- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
HIV-1 Fusion Assay
Published: Vol 4, Iss 16, Aug 20, 2014 DOI: 10.21769/BioProtoc.1212 Views: 16415
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1896 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2460 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3038 Views
Abstract
The HIV-1 fusion assay measures all steps in the HIV-1 life cycle up to and including viral fusion. It relies on the incorporation of a β-lactamase–Vpr (BlaM-Vpr) protein chimera into the virion and the subsequent transfer of this chimera into the target cell by fusion (Figure 1). The transfer is monitored by the enzymatic cleavage of CCF2, a fluorescent dye substrate of β-lactamase, loaded into the target cells. Cleavage of the β-lactam ring in CCF2 by β-lactamase changes the fluorescence emission spectrum of the dye from green (520 nm) to blue (447 nm). This change reflects virion fusion and can be detected by flow cytometry (Figure 2).
Materials and Reagents
- 293T cells (ATCC)
- pAdVAntage (Promega Corporation, catalog number: E1711 )
- pCMV4-BlaM-Vpr (Addgene, catalog number: 21950 )
- pNL4-3 proviral DNA (NIH AIDS Reagent Program) or TN6-GFP encoding primary Env (available upon request to Warner Greene)
- DMEM (Mediatech, Cellgro®, catalog number: 10-013-CV )
- RPMI 1640 (Mediatech, Cellgro®, catalog number: 15-040-CV )
- 1x phosphate-buffered saline (PBS)
- Fetal bovine serum (FBS)
- 100 U/ml of penicillin and 100 U/ml of streptomycin (Life Technologies, Gibco® catalog number: 15140-122 )
- 2 M CaCl2
- Alliance HIV-I p24 ELISA kit (PerkinElmer, catalog number: NEK050B001KT ) or FlaQ assay reagents (Hayden et al., 2003)
- Peripheral blood lymphocytes (PBLs)
- CCF2-AM substrate and loading solutions (Life Technologies, catalog number: K1032 )
- CO2-independent media (Life Technologies, Gibco®, catalog number: 18045-088 )
- Probenecid (Sigma-Aldrich, catalog number: P8761 )
- Mouse antihuman CD3 conjugated to APC-Cy7 and mouse antihuman CD4 conjugated to PE-Cy7 (BD Biosciences, catalog numbers: 557832 and 557852 )
- BD CompBeads (BD Biosciences, catalog number: 552843 )
- 16% Paraformalehyde (Electron Microscopy Sciences, catalog number: 15710 )
- HBSS (see Recipes)
- Dulbecco’s modified Eagle medium (DMEM) culture media (see Recipes)
- Roswell Park Memorial Institute (RPMI) culture media (see Recipes)
- CCF2 loading solution (see Recipes)
- Stock solution of probenecid (250 mM) prepared in 250 mM NaOH (see Recipes)
- Development media (see Recipes)
Equipment
- T175 cm2 culture flasks
- 96-well V-bottom plate (Corning Incorporated, catalog number: 3363 )
- 5, 10, 25 ml pipettes
- 50-ml Falcon tube
- 0.22 μm poresize Steriflip (EMD Millipore, catalog number: SE1M179M6 )
- 2 ml Nalgene tubes (Thermo Fisher Scientific, Nalgene®, catalog number: 5000-0020 )
- Clear ultracentrifuge tubes (BD, catalog number: BD340437 )
- 37 °C, 5% CO2 incubator set at 90% humidity
- Ultracentrifugation equipment with SW28 rotor
- Flow cytometer
Note: The fusion assay alone requires a flow cytometer equipped with a violet laser excitation (405 nm) and two measurement parameters. The photomultiplicator tube (PMT) with a 450/50 nm band pass filter, commonly used for the detection of Pacific Blue, is used for the detection of the cleaved CCF2 substrate. The other PMT with a 515/20 nm band pass filter, commonly used for Amcyan detection, is used for the detection of the uncleaved CCF2 substrate. Additional PMTs are necessary for the measurement of the fluorochromes associated with the CD3 and CD4 antibodies. APC-Cy7 is excited by 633 nm red laser and detected with a PMT with a 755 long pass filters. PE-Cy7 is excited by a 531 nm yellow-green laser and detected with the PMT with a 755 long pass filter.
Software
- FlowJoX software (Tree Star) or other FACS analysis software
Procedure
- Plate 1.5 x 107 293T cells in a T175-cm2 tissue culture flask with 20 ml of DMEM culture media and culture overnight at 37 °C in a 5% CO2 humidified incubator.
- Prepare 1.75 ml of H2O containing 60 μg of TN6-GFP proviral DNA, 20 μg of pCMV-BlaM-Vpr, and 10 μg of pAdVAntage vectors.
- Slowly add 2 ml of 2x HBSS and mix gently by pipetting up and down.
- Add 250 μl of 2 M CaCl2 drop by drop.
- Incubate 10 min at room temperature to precipitate the DNA.
- Replace the 293T cell culture media with 40 ml of fresh DMEM culture media pre-warmed to 37 °C.
- Slowly add 4 ml of DNA precipitate and incubate for 16 h at 37 °C.
- Replace the media with 40 ml of fresh DMEM culture media pre-warmed to 37 °C.
- Incubate for 24 h at 37 °C.
- Harvest the supernatant of the transfected 293T cells into a 50-ml Falcon tube.
- Centrifuge the supernatant at 365 x g at room temperature for 10 min to remove the cellular debris.
- Filter the clarified supernatant through Steriflip.
- Transfer 36 ml of virion-containing supernatant to Ultra-Clear centrifuge tubes.
- Place the tube in the bucket of the SW28 rotor and balance the tube with DMEM culture media if necessary.
- Ultracentrifuge (72,000 x g, 90 min) at 4 °C without using brakes.
- Remove the supernatant, resuspend the viral pellet in 1 ml of DMEM, divide into 100-μl aliquots, and store at -80 °C.
- Quantify p24Gag content of the viral preparation by Enzyme-Linked Immunosorbent Assay or FlaQ Assay (Maiti et al., 2014).
- Wash the PBLs with RPMI culture media.
- Count PBLs and make a suspension 2 x 107 cells/ml in RPMI culture media.
- Divide the cell suspension in aliquots of 100 μl (2 x 106 cells) per condition to be tested in a V-bottom 96-well plate. Distribute two additional aliquots of cells into two wells to serve as compensation controls. One well will be loaded with CCF2 substrate while the other will remain unloaded. These two control wells will not be stained with anti-CD3-APC-Cy7 and anti-CD4-PE-Cy7 antibodies.
- Add a quantity of HIV-1 virions containing BlaM-Vpr equivalent to 400 ng of p24Gag to all wells except the “non infected control” and the two compensation controls. Incubate for 2 h at 37 °C.
- Collect the cells by centrifugation at 365 x g for 5 min at room temperature.
- Wash the cells once with 200 μl of CO2-independent media and centrifuge at 365 x g for 5 min at room temperature.
- Resuspend the pellet in 100 μl of CCF2-AM loading solution and incubate for 1 h at room temperature in the dark. Make sure to keep one well unstained by resuspending with CO2 independent media only.
- Collect the cells by centrifugation at 365 x g for 5 min at room temperature.
- Wash the cells with 200 μl of development media and centrifuge at 365 x g for 5 min at room temperature.
- Resuspend the pellet in 200 μl of development media and incubate the cells at room temperature for 16 h in the dark.
- In two empty wells, add one drop of each of BD compensation beads vials (negative control and Anti-mouse IgK).
- Collect the cells and beads by centrifugation at 365 x g for 5 min at 4 °C.
- Wash the cells and beads once by addition of 200 μl of FACS staining buffer.
- Collect the cells by centrifugation at 365 x g for 5 min at 4 °C and resuspend the pellet in 100 μl of immunostaining solution (in FACS staining buffer) containing a 1/100 dilution of anti-CD3-APC-Cy7 and a 1/50 dilution of anti-CD4-PE-Cy7. Do not stain the 2 compensation controls nor the beads. Stain the CompBeads with a FACS staining buffer containing either a 1/100 dilution of anti-CD3-APC, or a 1/50 dilution of anti-CD4-PE-Cy7.
- Incubate for 30 min at 4 °C.
- Collect the cells and beads by centrifugation at 365 x g for 5 min at 4 °C.
- Wash the cells and beads with 200 μl of FACS staining buffer.
- Fix the cells in FACS staining buffer supplemented with 1.2% paraformaldehyde for 24 h at 4 °C.
- Acquire the samples on a FACS instrument. The set of samples should include: unloaded unstained cells, the CCF2 loaded unstained cells, the CD3-APC-Cy7 stained beads, the CD4-PE-Cy7 stained beads, the non-infected control loaded with CCF2 and immunostained, and the infected samples loaded with CCF2 and immunostained. Make sure to acquire emission in the FITC channel in addition to the parameters of the assay (uncleaved and cleaved substrate, APC-Cy7 and PE-Cy7). The voltage should be set using the non-infected, immunostained sample.
- Import the fcs files into the FlowJo software. Use the fcs files corresponding to the CompBeads to compensate for APC-Cy7 and PE-Cy7, use the CCF2 loaded and unloaded samples to compensate in the FITC channel. Apply the compensation to all samples.
- Gate the cells as shown Figure 2.
Representative data
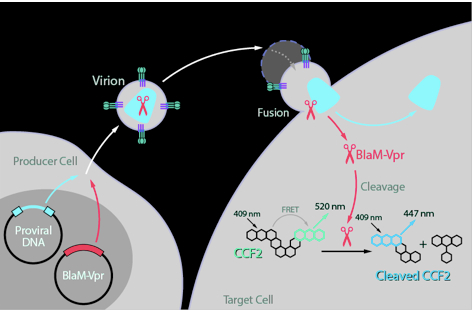
Figure 1. Principle of the fusion assay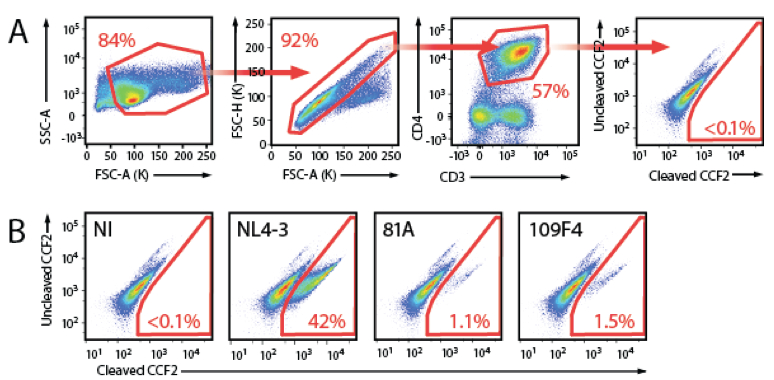
Figure 2. Representative data. A. Gating strategy to measure HIV-1 fusion to resting PBL cultures. The population of cells was first gated on Forward Scatter area (FSC-A) and Side Scatter area (SSC-A). Single cells were then gated on a FACS plot with FSC-A and FSC-H. The population of CD4 T cells was gated on APC-Cy7 and PE-Cy7. The fusion gate was drawn on the non infected sample in a FACS plot representing cleaved CCF2 versus uncleaved CCF2. B. Representative fusion FACS plot obtained for non-infected sample (NI), the X4-tropic NL4-3, the R5-tropic 81A and a primary HIV-1 envelope (Env) subcloned into TN6-GFP (Cavrois et al., 2011; Cavrois et al., 2014).
Notes
- While there is donor-to-donor variability in the susceptibility to HIV-1 fusion, normalization by 81A shows very good assessment of the relative ability of a given primary Env to mediate fusion (Cavrois et al., 2014).
Recipes
- HBSS
280 mM NaCl
10 mM KCl
1.5mM Na2HPO4
12 mM dextrose
50 mM N-(2-hydroxyethylpiperazine)-N′-(2-ethanesulfonic acid) (HEPES) (pH 7.05)
Stored at -20 °C - Fluorescence-activated cell sorting (FACS) staining buffer
1x phosphate-buffered saline (PBS)
2% fetal bovine serum
Stored at 4 °C - Dulbecco’s modified Eagle medium (DMEM) culture media
DMEM
10% fetal bovine serum
100 U/ml of penicillin
100 μg/ml of streptomycin - RPMI culture media
RPMI 1640
10% fetal bovine serum
100 U/ml of penicillin
100 μg/ml of streptomycin - CCF2 loading solution
- Resuspend CCF2-AM in dimethylsulfoxide (solution A of the CCF2 kit) to generate a stock solution (1 mM CCF2-AM). Divided into aliquots and stored in the dark at -80 °C.
- Mix by vortex-mixing 1 μl of 1 mM CCF2-AM with 9 μl of a solution containing 100 mg/ml of Pluronic-F127 and 0.1% acetic acid (solution B of the CCF2 kit).
- Add 1 ml of CO2-independent media and vortex again.
- Resuspend CCF2-AM in dimethylsulfoxide (solution A of the CCF2 kit) to generate a stock solution (1 mM CCF2-AM). Divided into aliquots and stored in the dark at -80 °C.
- Stock solution of probenecid (250 mM) prepared in 250 mM NaOH
Divided into aliquots and store at -20 °C - Development media
2.5 mM probenecid
10% fetal bovine serum in CO2-independent media
Acknowledgments
This assay was first published in (Cavrois et al., 2002) and the protocol described in detail in (Cavrois et al., 2004). The fusion assay also allows studies of fusion mediated by other viruses, including Ebola (Yonezawa et al., 2005) and henipavirus (Wolf et al., 2009). We would like to thank the team lead by Prof. Tsien for developing CCF2, the BlaM substrate (Zlokarnik et al., 1998).
References
- Cavrois, M., De Noronha, C. and Greene, W. C. (2002). A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 20(11): 1151-1154.
- Cavrois, M., Neidleman, J., Bigos, M. and Greene, W. C. (2004). Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol Biol 263: 333-344.
- Cavrois, M., Neidleman, J., Galloway, N., Derdeyn, C. A., Hunter, E. and Greene, W. C. (2011). Measuring HIV fusion mediated by envelopes from primary viral isolates. Methods 53(1): 34-38.
- Cavrois, M., Neidleman, J., Santiago, M. L., Derdeyn, C. A., Hunter, E. and Greene, W. C. (2014). Enhanced fusion and virion incorporation for HIV-1 subtype C envelope glycoproteins with compact V1/V2 domains. J Virol 88(4): 2083-2094.
- Hayden, M. S., Palacios, E. H. and Grant, R. M. (2003). Real-time quantitation of HIV-1 p24 and SIV p27 using fluorescence-linked antigen quantification assays. AIDS 17(4): 629-631.
- Maiti, M., Grant, R. and Cavrois, M. (2014). Fluorescence-linked antigen quantification (FLAQ) assay for fast quantification of HIV-1 p24Gag. Bio-protocol 4(24): e1366.
- Wolf, M. C., Wang, Y., Freiberg, A. N., Aguilar, H. C., Holbrook, M. R. and Lee, B. (2009). A catalytically and genetically optimized beta-lactamase-matrix based assay for sensitive, specific, and higher throughput analysis of native henipavirus entry characteristics. Virol J 6: 119.
- Yonezawa, A., Cavrois, M. and Greene, W. C. (2005). Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol 79(2): 918-926.
- Zlokarnik, G., Negulescu, P. A., Knapp, T. E., Mere, L., Burres, N., Feng, L., Whitney, M., Roemer, K. and Tsien, R. Y. (1998). Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279(5347): 84-88.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cavrois, M., Neidleman, J. and Greene, W. C. (2014). HIV-1 Fusion Assay . Bio-protocol 4(16): e1212. DOI: 10.21769/BioProtoc.1212.
Category
Microbiology > Microbe-host interactions > Virus
Immunology > Host defense > Human
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











