeloid cell dynamics in wounded corneas of c-fms-EGFP mice (Chan et al., 2013). Macrophage colony-stimulating factor (CSF-1) regulates the differentiation and proliferation of cells of the mononuclear phagocyte system. The activity of CSF-1 is mediated by the CSF-1 receptor that is encoded by c-fms (Csf1r) protooncogene. The c-fms gene is expressed in macrophage, trophoblast cell lineages, and to some extent granulocytes. In the c-fms-EGFP mice EGFP, enhanced green fluorescent protein, is driven under the Csf1r, colony stimulating factor 1 receptor, promoter and highlights myeloid cells (Sasmono et al., 2003). This protocol can be further adapted to image other transgenic mice expressing fluorescent proteins.
Materials and Reagents
- c-fms-EGFP mouse whose cornea was wounded by chemical injury 1 day prior (Chan and Werb, 2015)
- Avertin (Sigma-Aldrich, catalog number: T4, 840-2 )
- 15.5 ml tert-amyl alcohol (2-methyl-2-butanol) (Thermo Fisher Scientific, catalog number: A730-1 )
- Saline 0.9%
- GenTeal gel (0.3%, eye gel) (Novartis, NDC: 0078-0429-47 )
- Avertin stock solution (see Recipes)
- Avertin working solution (1.2%) (see Recipes)
Equipment
- 25 Gauge needle
- 5 ml and 50 ml syringes
- 1.5 ml microcentrifuge tubes
- 0.22 micron filter
- Head-holding adapter (Narishige, catalog number: SG-4N )
- Dumont no. 5 forceps (Fine Science Tools, catalog number: 11251-10 )
- UST-2 Solid Universal Joint (Narishige, catalog number: UST-2)
- Polyethylene tubing (0.28 mm i.d., 0.61 mm o.d.) (Warner Instruments, catalog number: 64- 0750 )
- Heating pad
- Nikon Spectral Confocal (C1Si) equipped with a GFP filter, an argon laser, a 40x objective (NIR/ 0.8 NA W lens) and camera (Nikon D-eclipse C1Si) (Figure 1)

Figure 1. Confocal setup. The imaging setup includes a confocal microscope, camera, argon laser, and computer.
Software
- Nikon Confocal Software (Nikon, Ez-C1 Gold version 3.8)
- Imaris Software (Bitplane, version 7.3.1 for Windows X64)
Procedure
Ethical statement: All procedures discussed here are in accordance with and were approved by the University of California, San Francisco Institutional Animal Care and Use Committee.
- Setup the Confocal Microscope.
- Turn on the argon laser.
- Open imaging software Ez-C1 Gold Ver. 3.8.
- Turn on the argon laser.
- Setup the head-holding adapter (Figure 2).
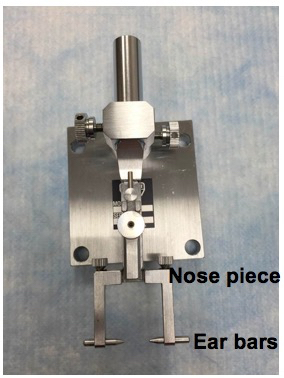
Figure 2. Head-holding adapter setup. The mouse head is stabilized by fixing the nose and ears in place using the nose piece and ear bars. - Setup the eye stabilizer (Figure 3).
- Attach the no. 5 Dumont forceps to the UST-2 Solid Universal Joint.
- Cut an 18 mm piece of polyethylene tubing. The polyethylene tubing serves to isolate and stabilize the whole eye. Cover the tips of the forceps with the tubing, creating a loop between the tips.
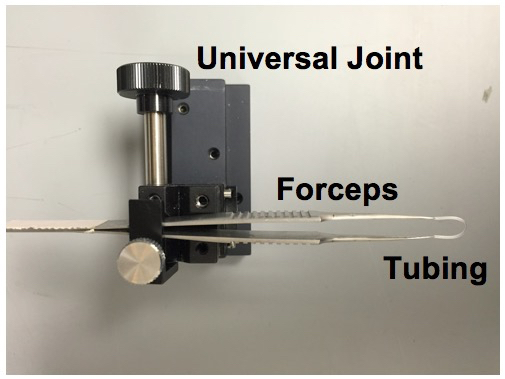
Figure 3. Eye stabilizer setup. The mouse eye is stabilized by looping and tightening the tubing around the eye. - Attach the no. 5 Dumont forceps to the UST-2 Solid Universal Joint.
- Place the heating pad under the objective. The heating pad is used to maintain body temperature at 37 °C during the procedure.
- Anesthetize the mouse by injecting 1.2% Avertin solution using a dosage of 0.2 ml/10 grams body weight intraperitoneally with a 25 Gauge needle. It is critical to minimize breathing movements as much as possible so anesthetize the mouse until the breathing is very shallow and not rapid.
- After the mouse is fully anesthetized, place the mouse’s head in the head-holding adapter (Figure 4).
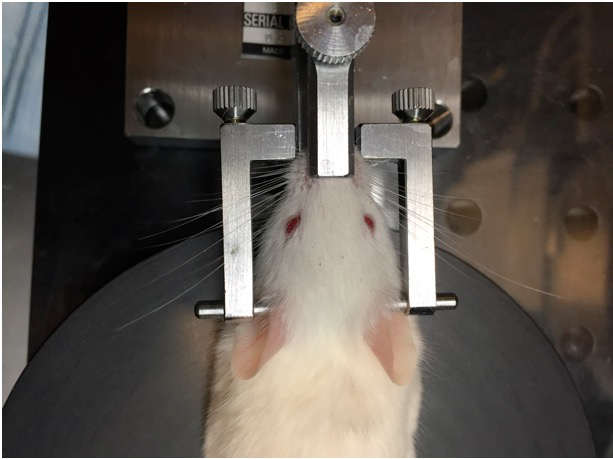
Figure 4. Placement of mouse in head-holding adapter. The mouse head is fixed in place using the nose piece and ear bars. - Place the head holder together with the mouse on a heating pad under the 40x microscope objective. Angle the head so that the cornea selected for imaging is facing upward towards the lens objective (Figure 5).
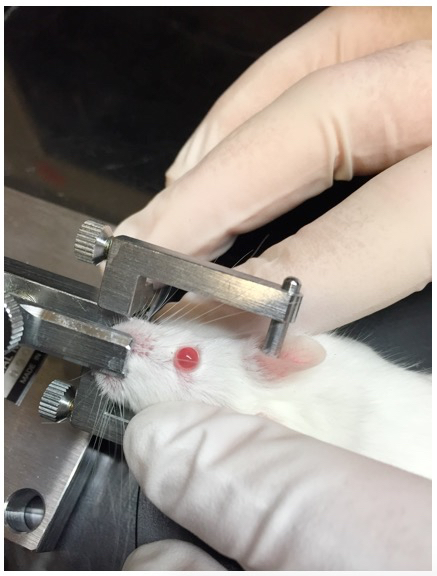
Figure 5. Rotation of mouse head in head-holding adapter. The mouse head is rotated so that the central cornea faces towards the ceiling when the eye is proptosed using periorbital pressure. Rotation of the head to an angle is achieved by manual tilting of the head holding adaptor. - Apply mild periorbital pressure to proptose the eye (Figure 5). Gently place the polyethylene tubing loop of the eye stabilizer below the corneoscleral junction (Figure 6, Figure 8). Be careful not to interrupt the breathing or blood circulation while holding the mouse. Adjust the forceps to tighten the loop just enough to stabilize the eye and keep it proptosed.

Figure 6. Stabilization of the eye. Polyethylene tubing looped around the eye below the corneoscleral junction and the forceps are tightened to keep the tubing in place. Final positioning of the mouse, head-holding adapter, and universal joint with forceps. - Squeeze the tube of GenTeal Gel gently and place a small amount of GenTeal Gel directly from the tube onto the cornea sufficient to use as an immersion liquid between the objective and the cornea (Figure 7).
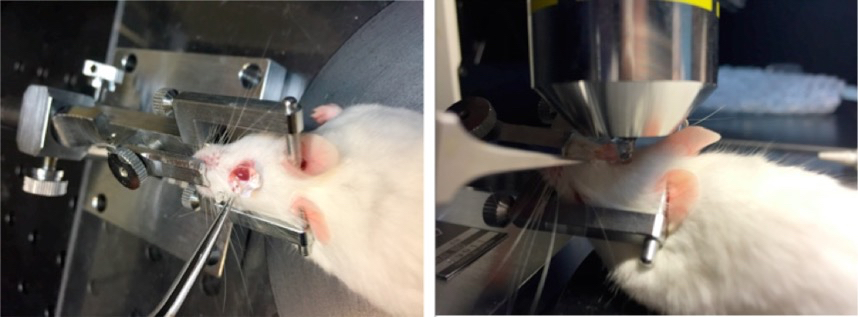
Figure 7. Application of GenTeal gel. The tube of Genteal Gel is squeezed and the ointment is applied directly onto the entire surface of the eye. A column of Genteal Gel extends from the mouse cornea to the microscope objective. - Use brightfield observation to focus on the central cornea (Figure 8).
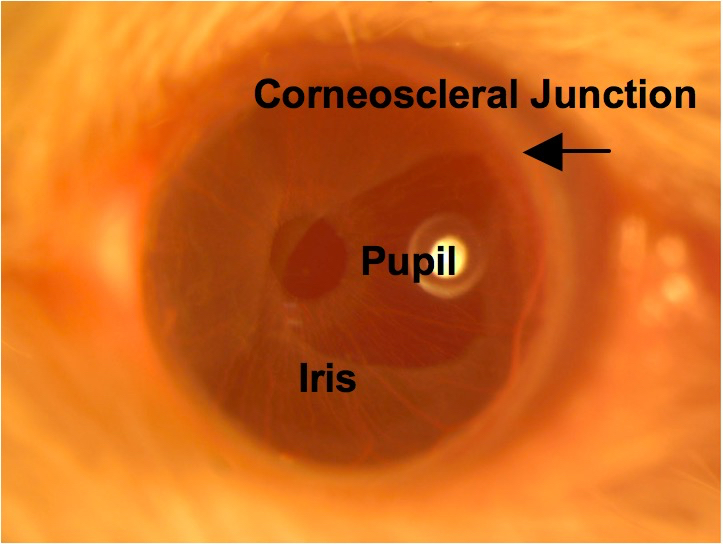
Figure 8. Central cornea. The transparent cornea is in focus using brightfield illumination, as are the iris tissue and round pupil. The corneoscleral junction is indicated by the arrow. - Turn off the brightfield lamp and turn on the epifluorescence lamp. Find the optimal corneal area and depth for imaging using the GFP filter. Try to image as close as possible to the central cornea because it is flatter than the paracentral cornea and allows for more cells to be in focus within the same plane.
- Switch the microscope setting from epifluorescence to confocal. Select a filter cube with 488 excitation. Open imaging software Ez-C1 Gold Ver. 3.8. and use the following acquisition settings. The below settings optimize the resolution of the myeloid cells while limiting the amount of photobleaching.
- Pinhole: large (may use a smaller pinhole if there is enough signal)
- Beam Splitter: 408/488/581or 488/561/638
- Excitation intensity: for 488 = up to 10%
- Lines per dimension: 512 x 512
- Gain: 180-190 (may adjust as needed)
- Pixel dwell: between 6 and 8
- Timelapse-as fast as possible
- Pinhole: large (may use a smaller pinhole if there is enough signal)
- Use timelapse acquisition to continuously capture images approximately every 3.22 sec for 20-90 min.
- Remove the mouse from the eye stabilizer and head-holding adaptor.
- Euthanize the mouse.
- Analysis of the data is a very complex issue, which cannot be covered here in its entirety. The type of data analysis performed depends on the research question being asked. A recent review highlights the various analyses that can be performed (Weigert et al., 2013). Several software and freeware programs are available to image analysis including Bitplane Imaris, ImageJ, and CellTrack. Figure 10 demonstrates an example track analysis of a single myeloid cell using the Bitplane Imaris software program. Detailed Imaris software instructions are available through the company website (http://www.bitplane.com/imaris). Axial fluctuations manifested by slow changes to specimen focus may occur over the course of time-lapse imaging. The Imaris software program can help correct for these fluctuations in focus through its drift correction function.
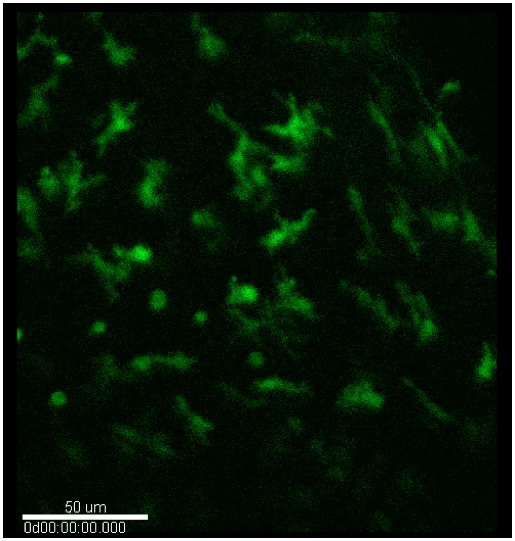
Figure 9. Confocal microscopy. Typical image of myeloid cells (green) in the stroma of un-injured and injured corneas.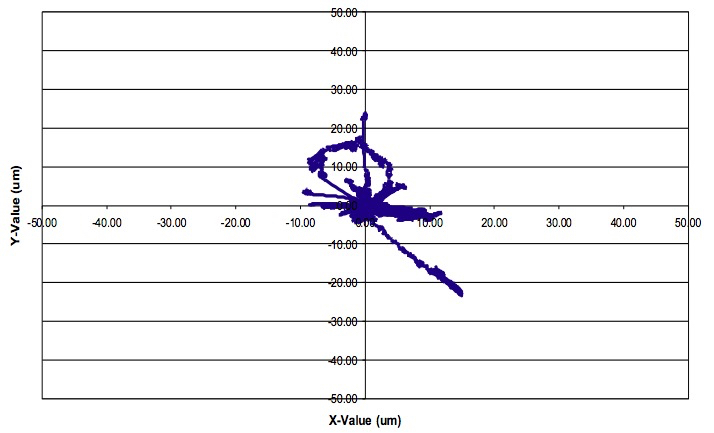
Figure 10. Image analysis. Sample motility analysis of a single myeloid cell over 90 minutes using the Bitplane Imaris software program.
Recipes
- Avertin stock solution
Mix 25 g Avertin (2, 2, 2-Tribromoethanol) with 15.5 ml tert-amyl alcohol (2-methyl-2-butanol) at room temperature for 12 h in a dark bottle
Make 0.5 ml stocks
Stored at -20 °C in the dark - Avertin working solution (1.2%)
Pre-warm 0.9% saline at 20 °C
Mix 0.5 ml Avertin stock with 39.5 ml pre-warmed 0.9% saline
Mix in an airtight dark or foil covered container
Filter the solution though a 0.22 micron filter into a dark or foil covered container
Stored at 4 °C
The working solution expires in 1 week
Acknowledgments
This protocol was adapted from Speier et al. (2008). This work was supported by grants from the National Institutes of Health (K08 EY018858 and R01 EY002162 to M.F.C. and R01 CA057621and P01 AI053194 to Z.W.).
References
- Chan, M. F. and Werb, Z. (2015). Animal models of corneal injury. Bio-protocol 5(13): e1516.
- Chan, M. F., Li, J., Bertrand, A., Casbon, A. J., Lin, J. H., Maltseva, I. and Werb, Z. (2013). Protective effects of matrix metalloproteinase-12 following corneal injury. J Cell Sci 126(Pt 17): 3948-3960.
- Lee, E. J., Rosenbaum, J. T. and Planck, S. R. (2010). Epifluorescence intravital microscopy of murine corneal dendritic cells. Invest Ophthalmol Vis Sci 51(4): 2101-2108.
- Sasmono, R. T., Oceandy, D., Pollard, J. W., Tong, W., Pavli, P., Wainwright, B. J., Ostrowski, M. C., Himes, S. R. and Hume, D. A. (2003). A macrophage colony-stimulating factor receptor–green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101(3): 1155-1163.
- Speier, S., Nyqvist, D., Cabrera, O., Yu, J., Molano, R. D., Pileggi, A., Moede, T., Kohler, M., Wilbertz, J., Leibiger, B., Ricordi, C., Leibiger, I. B., Caicedo, A. and Berggren, P. O. (2008a). Noninvasive in vivo imaging of pancreatic islet cell biology. Nat Med 14(5): 574-578.
- Speier, S., Nyqvist, D., Kohler, M., Caicedo, A., Leibiger, I. B. and Berggren, P. O. (2008b). Noninvasive high-resolution in vivo imaging of cell biology in the anterior chamber of the mouse eye. Nat Protoc 3(8): 1278-1286.
- Tan, Y., Abdulreda, M. H., Cruz-Guilloty, F., Cutrufello, N., Shishido, A., Martinez, R. E., Duffort, S., Xia, X., Echegaray-Mendez, J., Levy, R. B., Berggren, P. O. and Perez, V. L. (2013). Role of T cell recruitment and chemokine-regulated intra-graft T cell motility patterns in corneal allograft rejection. Am J Transplant 13(6): 1461-1473.
- Weigert, R., Porat-Shliom, N. and Amornphimoltham, P. (2013). Imaging cell biology in live animals: ready for prime time. J Cell Biol 201(7): 969-979.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chan, M. F. and Werb, Z. (2015). Confocal Imaging of Myeloid Cells in the Corneal Stroma of Live Mice. Bio-protocol 5(13): e1517. DOI: 10.21769/BioProtoc.1517.
Category
Immunology > Immune cell imaging > Confocal microscopy
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link