Advanced Search
Mono-allelic expression of HLA-I alleles for HLA-peptidomics analysis
Last updated date: Jan 24, 2023 Views: 538 Forks: 0
Adi Nagler1*, Shelly Kalaora2*, Chaya Barbolin3, Michal Alon3, Polina Greenberg3, Gal Yagel3, Aviyah Peri3, Yishai Levin4, Yardena Samuels3**
1 Dana-Farber Cancer Institute; Harvard Medical School, Boston, Massachusetts, United States
2 Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, United States
3 Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel
4 The de Botton Institute for Protein Profiling, The Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science, Rehovot, Israel
* Contributed equally to this work
** Corresponding author: yardena.samuels@weizmann.ac.il
Lead Contact: Prof. Yardena Samuels
Abstract
The identification of human leukocyte antigen (HLA)-bound peptides using HLA-peptidomics (also called “Immunopeptidomics”) analysis has provided an in-depth understanding of antigen presentation. A possible obstacle to such an analysis is the co-expression of multiple HLA alleles, thus reducing the specificity of this assay. Here we present a mono-allelic strategy for HLA class-I (HLA-I) expression, which can be further utilized for HLA peptidome profiling by liquid chromatography-tandem mass spectrometry (LC/MS-MS). Using this application combined with the overexpression of genes of interest has been demonstrated to enable the identification of SARS-CoV-2-derived HLA-I peptides. We thus demonstrate a strategy for systematic HLA- allele specific analysis of antigen presentation.
Keywords: HLA-I, mono-allele, lentivirus, peptides
Graphical Abstract
Background
The HLA-I molecules (HLA-A/B/C) are heterodimeric proteins presented on the surface of human nucleated cells, encoded by genes displaying a large degree of polymorphism with hundreds of allelic variations (Jin and Wang, 2003; Robinson et al., 2015). HLA-I bound peptides arise from intracellular proteins that are cleaved by the proteasome and displayed by the surface HLA-I proteins serving as an immunological signature for the detection by CD8+ T cells via their T cell receptor.
Characterization of the HLA-bound peptides by immunopurification and LC-MS/MS has revealed rules for peptide binding to the HLA molecules and has been performed on various HLA alleles.(Abelin et al., 2017; Bassani-Sternberg et al., 2015; Hickman et al., 2004; Rammensee et al., 1995; Vita et al., 2019) This unbiased powerful approach can elucidate novel peptides presented by the HLA-I alleles informing the rational design of CD8+ T cell inducing vaccines to control pathogen infections or to combat tumors.
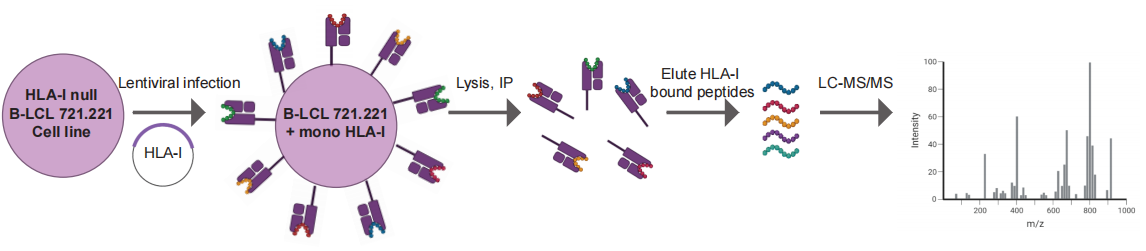
Here we provide a mono-allelic approach that has been used to detect the presentation of SARS-CoV-2 viral peptides (Nagler et al., 2021), bacterial peptides (Kalaora et al., 2021) and has also been demonstrated efficient in the detection of neoantigens recognized by CD8+ T cells (Bear et al., 2021; Peri et al., 2021). Briefly, the protocol can be broadly outlined as a three-step process. In the first step, DNA sequences coding for the HLA-I alleles are cloned into a lentivirus vector with neomycin selection and lentiviral particles are produced. In the second step, human B-LCL 721.221 (HLA-I null) cells are infected and selected with antibiotics. Productive over-expression of HLA-I is validated approximately two weeks post-infection via flow cytometry. In the third step, the 721.221 HLA-I cells are re-infected with lentivirus containing a gene of interest. This HLA-I monoallelic expression protocol can be further implemented for HLA-I peptidome analysis in which the HLA-I peptide complexes are purified and the HLA-I presented peptides are eluted off the complexes. These peptides are sequenced with LC-MS/MS and identified by a database search.
Materials and reagents
- dsDNA sequences coding for HLA-I alleles (IPD-IMGT/HLA, twist bioscience)
- pCDH-CMV-MCS-EF1α-Neo vector (SBI, #CD514B-1)
- PMD2.G (#12259, Addgene)
- psPAX2 (#12260, Addgene)
- HEK293T cells (ATCC)
- Human B-LCL 721.221 (ATCC)
- XbaI (NEB, #R0145L)
- NotI (NEB, #R0189L)
- T4 DNA ligase (NEB, #M0202L)
- Lipofectamine 2000 (Invitrogen, #11668027)
- Mouse monoclonal anti-pan HLA-I (clone W6/32, Purified from HB95 hybridoma cells)
- RPMI 1640 Medium(BI, 01-100-1A)
- Opti-MEM™ I Reduced Serum Medium (Gibco, 11058021)
- Steriflip-GP Sterile Centrifuge Tube Top Filter Unit (Millipore, SE1M179M6)
- Geneticin™ Selective Antibiotic (G418 Sulfate, ThermoFisher, 10131035)
- 50ml conical tubes
- 10 cm2 dishes
- 6 Well Cell Culture Plate
Equipment
- Tissue culture hood
- Incubator 37°C
- BD LSR II (BD Biosciences)
Software
- FACSDiva (BD, biosciences, https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software)
- FloJo (FlowJo, LLC, https://www.flowjo.com)
Procedure
- DNA sequences coding for HLA-I alleles were taken from IPD-IMGT/HLA database (https://www.ebi.ac.uk/ipd/imgt/hla/allele.html).
- Synthetic dsDNA of HLA-I alleles were purchase from Twist bioscience with flanking XbaI and NotI restriction sites.
- Coding sequence was cloned into pCDH-CMV-MCS-EF1α-Neo vector.
- Lentivirus production was performed by co-transfection of 4.4 µg of the HLA-I plasmid, with 2.2 µg of PMD2.G and 3.4 µg of psPAX2 into 3X106 HEK293T seeded on a 10 cm2 dish in RPMI 1640 medium, using Lipofectamine 2000, as described by the manufacturer.
- After 24 hours post-transfection, media was changed to Opti-MEM™ Reduced Serum Medium. 72 hours post-transfection, media was collected, filtered using Steriflip-GP Sterile Centrifuge Tube, aliquoted and stored at −80°C.
- 2 X106 Human B-LCL 721.221 were seeded in a 6 well cell culture plate, infected with 1 ml of HLA-I lentivirus and after 72 hours were selected with 800 µg/ml neomycin (G418) for 2 weeks.
- HLA-I expression was confirmed via flow cytometry, using the monoclonal antibody W6/32.
- The B-LCL 721.221 HLA-I expressing cells could be re-infected with a vector containing a gene of interest and an alternative antibiotic selection such as Puromycin.
- B-LCL 721.221 HLA-I expressing cells could be further expanded and used for HLA-peptidomics, which is based on the immunoaffinity purification of the HLA complexes followed by the extraction of the HLA-bound peptides and their further analysis by mass spectrometry (Kalaora et al., 2016; Kalaora and Samuels, 2019; Kalaora et al., 2018)
References
Abelin, J.G., Keskin, D.B., Sarkizova, S., Hartigan, C.R., Zhang, W., Sidney, J., Stevens, J., Lane, W., Zhang, G.L., Eisenhaure, T.M., et al. (2017). Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 46, 315-326.
Bassani-Sternberg, M., Pletscher-Frankild, S., Jensen, L.J., and Mann, M. (2015). Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics 14, 658-673.
Bear, A.S., Blanchard, T., Cesare, J., Ford, M.J., Richman, L.P., Xu, C., Baroja, M.L., McCuaig, S., Costeas, C., Gabunia, K., et al. (2021). Biochemical and functional characterization of mutant KRAS epitopes validates this oncoprotein for immunological targeting. Nat Commun 12, 4365.
Hickman, H.D., Luis, A.D., Buchli, R., Few, S.R., Sathiamurthy, M., VanGundy, R.S., Giberson, C.F., and Hildebrand, W.H. (2004). Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol 172, 2944-2952.
Jin, P., and Wang, E. (2003). Polymorphism in clinical immunology - From HLA typing to immunogenetic profiling. J Transl Med 1, 8.
Kalaora, S., Barnea, E., Merhavi-Shoham, E., Qutob, N., Teer, J.K., Shimony, N., Schachter, J., Rosenberg, S.A., Besser, M.J., Admon, A., et al. (2016). Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget 7, 5110-5117.
Kalaora, S., Nagler, A., Nejman, D., Alon, M., Barbolin, C., Barnea, E., Ketelaars, S.L.C., Cheng, K., Vervier, K., Shental, N., et al. (2021). Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138-143.
Kalaora, S., and Samuels, Y. (2019). Cancer Exome-Based Identification of Tumor Neo-Antigens Using Mass Spectrometry. Methods Mol Biol 1884, 203-214.
Kalaora, S., Wolf, Y., Feferman, T., Barnea, E., Greenstein, E., Reshef, D., Tirosh, I., Reuben, A., Patkar, S., Levy, R., et al. (2018). Combined Analysis of Antigen Presentation and T-cell Recognition Reveals Restricted Immune Responses in Melanoma. Cancer Discov 8, 1366-1375.
Nagler, A., Kalaora, S., Barbolin, C., Gangaev, A., Ketelaars, S.L.C., Alon, M., Pai, J., Benedek, G., Yahalom-Ronen, Y., Erez, N., et al. (2021). Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep 35, 109305.
Peri, A., Greenstein, E., Alon, M., Pai, J.A., Dingjan, T., Reich-Zeliger, S., Barnea, E., Barbolin, C., Levy, R., Arnedo-Pac, C., et al. (2021). Combined presentation and immunogenicity analysis reveals a recurrent RAS.Q61K neoantigen in melanoma. J Clin Invest 131.
Rammensee, H.G., Friede, T., and Stevanoviic, S. (1995). MHC ligands and peptide motifs: first listing. Immunogenetics 41, 178-228.
Robinson, J., Halliwell, J.A., Hayhurst, J.D., Flicek, P., Parham, P., and Marsh, S.G. (2015). The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43, D423-431.
Vita, R., Mahajan, S., Overton, J.A., Dhanda, S.K., Martini, S., Cantrell, J.R., Wheeler, D.K., Sette, A., and Peters, B. (2019). The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47, D339-D343.
- Nagler, A, Kalaora, S, Barbolin, C, Alon, M, Greenberg, P, Yagel, G, Peri, A, Levin, Y and Samuels, Y(2023). Mono-allelic expression of HLA-I alleles for HLA-peptidomics analysis. Bio-protocol Preprint. bio-protocol.org/prep2129.
- Nagler, A., Kalaora, S., Barbolin, C., Gangaev, A., Ketelaars, S. L., Alon, M., Pai, J., Benedek, G., Yahalom-Ronen, Y., Erez, N., Greenberg, P., Yagel, G., Peri, A., Levin, Y., Satpathy, A. T., Bar-Haim, E., Paran, N., Kvistborg, P. and Samuels, Y.(2021). Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep 35(13). DOI: 10.1016/j.celrep.2021.109305
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
