Advanced Search
Ion transport activity assay of microbial rhodopsin expressed in E. coli cells
Last updated date: Nov 12, 2025 Views: 5113 Forks: 0
[Abstract] Microbial rhodopsins have variable functions including a light-driven ion pump, a light-gated ion channel, a photosensor and a light-regulated enzyme. As the number of genes predicted to be rhodopsins has increased in recent years, the need for rapid identification of these functions has been increasing. The patch-clamp method is often used to investigate the ion transport mechanism of microbial rhodopsins in mammalian cells, but the method requires a dedicated system and advanced techniques. The ion transport assay using the E. coli expression system described here evaluates the ion transport capacity by monitoring the pH change in the E. coli suspension. If the target rhodopsin has light-dependent ion transport activity, a light-dependent pH change is observed. The pH increase and decrease correspond to proton release from the cell and proton uptake into the cell, respectively. This method can be used to evaluate ion transport capacity in a high-throughput manner using a combination of general-purpose equipment without requiring special techniques.
Graphic abstract:
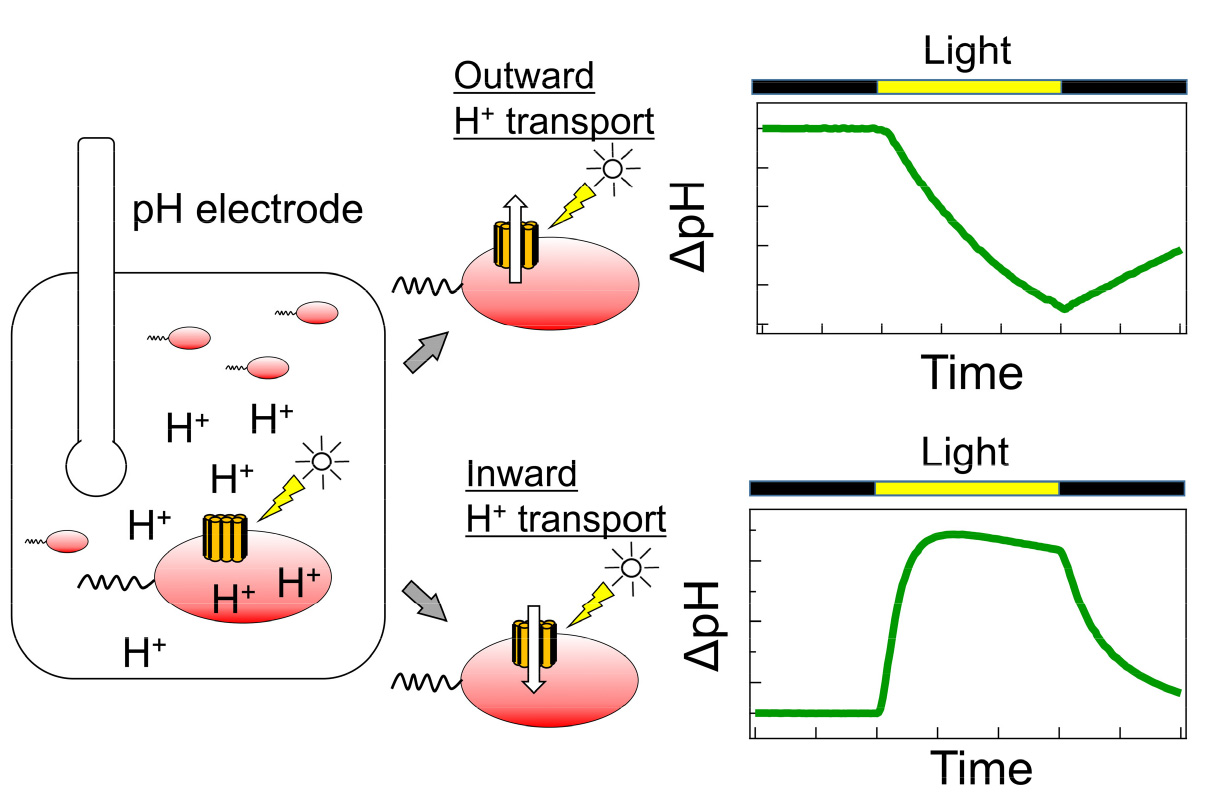
Schematic diagram of ion transport assay using rhodopsin-expressed E. coli cells.
Keywords: Microbial rhodopsin, Ion transport, Protein expression, Escherichia coli, Light-dependent pH change
[Background] Microbial rhodopsins are light-perceptive membrane proteins with all-trans retinylidine chromophore, so called retinal. The protein are widely spread among many types of organisms including bacteria, fungi and archaea and giant viruses (Ernst et al. 2014). With the accumulation of genetic information, the number of genes predicted to be microbial rhodopsins is increasing rapidly. On the other hand, many of their molecular properties remaining unknown.
Microbial rhodopsins have variable functions including a light-driven ion pump, a light-gated ion channel, a photosensor and a light-regulated enzyme. Especially, the rhodopsins which shows ion transport activity are attracting attention for their use as optogenetics tools, represented by optical control of neural activity, and rapid progress in gene screening is required. The patch-clamp method is often used to investigate the ion transport function and its mechanism of microbial rhodopsin in mammalian cells (Klapoetke et al. 2014). Also, proteoliposome into which purified rhodopsin protein is reconstituted is used to measure ion transport (Waschuk et al. 2005). But these methods require dedicated systems and advanced techniques.
On the other hand, the ion transport assay using the rhodopsin-expressed E. coli cells can also be availlable to evaluate the ion transport capacity. In this method, rhodopsin-expressing cells are suspended in a salt solution such as NaCl, KCl, or Na2SO4, and the light-dependent pH change of the cell suspension is monitored. This light-dependent pH change is due to H+ transport across the plasma membrane, which is coupled with ion transport by rhodopsin. In contrast to the patch clamp method, this method can be used to evaluate ion transport capacity in a high-throughput manner using a combination of general-purpose equipment without requiring special techniques. In fact, this method has contributed to the discovery of novel microbial rhodopsins with various ion transport capacities (Inoue et al. 2013; Yoshizawa et al. 2014; Harris et al. 2015; Hasemi et al. 2016; Inoue et al. 2016; Pushkarev and Béjà 2016; Needham et al. 2019; Inoue et al. 2020). The details of this method will be described here.
Materials and Reagents
1. E. coli C43 (DE3) cells (Lucigen, OverExpressTM strain, catalog number: 60446-1)
2. Target microbial rhodopsin gene cloned into pET21a(+) vector.
Note: The target gene was optimized its codon for E. coli and synthesized (Genscript).The synthesized gene was cloned between the NdeI and XhoI sites of the pET21a(+) vector (Sigma- Aldrich, Novagen, catalog number: 69740).
3. Yeast extract (Becton, Dickinson and company, catalog number: 212750)
4. Tryptone (nacalai tesque, catalog number: 35640-95)
5. Sodium chloride (FUJIFILM Wako Pure Chemical, catalog number: 191-01665)
6. Ampicillin sodium (FUJIFILM Wako Pure Chemical, catalog number: 014-23302)
7. Antifoam PE-L (FUJIFILM Wako Pure Chemical, catalog number: 013-17201)
8. Isopropyl-β-D-thiogalactoside (IPTG) (Sigma-Aldrich, catalog number:I5502)
9. All-trans retinal (Toronto Research Chemicals, catalog number: R240000)
10. Hydrochloric acid (FUJIFILM Wako Pure Chemical, catalog number: 080-01066)
11. Sodium hydroxide (FUJIFILM Wako Pure Chemical, catalog number: 192-15985)
12. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma-Aldrich, Merck, catalog number: C2759)
13. Dimethyl Sulfoxide (DMSO) (FUJIFILM Wako Pure Chemical, catalog number: 046-21981)
14. Disodium hydrogenphosphate (FUJIFILM Wako Pure Chemical, catalog number: 042-30055)
15. Hydroxylamine Hydrochloride (FUJIFILM Wako Pure Chemical, catalog number: 081-01471)
16. n-dodecyl-β-D-maltoside (DDM) (Anatrace, catalog number: D310)
17. Lysozyme, from Egg White (FUJIFILM Wako Pure Chemical, catalog number: 129-06723)
18. Deoxyribonuclease I, from Bovine Pancreas, Precrystalline (DNaseI) (FUJIFILM Wako Pure Chemical, catalog number: 043-26773)
19. Disposable pipette tips, 0.1–10 μL (e.g. M&S Instrument, catalog number: F161630)
20. Disposable pipette tips, 2.0–200 μL (e.g. M&S Instrument, catalog number: F161930)
21. Disposable pipette tips, 100–1000 μL (e.g. M&S Instrument, catalog number: F161670)
22. Disposable pipette tips, 500–5000 μL (e.g. M&S Instrument, catalog number: F161571)
23. Glass test tubes, 18 × 150 mm
24. Aluminum caps for glass tubes
25. 300 mL Shaking Erlenmeyer Flasks, with baffles
26. Silicone stoppers (AS ONE, BIO-SILICO®, N-42, catalog number: 5-1100-03)
27. Polypropylene conical centrifuge tubes (Thermofisher scientific, catalog number: 3009650)
28. Disposable cell for Near-Ultraviolet, Visible Light (AS ONE, Standard Type, catalog number: 1- 2848-01)
29. Sterile syringe filters, 0.2 μm (Pall, Acrodisc®, Product ID: 4612)
Equipment
1. Constant temperature incubator shaker (TAITEC, Bioshaker BR-43FL·MR, Code number: 0053027-000)
2. High-speed refrigerated centrifuge (Eppendorf himac technologies, CF15RN)
3. Fixed angle rotor (Eppendorf himac technologies, T15A42)
4. Micropipettors (e.g., 10- and 200-μL, and 5-mL; Gilson)
5. Vortex mixer (Electro Scientific Industries, GENIE2, SI-0286)
6. Digital colorimeter (TAITEC, miniphoto518R, catalog number: 0040889-000)
7. Tube rotator (AS ONE, ACR-100)
8. pH meter (HORIBA, LAQUA, F-55)
9. pH electrode (HORIBA, 9618S-10D)
10. Benchtop darkroom
11. Cover of pH electrode for light protection
Note: We made the cover for pH electrode by cutting the bottom of a 15 mL tube and covering it with black gummed cloth tape.
12. Water-jacketed glass cell vial
13. Magnetic stirrer
14. Low-temparature circulator bath (EYELA, NCB-1210)
15. 300-W xenon arc light source (ASAHI spectra, MAX-303)
16. Long pass filter (AGC Techno Glass, Y-52)
17. Heat-absorbing filter (SIGMAKOKI, HAF-50S-50H)
18. Ultrasonic homogenizer (TAITEC, VP-300N)
19. UV-visible spectrometer (JASCO, V-750)
20. Integral sphere unit for UV-visible spectrometer (JASCO, ISV-922)
21. Suction pump (AIR LIQUIDE, SP30)
Software
1. Data collection software for pH meter (HORIBA)
Procedure
A. Protein expression in E. coli cells
1. Inoculate the E. coli cells carryng the target rhodopsin gene-cloned plasmid into 4 mL of 2×YT broth containing 50 μg/μL ampicillin, and incubate at 37 °C with shaking (200 rpm) over night.
2. Inoculate 1.15 mL of overnight-culture into 115 mL of 2×YT broth containing 50 μg/μL ampicillin and 0.01 % (v/v) antiform, and incubate at 37 °C with shaking (200 rpm) till mid- log phase (OD660 of 0.5–0.7).
3. When the optical density reaches the target value, add IPTG and all-trans retinal stock solutions at final concentrations of 0.1 mM and 10 µM, respectively.
4. Incubate at 37 °C with shaking (200 rpm) for 4 hrs.
Note: The cultured cells can stored overnight at 4 °C in the dark (covered with aluminum foil to prevent light exposure).
B. Measurement of ion transport activity by monitoring extraceluller pH change.
1. Measure the OD660 of the cultured cells and dispense the cultured cells into a 15 mL plastic tube, which contains a cell amount equivalent to 15 OD·mL (OD660 × total volume (mL) of the cell susupension).
2. Collect the cells by centrifugation (4800×g, 2 min, 20 °C) and discard the supernatant.
3. Suspend the cells with 5 mL of unbuffered 100 mM NaCl solution and collect the cells by centrifugation (4800×g, 2 min, 20 °C)
4. Resuspend the cells with 5 mL of unbuffered 100 mM NaCl solution, mix the cells at room temperature with rotation for 10 min and then collected the cells by centrifugation (4800×g, 2 min, 20 °C).
5. Repeat step 4 three times.
6. Suspend the cells with 7.5 mL of unbuffered 100 mM NaCl solution.
7. Transfer the cell suspension to water-jacketed glass cell vial at 20 °C.
8. Immerse an pH electrode protected from the light to the cell vial and start to monitor the pH value of the cell suspension with stirring with a magnet.
9. Adjust the pH of the cell suspension to 6.9–7.0 with small amount of NaOH or HCl and wait for the pH fluctuation to stabilize.
10. Record the pH changes over time with the procedure described below.
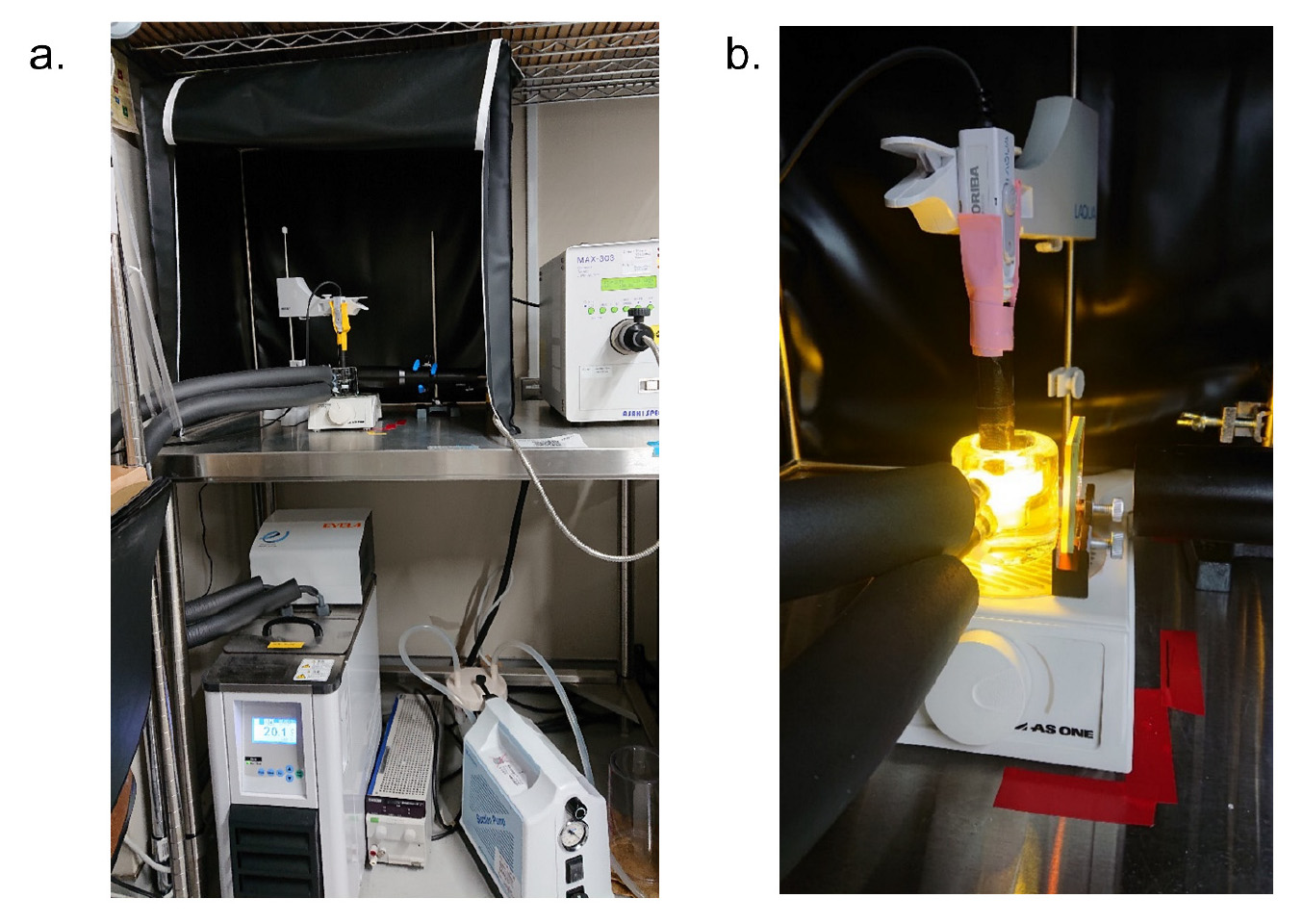
Figure 1. Setup for measuring light-dependent pH change. a. Overall view of the setup. b. Close-up view of the measurement part.
a. Record the baseline for 3 min in the dark.
b. Using the cut-off filter (Y-52) and infrared cutting filter, illuminate the sample with the light with a wavelength range that sufficiently covers the absorption wavelength of rhodopsin from a xenon lamp for 150 sec.
c. Turn off the light and wait the pH value to be back to the baseline level (it takes more than. 150 sec).
d. Record baseline again for 2 min in the dark.
e. Add 10 μL of 20 mM HCl and monitor the pH for 150 sec, in order to calibrate the buffering effect of the cell suspension.
f. Add 10 μL of 20 mM NaOH to bring the pH back to the baseline level.
Note: Representative trace is shown in Figure 2.
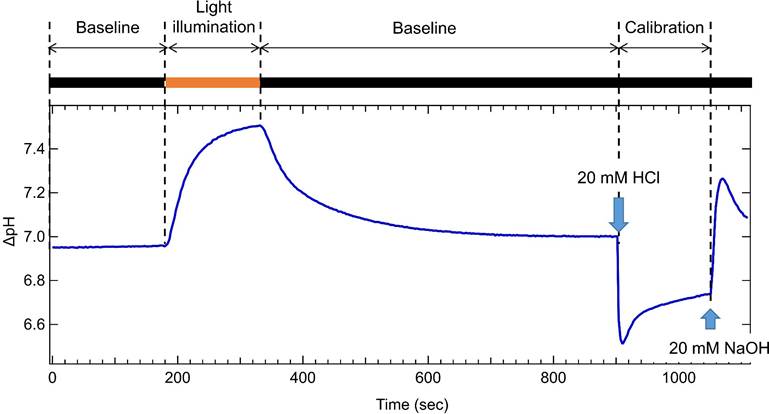
Figure 2. Representative trace of monitoring extraceluller pH change. Black and orange boxes indicate the dark (black) and light-illuminated (orange) conditions, respectively.
11. Add 2.5 μL of 30 mM CCCP and repeat step 10.
Note: Because CCCP is a protonophore, the pH change decreases when rhodopsin is directly transporting H+, but increases when secondary H+ transport associated with other ion transport, such as Na+ or Cl– (Inoue et al. 2013; Yoshizawa et al. 2014).
12. After completing the measurement, remove the cell suspention from the glass cell with micropipetter.
13. Wash the glass cell with ultrapure water five times.
Note: To remove the waste water from the glass cell completely, suck the waste water with a suction pump.
C. Quantification of expressed protein by hydroxylamine (HA) bleach method.
1. Sample preparation
a. Measure the OD660 of the cultured cells and dispense the cultured cells into a 15 mL plastic tube, which contains a cell amount equivalent to 20 OD·mL.
b. Collect the cells by centrifugation (4800×g, 2 min, room temperature) and discard the supernatant.
c. Stored the cell pellet at –80 °C until use.
d. Suspend the collected cells with 1 mL of bleaching buffer containing 1 mM lysozyme (M.W.=14,307) and adjust the volume up to 3 mL with bleaching buffer.
Note: Dissolve 1 mM lsozyme just before use.
e. Add a small amount of DNaseI on the tip of a spatula to the cell suspension. Then cover the tube with aluminum foil and mix by rotating at room temperature for 1 hr.
f. Place the cell suspention on ice and disrupt the cells by sonication until the lysate becomes less turbid. The sonication conditions are 5-sec pulses of ultrasound irradiation, 5-sec interval and 35 % output for a total of 10 min.
g. To solubilize the membrane protein, add 0.09 g of DDM to the sonicated cell suspension.
Final concentration of DDM is 3 %. Then cover the tube with aluminum foil and mix by rotating at 4 °C for overnight.
2. Measurement of the absorption spectrumNote: The absorption spectrum is measured with a UV-visible spectrometer with an integrating sphere mounted on. All operations are carried out in the dark or under the red-light (λ > 660 nm) condition.
a. Transfer the 2.7 mL of the solubilized sample to a disposable optical cuvette with a 10 mm optical path length. Then measure the absorbance before the addition of HA.
b. Add 0.9 mL of 2 M HA stock solution to the solubilized sample and mix well by pipetting. Final concentration of HA is 500 mM. Measure the absorption spectrum immediately after addition of HA in the dark (illumination time, t = 0).
c. Using a cut-off filter (Y-52) and heat-absorbing filter (HAF-50S-50H), illuminate the sample with light of λ > 500 nm wavelength from a xenon lamp for 1 minute (t = 1 min) and then measure the spectrum again.
d. Calculate the difference spectrum of before and after light illumination.
e. Illumination time is prolonged in the geometric progression manner (t = 2, 4, 8, 16, 32 min, …) and repeat the step c and d until the bleaching signal of rhodopsin is completely saturated.
f. Calculate the molar extinction coefficient of rhodopsin (ε) from the ratio between the absorbance of bleached rhodopsin and that of the generated retinal oxime (ε = 33,600 M–1 cm–1). The molar amount of rhodopsin expressed in E. coli cells is calculated by dividing the absolute value of the absorbance of bleached rhodopsin by ε.
Data analysis
1. When comparing the activity for multiple molecular species, normalize each trace by the amount of pH change after 150 sec from the addition of HCl as shown in step B.10.e-f of “Procedure” and the amount of protein expression as shown in step C.2.of “Procedure”.
Notes
1. By changing the type of salt used in the measurement, it is possible to evaluate the transport capacity of various ion species; for example, KCl, CsCl, NaNO3 and Na2SO4.
Recipes
1. 2×YT medium
| Component | Concentration (g/L) |
| Tryptone | 16 |
| Yeast extract | 10 |
| Sodium chloride | 5 |
a. Dissolve all components in ultrapure water and adjust the volume.
b. Dispense the medium to 115 mL in 300 mL flasks topped with silicone stoppers.
c. Autoclave the medium at 121 °C for 20 min.
d. Leave it until it cools down.
2. 50 mg/mL Ampicilin stocks
a. Add 10 mL of ultrapure water to 0.5 g of ampicillin powder and dissolve it completely.
b. Filter and sterilize the solution using a filter with a pore size of 0.2 μm.
c. Dispense the solution to 1 mL in sterile 1.5 mL tubes.
d. Store the stocks at –20 °C.
3. 30 mM CCCP stocks
a. Add 16.3 mL of DMSO to 100 mg of CCCP powder and dissolve it completely.
b. Dispense the solution to 100 μL in 1.5 mL tubes.
c. Store the stocks at –20 °C.
4. 2 M Hydroxylamine stocks
a. Dissolve 13.898 g of hydroxylamine hydrochloride in ultrapure water.
b. Adjust the pH to 8 with NaOH solution and then adjust the volume up to 100 mL with ultrapure water.
c. Dispense the solution to 1 mL in sterile 1.5 mL tubes.
d. Store the stocks at –20 °C.
5. Bleaching buffer
| Component | Concentration (mM) |
| NaCl | NaCl |
| Na2HPO4 | 66 |
a. Dissolve all components in ultrapure water.
b. Adjust the pH to 8 with NaOH solution and then adjust the volume with ultrapure water.
c. Transfer the solution into a glass-screw capped bottle and store the buffer at room temperature.
Acknowledgements
This work was supported by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) for Scientific Research to K.I. (KAKENHI grant Nos. 17H03007, 20K21383, and 20H05758) and H.K. (KAKENHI grant No. 18H03986), and CREST, the Japan Science and Technology Agency (JST) to H.K. (JPMJCR1753) and PRESTO, JST to K.M. (JPMJPR1903). This protocol was adopted from our original research paper (Inoue et al. 2020).
References
Ernst, O. P., Lodowski, D. T., Elstner, M., Hegemann, P., Brown, L. S. and Kandori, H. (2014) Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem Rev 114(1): 126-163.
Harris, A., Ljumovic, M., Bondar, A. N., Shibata, Y., Ito, S., Inoue, K., Kandori, H. and Brown, L. S. (2015) A new group of eubacterial light-driven retinal-binding proton pumps with an unusual cytoplasmic proton donor. Biochim Biophys Acta 1847(12): 1518-1529.
Hasemi, T., Kikukawa, T., Kamo, N. and Demura, M. (2016) Characterization of a cyanobacterial chloride- pumping rhodopsin and its conversion into a proton pump. J Biol Chem 291(1): 355-362.
Inoue, K., Ito, S., Kato, Y., Nomura, Y., Shibata, M., Uchihashi, T., Tsunoda, S. P. and Kandori, H. (2016) A natural light-driven inward proton pump. Nat Commun 7: 13415.
Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa, S., Ito, H., Kogure, K. and Kandori, H. (2013) A light-driven sodium ion pump in marine bacteria. Nat Commun 4: 1678.
Inoue, K., Tsunoda, S. P., Singh, M., Tomida, S., Hososhima, S., Konno, M., Nakamura, R., Watanabe, H., Bulzu, P. A., Banciu, H. L., Andrei, A., Uchihashi, T., Ghai, R., Béjà, O. and Kandori, H. (2020) Schizorhodopsins: A family of rhodopsins from asgard archaea that function as light-driven inward H+ pumps. Sci Adv 6(15): eaaz2441.
Klapoetke, N. C., Murata, Y., Kim, S. S., Pulver, S. R., Birdsey-Benson, A., Cho, Y. K., Morimoto, T. K.,
Chuong, A. S., Carpenter, E. J., Tian, Z., Wang, J., Xie, Y., Yan, Z., Zhang, Y., Chow, B. Y., Surek, B., Melkonian, M., Jayaraman, V., Constantine-Paton, M., Wong, G. K. and Boyden, E. S. (2014) Independent optical excitation of distinct neural populations. Nat Methods 11(3): 338-346.
Needham, D. M., Yoshizawa, S., Hosaka, T., Poirier, C., Choi, C. J., Hehenberger, E., Irwin, N. A. T., Wilken, S., Yung, C. M., Bachy, C., Kurihara, R., Nakajima, Y., Kojima, K., Kimura-Someya, T., Leonard, G., Malmstrom, R. R., Mende, D. R., Olson, D. K., Sudo, Y., Sudek, S., Richards, T. A., DeLong, E. F., Keeling, P. J., Santoro, A. E., Shirouzu, M., Iwasaki, W. and Worden, A. Z. (2019) A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc Natl Acad Sci USA 116(41): 20574-20583.
Pushkarev, A. and Béjà, O. (2016) Functional metagenomic screen reveals new and diverse microbial rhodopsins. ISME J 10(9): 2331-2335.
Waschuk, S. A., Bezerra, A. G., Jr., Shi, L. and Brown, L. S. (2005) Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci USA 102(19): 6879-6883.
Yoshizawa, S., Kumagai, Y., Kim, H., Ogura, Y., Hayashi, T., Iwasaki, W., DeLong, E. F. and Kogure, K. (2014) Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc Natl Acad Sci USA 111(18): 6732-6737.
Related files
 Bio-protocol_ion_transport_assay.pdf
Bio-protocol_ion_transport_assay.pdf - Konno, M, Inoue, K and Kandori, H(2021). Ion transport activity assay of microbial rhodopsin expressed in E. coli cells. Bio-protocol Preprint. bio-protocol.org/prep800.
- Inoue, K., Tsunoda, S. P., Singh, M., Tomida, S., Hososhima, S., Konno, M., Nakamura, R., Watanabe, H., Bulzu, P., Banciu, H. L., Andrei, A., Uchihashi, T., Ghai, R., Béjà, O. and Kandori, H.(2020). Schizorhodopsins: A family of rhodopsins from Asgard archaea that function as light-driven inward H+ pumps . Science Advances 6(15). DOI: 10.1126/sciadv.aaz2441
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

