Advanced Search
For High Throughput Screening: A Quick DNA Extraction Method for Gram-positive Bacteria
Last updated date: Apr 27, 2022 Views: 1466 Forks: 0
For High Throughput Screening: A Quick DNA Extraction Method for Gram-positive Bacteria
Nuo Chen1, 2*, Xiaoming Yuan1, 2
1Department of Food Science & Technology, Institute of Food Safety and Nutrition, Jinan University, Guangzhou, 510632, China, 2 Institute of Microbiology, Guangdong Academy of Sciences, State Key Laboratory of Applied Microbiology Southern China, Key Laboratory of Agricultural Microbiomics and Precision Application, Ministry of Agriculture and Rural Affairs; Guangdong Provincial Key Laboratory of Microbial Safety and Health, Guangzhou 510070, China
*For correspondence: chennuo7676@163.com
[Abstract] In this study, an ultrasonic based DNA extraction method, by using which the whole process can be finished within 10 min, was developed. This method is almost no cost and timesaving, which is useful for high throughput screening, especially in the screening of mutants generated in random mutagenesis.
Keywords: Gram-positive Bacteria, DNA Extraction, Ultrasound, High Throughput Screening, PCR
[Background] Colony PCR, which is commonly used to verify the expected DNA sequence in Escherichia coli, but it failed in Gram- positive bacteria like Bacillus subtilis even treated by chemical reagents (Azevedo et al., 2017; Packeiser et al., 2013). The reason why colony PCR failed in these cases is because the thick cell wall that exist in most Gram-positive bacteria impedes cell lysis and subsequent DNA release. Enzymes like lysozyme, proteinase K and other chemical reagents are used to lyse the Gram- positive bacterial cell wall; however, these methods are costly, time consuming, and not eco-friendly for DNA extraction of huge quantity of samples. For example, it is very difficult to achieve a fast screen towards thousands of mutants generated in random mutagenesis. It is therefore necessary to set up a convenient and economical method.
Ultrasound is a common physical disruption method employed to extract DNA from microbes. In fish intestinal microflora and compost microbes, ultrasonic lysis method combined with lysozyme is used to extract DNA (Z et al., 2018; ZH et al., 2006). In this study, an ultrasonic based DNA extraction method was developed and it can screen large samples in just one day. Although purity is not a patch on commercial kit, this method can be used for high throughput screening purpose, which needs to extract thousands of DNA, in Gram- positive bacteria like B. cereus,Bacillus. Thuringiensis, Bacillus subtilis, and Listeria monocytogenes.
Materials and Reagents
- Strains
- Bacillus cereus ATCC 14579 (available from ATCC, catalog number: NZ_CP053931.1)
- Bacillus. thuringiensis ATCC 10792 (available from ATCC, catalog number: GCF_002119445.1)
- Bacillus subtilis ATCC6633 (available from ATCC, catalog number: NZ_CP034943.1)
- Listeria monocytogenes ATCC 19115 (available from ATCC, catalog number: GCF_001652935.1)
- Reagents
- Nutrient agar plate
- Taq DNA Polymerases (Thermo Fisher Scientific, cat No. 10966018)
- Marker D (100~2000 bp, Sangon Biotech, cat No. B600335-0050)
- GoldView™ (Coolaber, cat No. GD001-1ml*5)
Equipment
- Incubator
- Benchtop centrifuge (Eppendorf)
- Ultrasonic cleaning machine (40KHz, 20°C)
- Thermal cycler (C1000 Touch, Bio-Rad)
Software
Image Lab (Bio-Rad)
Procedure
- Suspend a single colony into 30 μl ddH2O in a 1.5 ml tube.
Note: the size of B. cereus ATCC 14579, B. thuringiensis 10792, B. subtilis ATCC 6633 and L. monocytogenes ATCC 19115 is recommended in Table 1.
- Ultrasonic treat the bacterial suspension at 40 KHz at 20°C for 5 min and then centrifuge at 10,000 × g at room temperature for 1 min.
- Carefully transfer the upper aqueous phase to a new 1.5 ml tube without disturbing the pellet.
- The DNA samples are tested by polymerase chain reaction (PCR). Primers and parameters for PCR are listed in Table 2 and 3. After electrophoresis on 1.2% agarose gels (in 1x TAE), all amplicons are visualized under UV light with GoldView™ staining. The results are shown in Figure 2.
Note: the bands are brighter if you increase the PCR reaction cycles.
Table 1. Colony size of B. cereus, B. thuringiensis, B. subtilis and L. monocytogenes.
| Microorganism | Colony Size |
| B. cereus ATCC 14579 | 4 - 5 mm |
| B. thuringiensis 10792 | 4 - 5 mm |
| B. subtilis ATCC 6633 | 3.5 - 4 mm |
| L. monocytogenes ATCC 19115 | 1.2 - 2 mm |
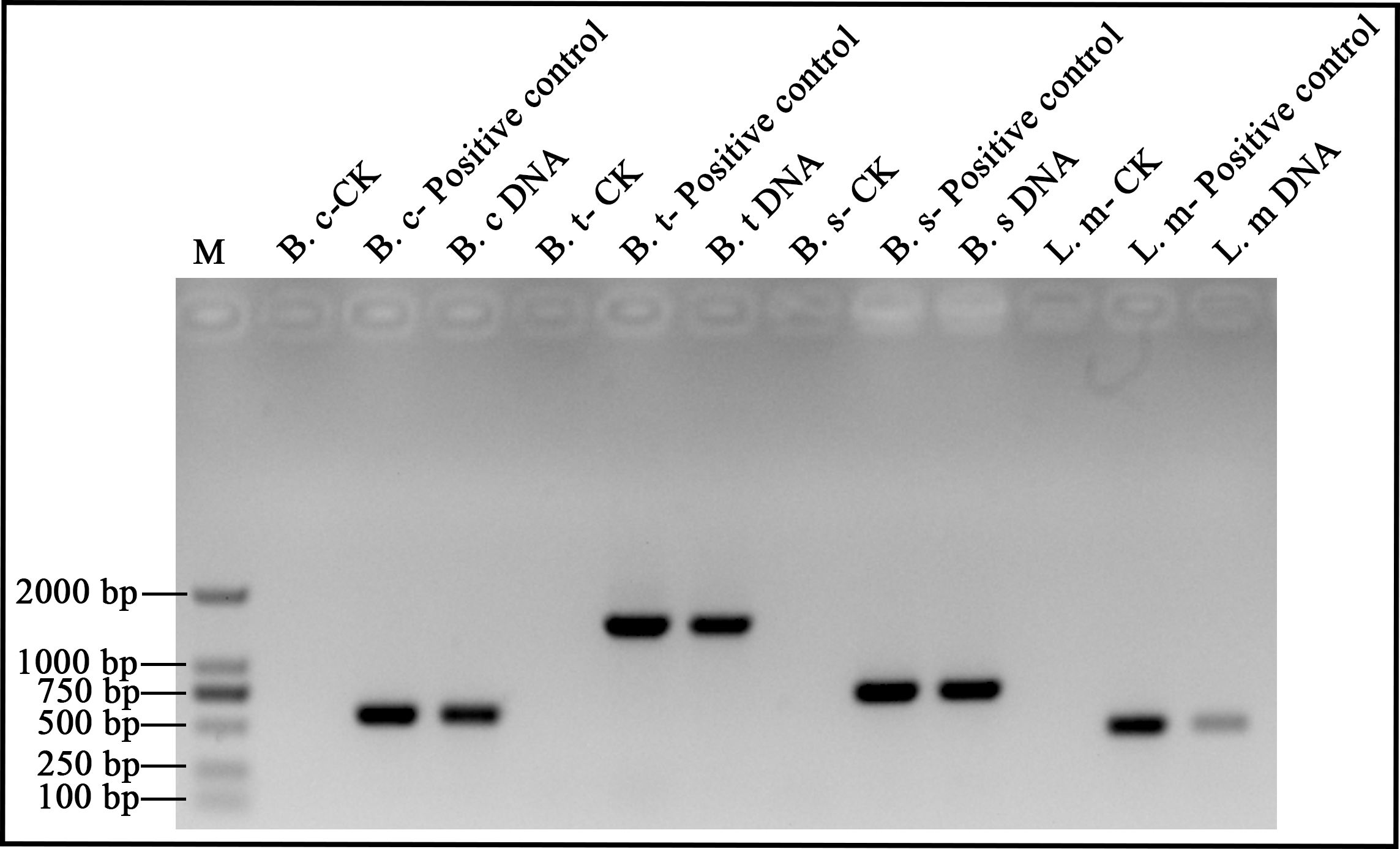
Figure 1. B. cereus, B. thuringiensis, B. subtilis and L. monocytogenes DNA analyzed by electrophoresis. lane M: 2000-bp DNA ladder. B. c: B. cereus, B. t: B. thuringiensis, B. s: B. subtilis, L. m: L. monocytogenes. DNA: DNA extracts by ultrasonic method for 5 min. Positive control: DNA extracted by commercial kit. CK: Check control.
Table 2. Primer sequences used in B. cereus, L. monocytogenes and B. subtilis, respectively.
| Microorganism | Target gene | Primer sequence |
| B. cereus ATCC 14579 | gmk | F-TTAAGTGAGGAAGGGTAGG R-AATGTTCACCAACCACAA |
| B. thuringiensis 10792 | pycA | F-GTGAAAGCAAGAACACAAGC R-ATAGTTTTTGTATCCAACTGCG |
| B. subtilis ATCC 6633 | SigB | F-ATGACACAACCATCAAAAACTACGA R-TTACATTAACTCCATCGAGGGATCT |
| L. monocytogenes ATCC 19115 | prfA | F-GTCAAAACATACGCTCTTATC R-ACATAATCAGTCCAAAGTAGATGC |
Initial denaturation and denaturation | Annealing | Extension and final extension | Number of circles |
95°C, 3 min and 95°C, 30 s | 55°C, 30 s | 72°C, 1 min and 72°C, 10 min | 25 × |
Table 3. PCR conditions used to test sample quality.
Acknowledgements
This research was supported by Guangdong Major Project of Basic and Applied Basic Research (2020B0301030005), Guangdong Provincial Key Laboratory (2020B121201009), and Guangdong Province Academy of Sciences Special Project for Capacity Building of Innovation Driven Development (2020GDASYL-20200301002).
Reference
- Azevedo, F., Pereira, H., & Johansson, B. (2017). Colony PCR. Methods in molecular biology (Clifton, N.J.), 1620,129–139.
- Han, Z., Sun, J., Lv, A., Sung, Y., Sun, X., Shi, H., Hu, X., Wang, A., & Xing, K. (2018). A modified method for genomic DNA extraction from the fish intestinal microflora. AMB Express, 8(1), 52.
- Mertens, K., Freund, L., Schmoock, G., Hänsel, C., Melzer, F., & Elschner, M. C. (2014). Comparative evaluation of eleven commercial DNA extraction kits for real-time PCR detection of Bacillus anthracis spores in spiked dairy samples. International journal of food microbiology, 170, 29–37.
- Packeiser, H., Lim, C., Balagurunathan, B., Wu, J., & Zhao, H. (2013). An extremely simple and effective colony PCR procedure for bacteria, yeasts, and microalgae. Applied biochemistry and biotechnology, 169(2), 695–700.
- Yang ZH, Xiao Y, Zeng GM, Liu YG, Deng JH (2006) DNA extraction methods of compost for molecular ecology analysis. Environmental Science, 27(8), 1613–1617. In Chinese.
- Chen, N and Yuan, X(2022). For High Throughput Screening: A Quick DNA Extraction Method for Gram-positive Bacteria. Bio-protocol Preprint. bio-protocol.org/prep1644.
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
