Advanced Search
Biodiversity online databases: An applied R protocol to get and curate spatial and climatic data
Last updated date: Mar 8, 2022 Views: 1520 Forks: 0
Biodiversity online databases: An applied R protocol to get and curate
spatial and climatic data
Coca-de-la-Iglesia, M.1* Valcárcel, V.1, 2 and G. Medina, N.1,2
1Departamento de Biología, Universidad Autónoma de Madrid (UAM), Madrid, Spain; 2Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Madrid, Spain; $Current/Present address: Dept/Center, Institution name, City, Country
*For correspondence: marina.coca@inv.uam.es
[Abstract]
Ecological and evolutionary studies often require high quality biodiversity data. This information is easy to access through the many online databases that have compiled biodiversity data from herbaria, museums, and human observations. However, the process to get this information ready for analyses is complex and time-consuming. In this study, we have developed a protocol in R language to process spatial data (download, merge, clean and correct) and extract climatic data, using some genera of the ginseng family (Araliaceae) as example. The protocol provides an automatic way to process spatial and climatic data for numerous taxa independently and from multiple online databases. The script uses GBIF, BIEN and WorldClim as the online data sources, but it is easy to adapt to include other online databases. Also, the script uses genera as the sample unit, but it provides the way to use species as the target. The cleaning process incorporates a filter that removes occurrences outside the natural range of taxa, gardens and other human environments, and erroneous locations and a spatial correction for misplaced occurrences (i.e, occurrences within a distance buffer from the coastal limit). Additionally, each step of the protocol can be run independently. Thus, the protocol can be started on the data cleaning, if the database is already compiled, or on the climatic data extraction, if the database is already parsed. Every line in the R script is commented so that it can also be run by users with little knowledge on R.
Keywords: BIEN, Data cleaning, GBIF, Online biodiversity databases, R language, WorldClim.
[Background] Our knowledge on species distributions is central to biogeographers, but also to phylogenetists and ecologists. Indeed, species ranges are needed to perform phylogenetic climatic reconstructions, species niche characterizations or species distribution models, and address multiple evolutionary questions. However, achieving accurate spatial information on the species distributions requires from good-quality occurrence databases with high geographic coverage that are difficult to gather.
The principal sources of geographical information are field inventories and biodiversity collections (museums and herbaria), for which accessibility was a serious limitation until recently. The digitization effort done during the last decades has facilitated the access to tons of biodiversity data that was previously scattered in different institutions across the world, through online databases such as the Global Biodiversity Information Facility (GBIF; GBIF.org, 2021). As a result, we have now available large amount of biodiversity data, which provides an unprecedented opportunity to take advantage of centuries of naturalist’s observations across the world. However, the use of this valuable information is limited because persistent gaps of (i) knowledge and technical limitations (ii). On the one hand (i), our knowledge on species distribution is still poor, biased or imprecise (Hortal et al., 2007) and this is reflected on the information gathered in biodiversity databases that is not uniform across lineages or across regions. This biases lead to groups of organisms and regions of the world that have scarce information while others concentrate large amount of data (Hortal & Lobo, 2005). On the other hand (ii), the complexity of the process to get online data parsed and ready for analyses is high. For example, it is frequent that online repositories include records with imprecise or erroneous spatial information (such as land organisms falling into the sea) or with outdated taxonomic nomenclature (Soberón & Peterson, 2004). Thus, every study based on online data requires from an initial step of cleaning and parsing to remove or minimize the impact of these sources of uncertainty (persistent gaps of knowledge and technical limitations) on further analyses (Hortal et al., 2007).
In parallel with the international digitization effort done in the last decades, several methodologies and pipelines have been conceived to deal with these sources of uncertainty and simplify the different steps when working with online biodiversity data. Some of the most relevant protocols have been developed in R (R Core Team, 2018) and include geographic, taxonomical or temporal data cleaning (see for example: bRacatus, Arlé et al., 2021; BDcleaner, Jin & Yang, 2020; plantR, Lima et al., 2021; Biogeo, Robertson et al., 2016; SpeciesGeoCorder, Töpel et al., 2016; CoordinateCleaner, Zizka et al., 2019). However, none of them deals with the uncertainty introduced both by the spatial gaps of knowledge and the technical limitations. Also, most of them are focused in one or a few steps of the process. Thus, to complete the process (from the initial download of rough occurrences to the climatic data extraction of the cleaned and parsed spatial database) users need to deal with different protocols, some of which require programming skills or deep R background.
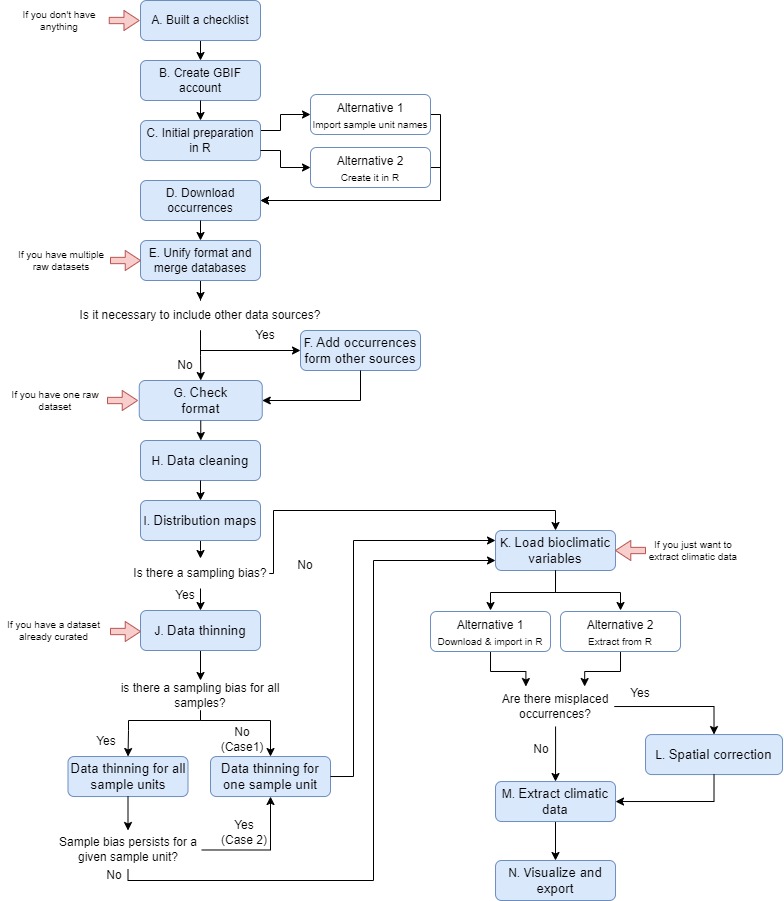
The R protocol that we present here is designed to create reliable databases of species occurrences and climatic data from online repositories. It provides an automatic procedure to deal with the most frequent sources of spatial uncertainty of online biodiversity databases. It also includes an automatic script to run each sample (species, genus, family, etc.) separately, which allows for an easy and fast way to process hierarchical databases. The script also includes a post-processing code to run after the spatial pipeline and extract the climatic data. The protocol describes step-by-step how to download, parse, clean and merge spatial and climatic data from three online databases (Figure 1; GBIF, GBIF.org, 2021; BIEN, Maitner, 2020; and Worldclim, Fick & Hijmans, 2017). However, the protocol can be easily adapted to include any other online biodiversity database that may be of interest. The cleaning steps include how to automatically update nomenclatural information, identify and remove records outside the natural distribution of taxa, records from gardens and other human environments or geographically inaccurate records. To explain the protocol, we used the Asian Palmate Group (AsPG) of Araliaceae as case study, using genera as the sample unit. To speed up the protocol execution process we selected 16 of the AsPG. The selection of genera was done as to display uneven spatial information across genera and across areas of the world (to address the issue derived from gaps of knowledge as source of spatial uncertainty), and that are largely affected by erroneous and misplaced records (to address the issue derived from technical limitations as source of spatial uncertainty).
To summarize, the main advantages of this protocol are that it: (1) can be applied to all groups of organisms (as long as they have information available in GBIF or BIEN databases) and at any taxonomic rank, not only at the species level; (2) provides an automatic way to process hierarchical databases, which is very helpful when studying highly diversified groups (genera with high number of species, families with high number of genera, etc.; (3) provides a complete pipeline from spatial data download (including multiple databases merging) to climatic data extraction; (4) deals with uncertainty coming from technical limitations (such as wrong records), but also with the uncertainty derived from persistent gaps of knowledge (such as spatial biases across different parts of the world and across lineages); (5) provides an easy way of filtering records outside natural range; (5) applies a spatial correction for erroneous occurrences outside the coastal limit; (6) includes independent steps for each part of the process that can be run separately; and (7) can be easily used and modified by any kind of users, from undergraduate students to professors, irrespective of their ability, knowledge or background on R, because it is accompanied by instructions to guide the user.
Equipment
1.Computer with Microsoft® Windows® XP or Mac® OS X® 10.4 operator system or later versions of both.
Software
1.R version 3.5.1 (https://r-project.org/).
Packages: “BIEN“, “countrycode“, “data.table“, “devtools”, “dpyr“, “plyr”, “raster“, “readr“, “rgbif“, “rgdal“, “spocc“, “spThin“, "SEEG-Oxford/seegSDM" and “tidyr”.
2.RStudio version 1.1.456 (https://rstudio.com/products/rstudio/)
The use of RStudio is optional. RStudio is an interface that improves the use of R.
3.Microsoft® Excel® 2016.
4.Any text-editing program capable of exporting files in .txt, like WorldPad, Microsoft® Word®, Notepad++, etc.
Procedure
The R script can be freely downloaded from GitHub (link: https://github.com/NiDEvA/R-protocols.git; Note 1). The pipeline of the procedure coincides with the steps of the R script of this protocol (Figure 1). First, you have to create two working folders, one named “input” (it contains the information needed to run the R script) and another one named “output” (it will contain the resulting files after running the R script). In this protocol we used the genus rank as the sample unit but the script also includes commented lines (those preceded with “#”) with the functions needed if you want to use species as the sample unit. Besides, it can be easily modified to use family or any other higher taxonomic level as the sample unit if needed. Also, we cleaned the data by removing records outside the natural distribution of genera, from gardens and other human environments or geographically inaccurate, but it can also be easily modified to meet any particular data cleaning requirements.
A. Build checklist of taxa native range. It is necessary to know the countries for which the taxa are native. For plants this information can be found in World Checklist of Selected Plant Families (WCSP, Govaerts et al., 2008). WCSP is a database that compile checklists of a 200 seed plant families. The database is updated frequently, each new name published by International Plant Name Index (IPNI; International Plant Names Index, 2020), is reviewed and added to WCSP. Other sources of information on the natural distribution range of other organisms are available in ASM Mammal Diversity Database (https://www.mammaldiversity.org/index.html, Mammal Diversity Database, 2020), Avibase - The World Bird Database (https://avibase.bsc-eoc.org/avibase.jsp, Lepage et al., 2014), Catalogue of Life (https://www.catalogueoflife.org/, Bánki et al., 2022), Checklist of Ferns and Lycophytes of the World (Hassler, 2022a), Global Assessment of Reptile Distributions (http://www.gardinitiative.org/, GARD, 2022), Reptile Database (http://www.reptile-database.org/, Uetz et al., 2021), USDA Plants Database (https://plants.usda.gov, USDA, NRCS, 2022), World Plants (https://www.worldplants.de, Hassler, 2022b). To do so:
1.Create a txt file with the names of all taxa separated by “Enter” in a plain text editor (e.g., Notepad++, BBedit). Save it as “Natural_Distribution_Checklist_TDWG.txt”.
2. Visit the website https://wcsp.science.kew.org/home.do (or the correspondent webpage, see above) and introduce the taxon name in the search engine. WCSP uses two ways to describe the distribution of taxa, one in narrative form and the other one through international codes (Figure 2). The international code used in WCSP is the third level of geographical codes of the Taxonomic Databases Working Group (TDWG, Brummitt, 2001) (Note 2).
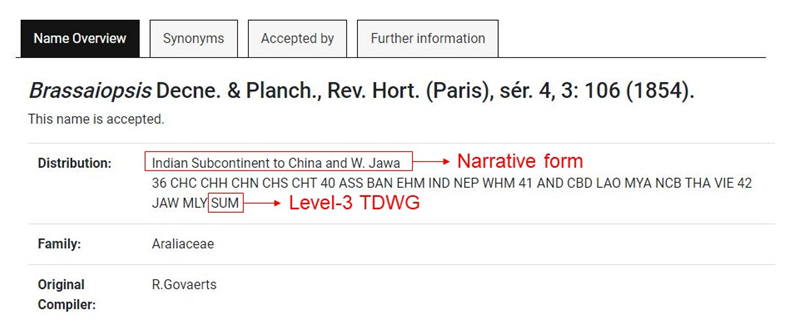
3.Copy each code of three capital letters (just the codes, not the numbers that appear at the begin of the country code line) and paste in “Natural_Distribution_Checklist_TDWG.txt” in the same line right after the corresponding taxa name separated by “;”. In some cases, symbols (“?”, “(?)”, “+”, ”†”) or lowercase letters may appear in distribution. According to TDWG, “?” is used when the presence of a taxon in a given area is not certain. If this symbol is used within brackets is because there is no exact location known within a country. When a taxon is extinct or may be extinct in an area the symbol ”†” is placed after the country code. When the country code is not known “+” is used. Lowercase letters for the country code indicate naturalization. For this protocol, we have only used the codes with three capital letters that do not have any symbol. For more information, consult the "about checklist" section on the WCSP website.
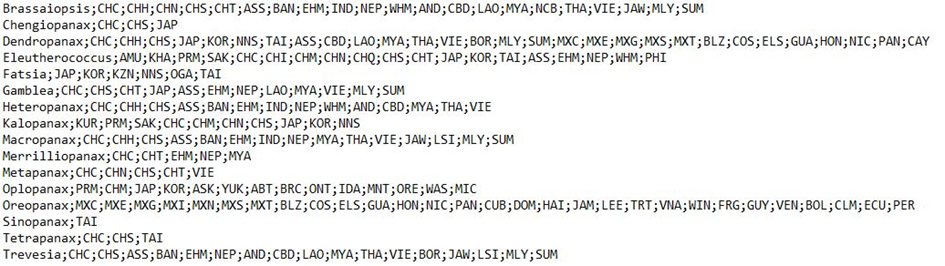
4.Repeat steps 2 and 3 until all taxa are completed and save the document in the “input” folder. The format of the resulting txt file should look as in Figure 3. It is advisable to sort the taxa alphabetically in the text file.
B. Create an account in GBIF database.
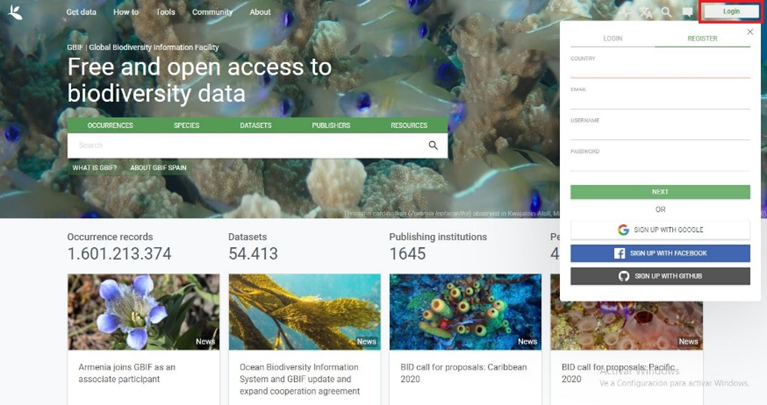
1.Visit the website https://www.gbif.org/. Click on “Login” located in the upper right corner of the web, and then on “REGISTER” (Figure 4).
2.Fill the “COUNTRY”, “EMAIL”, ”USERNAME”, and ”PASSWORD” fields, click on next and follow the instructions to create the account. It is also possible to create the account through Google, Facebook or Github. Important to remember: do not forget the information filled in the email, username and password, because it will be used later in the R script.
C. Initial preparation in R.
- Open RStudio (Note 3).
- Create the paths in R for the input and output data. These paths correspond with “input” and “output” folders. Replace the example path with the path of the input folder.
- Install the packages needed to run the R script. This packages are “BIEN” (Maitner et al., 2018), “countrycode” (Arel-Bundock et al., 2018), “data.table” (Dowle et al., 2019), “devtools” (Wickham et al., 2021),“dplyr” (Wickham et al., 2020), “plyr” (Wickham, 2020), “raster” (Hijmans et al., 2020), “readr” (Wickham et al., 2018),“rgbif” (Chamberlain et al., 2020a), “rgdal” (Bivand et al., 2020), “seegSDM” (Golding & Shearer, 2021), “spocc” (Chamberlain et al., 2020b), “spThin” (Aiello-Lammens et al., 2019) and “tidyr” (Wickham et al., 2022). The packages can be download directly into RStudio with “install.packages” function, except for “seegSDM” package which is installed through the “devtools” package as it is specified in L48 of the R script. In this protocol we use BIEN and GBIF as the online biodiversity databases to obtain the spatial data, if you want to include any other online database to obtain records for your case study, you will need to look for the correspondent R package and install them at this step of the protocol.
- Create a vector with the names of the taxa (sample unit). The vector can be created in R (see “Alternative 1” in the R script) or it can be imported from a file (.csv, .txt, .xls; see “Alternative 2” in the R script). For this protocol, we use “Alternative 2”. Be aware that the names of sample units must be exactly the same as in the “Natural_Distribution_Checklist_TDWG.txt” file and it must also be arranged in the order as in the “Natural_Distribution_Checklist_TDWG.txt” file (see above step A-1).
- Read the natural distribution text file built in step A named “Natural_Distribution_Checklist_TDWG.txt”. First, load in R the txt file that contains the natural distribution of sample units as vector. Then, convert the vector into a list. Each item in the list corresponds to a genus (or the correspondent sample unit: species, family, etc.) followed by its corresponding Level-3 TDWG country codes, separated by “;”.
D. Download the occurrence data from online databases (GBIF and BIEN, or the desired database).
- Use the package “rgbif“ to download the records from the GBIF database.
- Indicate the username, email and password created in step B-2. Replace de “XX” with your credentials (important do not remove the “” characters).
- Search the taxon keys of each taxon. GBIF has a key to identify each taxon in the database. In case you use species or families instead of genera as the sample unit, replace rank=”genus” in name_backbone function with rank=”species” or rank=”family” respectively.
- Prepare the download request and download data.
- Create the path to a folder that will contain the files downloaded from GBIF and create a folder inside the “input” folder and name it as “download_GBIF”. This folder will contain the files downloaded from GBIF. Indicate the number of taxa included in the txt file with the checklist of taxa native range.
- Prepare and download the occurrences. The download request is executed with the “occ_download” function which allows you to establish criteria for filtering the data downloaded. In this protocol, as we wanted to keep only native records with coordinates, we used the arguments explained in Table 1. The “occ_download_wait” function indicates the status of the download and the code continues running until the status changes to “succeeded”. The download is performed with the “occ_download_get” function and will result in as many zip files inside “download_GBIF” folder as taxa included in your request. The "occ_download_import" function imports into R the dataset downloaded from the "download_GBIF" folder (Note 4).
Table 1. Necessary arguments of occ_download function from “rgbif“ package.
| Argument | Description | To download |
| pred | Downloads only the occurrences equal to unique condition | Select “taxon” for “taxonKey” and TRUE for “hasCoordinate” |
| pred_not | Downloads only the occurrences not equal to the condition | Select "INTRODUCED ",”INVASIVE”, “MANAGED” and “NATURALISED” for “establishmentMeans” |
| pred_in | Downloads the occurrences equal to multiple conditions | Select "taxon.keys" for “taxonKey" |
2. Use the BIEN_occurrence_genus function from R package “BIEN“ to download the records from the BIEN database version 4.1.1 (Note 5). It is necessary to indicate some arguments to start the download (Table 2). We will refer to the resulting dataset as “raw.BIEN.dataset” onwards. If there are no records in "raw.BIEN.dataset" go directly to step E-3 to replace the column names and see the Note 6.
Table 2. Necessary arguments of BIEN_occurrence_genus function from “BIEN“ package. If you use species as sample unit, then you will need to use BIEN_occurrence_species function, replace the argument “genus” by “species” and the remaining arguments stay the same.
Argument | Description | For download |
| genus | Name of genus | This argument corresponds with names vector of taxa created in R (taxa.names) |
| cultivated | If TRUE, it also returns cultivated occurrences | Select FALSE (is selected for default) |
| all.taxonomy | If TRUE, it returns all taxonomic information | Select TRUE (FALSE is selected for default) |
| collection.info | If TRUE, it returns additional information about collection and identification | Select TRUE (FALSE is selected for default) |
| observation.type | If TRUE, it returns information on type of observation | Select TRUE (FALSE is selected for default) |
| political.boundaries | If TRUE, it returns information on political boundaries | Select TRUE (FALSE is selected for default) |
| natives.only | If TRUE, it returns only native species | Select TRUE (is selected for default) |
3. Save the R workspace with the downloaded data as "1_Workspace_Download.RData". It is very useful to save the objects created in the data download. If there is a problem in later steps, this workspace can be load and thus avoid making another download.
E. Unify the format of the downloaded databases and simplify the database by removing unnecessary columns. In order to join the information from the two databases the number of columns and their names have to be identical in “raw.GBIF.list” and “raw.BIEN.dataset”. Note that some columns from GBIF and BIEN have different names and yet contain the same information. In those cases, it is necessary to rename the columns (see below). Columns with information that will not be used in further analysis can be removed in this step also.
- Simplify raw GBIF dataset “raw.GBIF.list”.
- Create new columns for future merging between GBIF and BIEN raw datasets.
- Add “countryName” column to include full country names using the “countrycode“ R package and the “countryCode” column. The “countryCode” column includes 2-letter ISO 3166-1 standard for country codes and their subdivisions. This standard is used by GBIF to indicate the country in which the occurrence was recorded while BIEN uses the full name. The “countrycode” function transforms the “countryCode” column in country names (Note 7).
- Add “dataOrigin” column filled with GBIF. This column indicates if the record belongs to GBIF or BIEN, in this case it will be filled with “GBIF” for all records.
- Select useful columns to simplify the dataset before merging with BIEN. The number of columns of “raw.dataset” is about 50, it is advisable to reduce this number. To our purpose, the useful information is inside the following columns: "gbifID", "dataOrigin”, ”basisOfRecord", "genus", "species", "scientificName", "decimalLongitude", "decimalLatitude", "elevation", "countryName", "countryCode", "locality", "eventDate", "institutionCode", "collectionCode", "catalogNumber". Therefore, we will only keep these columns. We will refer to the resulting dataset as “simple.GBIF.dataset” onwards.
- Export “simple.GBIF.dataset” as csv file.
- Create new columns for future merging between GBIF and BIEN raw datasets.
- Simplify raw BIEN dataset “raw.BIEN.dataset”.
- Remove duplicated columns. In the download, the “data_collected” column is duplicated. This is because the “collection.info=TRUE” argument in the download adds the column “data_collected” in addition to other variables. However, if we select “collection.info=FALSE” we would lose other variables related to collection information and, thus, it is necessary to use "TRUE” (Note 8).
- Remove records that are also from the GBIF database to avoid replicated data. BIEN data source may contain occurrences that are also in GBIF, this information is available in the “datasource” column of “raw.BIEN.dataset”.
- Create necessary columns for merging with GBIF. Add the country code and elevation variables.
- Add the country codes assigned by the 2-letter ISO 3166-1 standard from the name of the country available in “country” column of “simple.BIEN.dataset”.
- Add an empty elevation column with “NA“ (Not Available information). The elevation is not included in the downloaded information from BIEN, but it is included in GBIF and we do not want to loss this information when merging the two databases.
- Add an empty “ID_Origin” column filled with “NA”. This information is not available in the BIEN database, but it is included in GBIF and we do not want to loss this information.
- Add “dataOrigin” column filled with BIEN. This column indicates if the record belongs to GBIF or BIEN, in this case it will be filled with BIEN for all records.
- If your sample unit is species and you have used “BIEN_occurrence_species” to download the data, then you will not have any column indicating the name of the genus. Since this column may be of interest, then it is desirable to run this function to add a column with the name of the genera, named “scrubbed_genus”. This line is commented in the script and, thus, it is not performed in the protocol unless you uncomment it (Note 9).
- Select useful columns from all available variables. To our purpose, the useful information is inside the following columns: "ID_Origin", "dataOrigin", "observation_type", "scrubbed_genus", "scrubbed_species_binomial", "verbatim_scientific_name", " longitude ", "latitude", "country", "country_ISOcode", "locality", "date_collected", "datasource", "collection_code", "catalog_number". Therefore, we will only keep these columns. We will refer to the resulting dataset as “simple BIEN dataset” onwards. Note that the R object “ID_Origin” contains the code of the record in the original database but transformed as exponential. To look for the exact original code go to that field in the exported ".cvs”.
- Export “simple BIEN dataset” as csv file.
- Match columns between “simple.GBIF.dataset” and “simple.BIEN.dataset”. To merge the two datasets you need to rename columns in both objects to match the following extract names ("ID_Originin", "Data_Origin", "Basis_of_Record", "Genus", "Spp", "Scientific_name", "Longitude", "Latitude", "Elevation", "Country_Name", "Country_ISOcode", "Locality", "Date", "Institution_code", "Collection_code", "Catalog_number") and following the equivalences indicated in Table 3. If there are no records in “raw.BIEN.dataset” it is important that you see Note 10.
- Merge “simple.GBIF.dataset” and “simple.BIEN.dataset”. Build the new dataset from the equivalence information of the “simple.GBIF.dataset” and “simple.BIEN.dataset” objects specified in Table 3. The resulting table has the information of both datasets and it has 16 variables, “merged dataset“ onwards. If there were no records in "raw.BIEN.dataset", you still need to rename your “simple.GBIF.dataset” by “merged.dataset”, because it is the name used onwards in the R script (Note 10).
Table 3. Equivalences between information of GBIF and BIEN simple datasets needed for merging datasets. Names of selected columns of “simple.GBIF.dataset” and “simple.BIEN.dataset”, and their corresponding name in the merged dataset.
| GBIF | BIEN | Merged dataset |
| ID_Originin (new) | ID_Originin | ID_Originin |
| Data_Origin (new) | Data_Origin | Data_Origin |
| genus | scrubbed_genus | Genus |
| species | scrubbed_species_binomial | Spp |
| scientificName | verbatim_scientific_name | Scientific_name |
| decimalLatitude | latitude | Longitude |
| decimalLongitude | longitude | Latitude |
| elevation | Not available (Later created as “elevation”) | Elevation |
| countryName (new) | country | Country_Name |
| countryCode | Not available (Later created as “country_code”) | Country_code |
| locality | locality | Locality |
| eventDate | date_collected | Date |
| institutionCode | datasource | Institution_code |
| collectionCode | collection_code | Collection_code |
| catalogNumber | catalog_number | Catalog_number |
| basisOfRecord | observation_type | Basis_of_Record |
5. Save “merged.dataset“ as csv file named “2_merged_dataset.csv”. This file contains all the simplified GBIF and BIEN information. Or only GBIF data, in case on record was downloaded from BIEN.
F. Add occurrences form other sources. This step is only necessary if the data from GBIF and BIEN is incomplete (that is, they do not completely reflect the distribution range of the study case) and the author deems necessary to include other data sources (such as additional online databases, herbaria specimens or citations in the literature) to complete taxa ranges. If this is not the case, skip this step and go to G.
- For additional online databases. In this protocol we will use only GBIF and BIEN occurrences downloaded using “rgbif“ and “BIEN“ R packages. This step is commented in the R script because it has not been accomplished in this protocol. However, we provided an optional example using package “spooc” in R. This package can download occurrences from a diverse set of data sources, including Global Biodiversity Information Facility (GBIF; GBIF.org, 2021), USGS Biodiversity Information Serving Our Nation (BISON, 2021), iNaturalist (iNaturalist, 2021), Berkeley Ecoinformatics Engine (Berkeley Ecoinformatics Engine, 2021), eBird (Sullivan et al., 2009), Integrated Digitized Biocollections (iDigBio, 2021), VertNet (VertNet, 2021), Ocean Biogeographic Information System (OBIS; Grassle, 2000), and Atlas of Living Australia (ALA, 2021). The procedure with this package is very similar to the one already done in this protocol with BIEN and GBIF. The result of the download is a list with as many elements as taxa in the “taxa.names vector”. Each of these elements contains information on the given taxon in the eight databases. Once the “spooc” R package has been run, you will need to reformat and simplify the downloaded databases so that they match for the column names and structure specified in step E-3. To do this, first go to step E-2-b and adapt the script onwards to remove GBIF replicates (E-2-b), create match column to merge the databases (E-2-c), adapt the script in E-2-d to identify the columns of interest and in E-2-e export the results in a file name “simple.spooc.dataset”. Then adapt E-3 to rename columns in “simple.spooc.dataset” as already done for GBIF and BIEN. Finally, go to E-4 and adapt it to merge the three databases (“simple.spooc.dataset”, “simple.GBIF.dataset” and “simple.BIEN.dataset”).
- For georeferenced herbaria specimens and literature occurrences. The exported csv file “merged_dataset.csv“ is used to manually add the records obtained from herbaria and literature.
- Open Microsoft Excel.
- Import “merged dataset” csv file exported in step E-5.
- Select “Data > Get External Data > From text” and open de “merged_dataset.csv” file. A new window, called “Text Import Wizard”, will appear.
- Select “Delimited” in “Original Data Type” and click on “Next”.
- Select “Comma” in “Delimiters” and click on “Next”.
- It is advisable select “Text” in “Column data format” for all columns of file. Click on “Next”. This is done to avoid problems with cells format in Excel that occur in columns that have special characters and symbols and, also (and most importantly), in the columns that include the coordinates.
- Add new occurrences filling all the fields of the variables of the table. When no information is available, complete the field with NA. This format will make the importation of the database to R easier (Note 11).
- Export the table as csv.
- Select “File > Save as”.
- A new window will appear. Choose the “input” folder as the destination folder for the file. Introduce in file name “merged_dataset_Version2” and “CSV (Comma delimited) (*.csv)” in save as type. Click in “Save”. If you are using Excel for MacOS, then export as “MS-DOS Comma Separated (.csv)” and check that the separator are commas. If that is the case then run line 258. If separators are semicolons, run the line 257 in R script.
- A new window will appear to remind you that only the active spreadsheet will be saved. Select “OK”. Select “OK” in the next window that appears.
G. Check the "merged.dataset” object. It is necessary to check that the dataset has the correct format.
- If you come from step F-2, return to RStudio and import the csv file obtained in step F-2-d-iii, “merged_dataset_Version2.csv”. This step is commented in the R script because it has not been done in this protocol. If you come from step E or F-1, go straight to step G-2.
- Visualize “merged_dataset”. Check that the number of columns in the dataset is 16. The structure must be a data.frame. “Latitude”, “Longitude” and “Elevation” must be numeric format, and the rest of the columns must be character format (Note 12).
H. Data cleaning. This step is focused on cleaning the most common errors.
- Remove the records that lack decimal coordinates. This was already done for GBIF records during the import in step D-1 but is necessary for records import from the BIEN database (or any other database if you run F-1, Note 12).
- Remove the records with invalid coordinates values (that is, Longitude or Latitude= 0). This is common in records from GBIF.
- Remove the records in which coordinates have low precision (for the geographical scale and our purposes, we have removed the coordinates that had less than 2 decimals, this can be modified in the script to meet the precision you need as indicated in the R script).
- Remove replicated records among databases. Delete the rows that have the same information for the fields: “Genus”, “Spp”, ”Date”, ”Locality”, ”Longitude”, ”Latitude”, ”Elevation”, ”basisOfRecord”, ”catalogNumber” and ”CountryCode”. Sometimes, the same record has been uploaded in different databases, avoiding redundant data is desirable to reduce processing time. Note that this filter is tied to the number of decimals.
- Remove records outside natural distribution of the taxa. Each record of the database after running steps H-1-4 (that is, R object “filtered.dataset”) is compared with a filtered version (“checklist.filtered”) of the checklist obtained in step A-5 (“checklist”), and those outside the natural distribution will be removed.
- Add level-3 TDWG code to “filtered data” for future comparison with the checklist created in step A-4.
- Download the world shapefile with the level-3 TDWG code used by WCSP available in GitHub. Go to https://github.com/tdwg/wgsrpd, click on “Clone” and “Download ZIP”. A zip folder named “wgsrpd-master.zip” will be downloaded. Unzip the folder and copy it into the “input” folder.
- Return to RStudio and open the level-3 TDWG code shapefile using “readOGR” function from “rgdal” package. Select the WGS84 projection to the shapefile using “crs” function from “raster” package.
- Extract level-3 TDWG code for the occurrences from “filtered.data” (Note 13).
- Remove records outside the limits of the level-3 TDWG shapefile. These records correspond to the ones that have “NA” values in the “Country_TDWGcode” column and, also, to those in which coordinates place the occurrence in the sea (because our study case is terrestrial). Remove “optional” column, that is created by default when using “extract” function (Note 14).
- Divide your dataset (“filtered.data.TDWG”) in sections according to your sample unit. Each section is a table with the records of one genus. Be aware that the sections of this table must correspond to the sample unit of your study (family, genus, species, etc.). Check that the taxa names appearing in “Name” column of the “checklist.filtered” that is cleaned in line 348 of the R script are the same as those indicated at the beginning of the “value” column of the same object.
- Filter your dataset (“filtered.data.TDWG”) to retain only natural records. Compare each genus (or the correspondent sample unit) with the countries of its natural distribution. The “Country_TDWGcode” column of a given sample unit is compared with the corresponding element of “filtered.checklist” created in step H-5-b from the original checklist created in step A-5. The resulting object is a list that contains all the filtered occurrences, and that is converted into a dataframe (“filtered.dataset.WCSP” onwards). Although GBIF database has a column that indicates if the record comes from cultivation, it is highly advisable to run this part of the script to remove naturalized records as well.
- Export as csv the “filtered.dataset.WCSP” object as “3_Cleaning_dataset_WCSP.csv” and save the workspace as “2_Workspace_Cleaning.RData”.
- Add level-3 TDWG code to “filtered data” for future comparison with the checklist created in step A-4.
I. Distribution maps. This step is focused on visualizing the global distribution of all taxa together and individual maps of each sample unit after data cleaning.
- Create global distribution map with all sample units as PDF named “4_global_distribution_map.pdf”. The occurrences of all sample units are colored in red and the surface of the world in grey.
- Create distribution maps for each sample unit as PDF named “4_distribution_maps.pdf”. Convert “filtered.dataset.WCSP” data.frame into a list in which, each element of the list belongs to the occurrences of a single sample unit. Each page of the PDF contains a map of each sample unit (expanding the geographic area in which it appears) titled with the name of the taxon. The occurrences of each sample units are colored in red and the surface of the world in grey.
J. Data thinning. The "spThin" package chooses an occurrence and randomly removes nearby occurrences according to the indicated distance in the buffer. This step is intended to remove the uncertainty when spatial data is unevenly distributed across your dataset and there are certain areas for all or a few sample units that are oversampled. To identify this sampling bias, visually inspect the maps created in step-I. If your sampling bias affects most or all of your sample units, then proceed with the thinning in step J-1-a, if the sampling bias affects only in one or two sample units, then proceed with the thinning in step J-1-b. If you detect sampling bias, the thinning is crucial to minimize errors in further spatial-based analyses such as avoid overestimation in the bioclimatic data and oversampled areas. If there is no bias in your dataset, then you can skip this step and go to the step K.
- Remove occurrences randomly with a given distance buffer (10 kilometers in this case) from “Latitude” and “Longitude” columns. You may need to modify this buffer based on the geographical scale of your case study. This is done in “thin.par” argument of “thin” function in the R script. It is very important to put the distance in kilometers. The random elimination of occurrences is done by sample unit. In our case study the sample unit is indicated in the “Genus” column, but you can modify it in the script if needed.
- Perform this step if you want to apply the thinning to all your sample units. The output of this step is a csv file for each sample unit that will be directly exported to the chosen directory (Note 15). If you want to apply the thinning to just one sample unit, then go to step J-1-b.
- Perform this step if you want to run the thinning just in one sample. In this step you will only apply the thinning to the sample unit that is affected by sampling bias. If you have not run step J-1-a, then select the proper buffer based on your geographical scale (Case 1). If you perform this step because a given sample unit is still affected by sampling bias after running step J-1-a, you must increase the buffer with respect to the one used in step J-1-a (Case2). This step has not been done in the protocol, but the procedure is included and commented in the R script (Note 16).
- Import csv files after data thinning using R package “readr” from “thin” folder.
- Import csv files for all taxa obtained in step J-1 if you only performed J-1-a or if you performed J-1-a and J-1-b (Note 17). Continue with step J-3-a.
- Perform this step only if the thinning has been performed only on one sample unit (that is, if you run step I-1-b directly without running step I-1-a). Import csv file obtained in I-1-b for the sample unit thinned. This step is commented in the R script because it has not been done in this protocol. Continue with step I-3-b.
- After data thinning, the exported files have only three columns: “Genus”, “Longitude” and “Latitude”. Therefore, if you run step I, add all the remaining 13 columns that contain the additional information of each record from the “filtered.dataset.WCSP” object to the dataset obtained after the thinning (“thin.occ”, in case you run step I-2-a; or “thin.taxon”, in case you run step I-2-b).
- If you come from step I-2-a, join columns of the “filtered.dataset.WCSP” to all taxa thinned imported in. Check that “thin.occ” has the same number of rows that “joined.dataset” (Note 18). Check that “joined.dataset” contains all the columns in “filtered.dataset.WCSP.
- If you come from step I-2-b, join columns of the “filtered.dataset.WCSP” to one taxon thinned and add to the rest of taxa of the “filtered.dataset.WCSP” object. This step is commented in the R script because it has not been done in this protocol (Note 18).
- Export the joined file “joined.dataset” as csv named “5_joined_dataset.csv" and save the workspace with thinned files as "3_Workspace_Thinning.RData".
K. Load bioclimatic variables from WorlClim version 2. This online climatic database contains 19 variables with the average values of 19 parameters that represent precipitation and temperature for the years between 1970 and 2000. There are two ways to obtain these bioclimatic variables. The alternative 1 is shown in step J-1-a, and it is available for all the resolutions available in Wordclim (10, 5, 2.5 minutes and 30 seconds). Alternative 2 is shown in step J-1-b, and it is only available for resolution of 10, 5, 2.5 minutes.
- Alternative 1. Download from WorldClim web and import in R. We use the 30 seconds resolution, (only available in website) because it is the highest resolution available and matches the minimum threshold of coordinates precision set in step H-3 (two decimals in our case study).
- Go to the following link: https://www.worldclim.org/data/worldclim21.html. Click in bio 30s, unzip the zip file downloaded named “wc2.1_30s_bio.zip” (Figure 5) and rename the folder as “Bioclimatic _variables_WC2”. Locate this folder inside the “input” folder.
- To avoid sorting problems in R, rename bioclimatic variables as follows: replace original names (“wc2.1_30s_bio_1.tif”) by adding a zero before the number or variable (“wc2.1_30s_bio_01.tif”). This is only done for variables 1 to 9.
c. Import bioclimatic variables to R. Remove “_” and “.tif” characters in column names.
2. Alternative 2. Download the standard WorldClim Bioclimatic variables directly from R using R package “raster”. This alternative is only for the resolution of 10, 5 and 2.5 minutes. In the argument “res” of “getData” function, indicate 10, 5 or 2.5 for the resolution selected. Thus, if you chose for a minimum threshold of two decimals in step H-3 and select this alternative be aware that you will be losing precision for your climatic analysis.
L. Spatial correction for terrestrial organisms. Despite the removal of all occurrences with inaccurate coordinates, it may happen that some of the occurrences may fall outside the limits of the earth's surface according to the limit of the cartographic base used as template. For terrestrial organisms these occurrences may be wrong (if the distance to the coastal limit is huge) or simply misplaced (if the distance to the coastal limit is small). Because we our ultimate goal is extract climatic data (see step L), we do not want to include wrong occurrences, but it is desirable that we do not loss misplaced records. Thus, we need to check for occurrences out of the Earth’s limit to remove wrong occurrences and apply a spatial correction for misplaced occurrences. If there are no occurrences outside Earth’s limits in your database, go to step M and proceed with climatic data extract. If there are occurrences outside the limits, then identify the occurrences that are between the coastal limit and 5 km from the coastal limit (misplaced occurrences) as established by bioclimatic variable 1 of WorldClim version 2 (same limit for the 19 available bioclimatic variables) and recalculate new coordinates so that the occurrence falls in the nearest climatic cell of the template. Occurrences located more than 5 km from the coastal limit (wrong occurrences) are eliminated (Note 19).
- Visualize the distribution of all the filtered occurrences of the case study available in the “joined.dataset” object, using bioclimatic variable 1 as template. The map will appear in the viewer in the lower right corner of the RStudio screen. If you want to extract these maps, uncomment lines 568 and 573 (the maps will be exported in a pdf file). If you have not performed the thinning process in step J, uncomment the line 561 of the R script only.
- Convert “joined.dataset” into the spatial object “joined.dataset.spatial”.
- Check if all joined.dataset.spatial points are within the boundaries of the bioclimatic variable layer. Occurrences located outside the layer boundaries are shown in red, located in the “outside_pts” object. If you want to extract these maps, uncomment lines 608 and 611 (the maps will be exported in a pdf file). If there are no occurrences in red, go to step M-1 (Note 20).
- Coordinate correction. Extract climatic data from bioclimatic variable 1 for all the occurrences of case study and identify which points have no climatic data (NAs, Note 19). The “nearestLand” function of the “seegSDM” package recalculates the coordinates of the occurrences that lies between the distance set in "dist" and the coastal limit to place it in the nearest climatic cell and thus obtain a climatic data. To check coordinate correction, maps with corrected points in green and uncorrected points in red appear in the viewer in the lower right corner of RStudio screen. The process is repeated by increasing the distance by 1 km until 5 km distance from the coastal limit is reached. Occurrences outside the 5 km are removed.
M. Extract climatic data from bioclimatic variable layers.
- Convert “joined.dataset” to spatial object and select WGS84 projection (Note 20).
- Extract bioclimatic data. We use “bilinear” method because this method returns values that are interpolated from the values of the four nearest raster cells. The “simple” method returns values in which the point falls. Using “bilinear” method, we assume that the coordinates of the dataset have a certain precision error, if we use the “simple” method this error is ignored. Be aware that this step can be time consuming (Note 21).
- Join the bioclimatic values to joined.dataset in a new data frame named “climatic.data” and remove the “optional” column created during the extraction of climatic data.
N. Visualize and export the final dataset as “6_Final_dataset.csv”. Save the final workspace as” 4_Workspace_Final_Data.RData”.
Notes
- Any text line preceded by the character “#” in the R script available from GitHub (“commented lines”) is only intended to guide the user or to provide alternative code to run under specific cases and will not be run when executed in the R console. If the specific case applies to you, and you need to run the functions in the commented line you need to uncomment the line (remove “#”) and execute the line. To comment a section of R script press “Ctrl+Shift+C” and it will not run.
- In case the WCSP geographical information for a given taxon is incomplete or not available, complete the natural distribution of the sample unit in other sources such as literature or other checklists and include it in the txt file using the level-3 TDWG code (available in Brummitt, R. K., 2001), as mentioned in steps A-3 and A-4.
- To run the lines of the script in RStudio, place the mouse cursor at the beginning of the line or selected regions of the code and press “Ctrl+Enter”. If the line is uncommented (that is, not preceded by “#”, it will be run”.
- Check in the "download_GBIF" folder that there is a zip file with the reference of the “gbif.download.key” object. The results are also available on your downloads user page of GBIF: https://www.gbif.org/user/download.
- Replace the function “BIEN_occurrence_genus” by “BIEN_occurrence_species” when sample unit is species and the argument “genus” by “species” inside the function.
- If there are no records in "raw.BIEN.dataset" only the object "raw.GBIF.dataset" will be further used in the R script. If this is the case, you must rename the columns in step E-3 and replace simple.GBIF.dataset. with merged.dataset in step E-4 and continue with the rest of the protocol.
- A warning may appear when creating “countryName” column (“Some values were not matched unambiguously: , ZZ”). This is because there are records with an ISO code equal to “ZZ”, which corresponds to an unknown country. The space before ZZ corresponds with empty values. The records with ZZ and empty values in ISO code will have NAs in the “countryName” column.
- If sample unit is species, replace raw.BIEN.dataset[,-28] by raw.BIEN.dataset[,-27].
- If sample unit is species, detach “scrubbed_species_binomial” column into “genus” and “temp.spp” to obtain the name of the genus in a separate column.
- If there are no records in “raw.BIEN.dataset” change only the column names of “raw.GBIF.dataset”, continue to step E-4 and replace merged.dataset <- rbind(simple.GBIF.dataset,simple.BIEN.dataset) by merged.dataset <- simple.GBIF.dataset.
- When the coordinate columns are filled, the separator for latitude and longitude columns must be a period. Avoid using “;” in fields since Excel exports csv separated by “;”.
- If you have added new occurrences in step F-2, replace the object “merged.dataset” by “merged.dataset.2” in the steps G-2 and H-1 of the R script.
- A warning may appear when all values of shape.level3TDWG are extracted (“In sp::proj4string(x) : CRS object has comment, which is lost in output”). It does not affect the result.
- Every time the function “extract()” is used, a new unnecessary column is created called “optional” and it is advisable to remove it.
- Depending on the number of occurrences of each sample unit (genus in this case), this step can be time consuming. Note that the number of cvs files created must be the same as the number of sample units.
- If you run step I-1-b after running step J-1-a, it is highly important that you delete the csv file of that given sample unit obtained with the previous buffer (step-J-1-a) to keep just one csv file per sample unit (i.e., the last csv file created for that sample unit in step J-1-b). The number of csv files in the thin folder must be equal to the number of sample units in taxa.names.
- When creating “thin.occ” the following warning appears “There were “number” warnings (use warnings() to see them)“. This warning means: “In bind_rows_(x, .id): binding character and factor vector, coercing into character vector”. Although this warning does not affect the results, check the object “thin.occ” to confirm that it contains as many all the information (three columns: “Genus” or “Spp”, “Longitude” and ”Latitude”) exported in csv files in step J-2 and that the unzip files are located in “unzip_GBIF_files” path.
- In case the number of rows in the “joined.dataset” is not the same as in “thin.occ”, it is possible that joining the two tables may generate duplicates. So, execute line 491: joined.dataset<-joined.dataset[!duplicated(joined.dataset[,c("Genus","Longitude","Latitude")]),]. This is done to remove duplicates and check the number of observations between “joined.dataset” and “thin.occ” again.
- We set a limit of 5 km to correct the coordinates but this value can be modified according to the needs in “success <- ifelse(dist > 5000, TRUE, success)” in the step J-5. Replace “5000” by the new distance. It is important that the new distance value is expressed in meters.
- When execute the step L-4, “Plot occurrences outside layer limits (NA values)” and occurrences outside limits are not obtained, go to step M-1 and uncomment the line 662.
- Depending on the number of occurrences of each sample units, this step can be time consuming. If the case study has many occurrences and many sample units, it is advisable to use a computer with a large RAM memory. If the extraction of climatic data of all occurrences takes a long time, replace method= "bilinear" by method= "simple".
Acknowledgments
This protocol was derived for the publication in pre print Coca-de-la-Iglesia et al. (2021), currently under review in American Journal of Botany. We also indebted to the people who are part of the Writing Workshop developed by the Biology and Ecology Departments of the Universidad Autónoma de Madrid, for all the comments and discussions that have helped to realize this work, specially to I. Ramos for helping us to correct code errors. This study was supported by the Spanish Ministry of Economy, Industry and Competitiveness 607 [CGL2017-87198-P] and the Spanish Ministry of Science an Innovation [PID2019-106840GA-608 C22]. M. Coca de la Iglesia was supported by the Youth Employment Initiative of European 609 Social Fund and Community of Madrid [PEJ-2017-AI-AMB-6636 and CAM_2020_PEJD-610 2019-11 PRE/AMB-15871].
Competing interests
We declare no competing interests.
References
Aiello-Lammens, M. E., Boria, R. A., Radosavljevic, A., Vilela, B., Anderson, R. P., Bjornson, R., & Weston, S. (2019). spThin: Functions for Spatial Thinning of Species Occurrence Records for Use in Ecological Models (0.2.0) [Computer software]. Website https://CRAN.R-project.org/package=spThin
Arel-Bundock, V., Enevoldsen, N., & Yetman, C. (2018). countrycode: An R package to convert country names and country codes. Journal of Open Source Software, 3(28): 848. https://doi.org/10.21105/joss.00848
Arlé, E., Zizka, A., Keil, P., Winter, M., Essl, F., Knight, T., Weigelt, P., Jiménez‐Muñoz, M., & Meyer, C. (2021). bRacatus: A method to estimate the accuracy and biogeographical status of georeferenced biological data. Methods in Ecology and Evolution, 12(9): 1609–1619. https://doi.org/10.1111/2041-210X.13629
ALA. (2021). Atlas of Living Australia – Open access to Australia’s biodiversity data. Website https://www.ala.org.au/ [accessed 7 October 2021].
Bánki, O., Roskov, Y., Döring, M., Ower, G., Vandepitte, L., Hobern, D., Remsen, D., Schalk, P., DeWalt, R. E., Keping, M., Miller, J., Orrell, T., Aalbu, R., Adlard, R., Adriaenssens, E., Aedo, C., Aescht, E., Akkari, N., Alonso-Zarazaga, M. A., et al. (2022). Catalogue of Life Checklist (Version 2022-01-14). Catalogue of Life. Website https://doi.org/10.48580/d4tp [accessed January 2021].
Berkeley Ecoinformatics Engine. (2021). Website https://ecoengine.berkeley.edu/ [accessed 7 October 2021].
BISON. (2021). Biodiversity Information Serving Our Nation. Website https://bison.usgs.gov/ [accessed 7 October 2021].
Bivand, R., Keitt, T., Rowlingson, B., Pebesma, E., Sumner, M., Hijmans, R., Rouault, E., Warmerdam, F., Ooms, J., & Rundel, C. (2020). rgdal: Bindings for the ‘Geospatial’ Data Abstraction Library (1.5-8) [Computer software]. Website https://CRAN.R-project.org/package=rgdal
Brummitt, R. K. (2001). World Geographical Scheme for Recording Plant Distributions. https://web.archive.org/web/20160125135239/http:/www.nhm.ac.uk/hosted_sites/tdwg/TDWG_geo2.pdf
Chamberlain, S., Oldoni, D., Barve, V., Desmet, P., Geffert, L., Mcglinn, D., & Ram, K. (2020a). rgbif: Interface to the Global ‘Biodiversity’ Information Facility API (2.3) [Computer software]. Website https://CRAN.R-project.org/package=rgbif
Chamberlain, S., Ram, K., Hart, T., & rOpenSci. (2020b). spocc: Interface to Species Occurrence Data Sources (1.0.8) [Computer software]. Website https://CRAN.R-project.org/package=spocc
Coca-de-la-Iglesia, M., Medina, N. G., Wen, J., & Valcárcel, V. (2021). Tropical-temperate dichotomy falls apart in the Asian Palmate Group of Araliaceae [Preprint]. bioRxiv. https://doi.org/10.1101/2021.10.20.465102
Dowle, M., Srinivasan, A., Gorecki, J., Chirico, M., Stetsenko, P., Short, T., Lianoglou, S., Antonyan, E., Bonsch, M., Parsonage, H., Ritchie, S., Ren, K., Tan, X., Saporta, R., Seiskari, O., Dong, X., Lang, M., Iwasaki, W., Wenchel, S., Vaughan, D. (2019). data.table: Extension of ‘data.frame’ (1.12.8) [Computer software]. Website https://CRAN.R-project.org/package=data.table
Fick, S. E., & Hijmans, R. J. (2017). WorldClim 2: New 1‐km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37(12): 4302–4315. https://doi.org/10.1002/joc.5086
GARD. (2022). Global Assessment of Reptile Distributions. Website http://www.gardinitiative.org/data.html [accessed 10 February 2022]
GBIF.org. (2021). GBIF: The Global Biodiversity Information Facility. Website https://www.gbif.org/ [accessed 14 July 2021].
Golding, N., & Shearer, F. (2021). SeegSDM: Streamlined Functions for Species Distribution Modelling in the SEEG Research Group [HTML]. spatial ecology and epidemiology group. Website https://github.com/SEEG-Oxford/seegSDM (Original work published 2013)
Govaerts, R., Dransfield, J., Zona, S., Hodel, D. R., & Henderson, A. (2008). World Checklist of Selected Plant Families: Royal Botanic Gardens, Kew. Published on the Internet; http://wcsp.science.kew.org/ Retrieved. Website https://wcsp.science.kew.org/home.do
Grassle, F. (2000). The Ocean Biogeographic Information System (OBIS): An On-line, Worldwide Atlas for Accessing, Modeling and Mapping Marine Biological Data in a Multidimensional Geographic Context. Oceanography, 13(3): 5–7. https://doi.org/10.5670/oceanog.2000.01
Hassler, M. (2022a). Checklist of Ferns and Lycophytes of the World. Website http://www.catalogueoflife.org/annual-checklist/2018/details/database/id/140 [accessed 9 February 2022]
Hassler, M. (2022b). World Plants. Synonymic Checklist and Distribution of the World Flora (12.9) [Computer software]. Website www.worldplants.de
Hijmans, R. J., Etten, J. van, Sumner, M., Cheng, J., Bevan, A., Bivand, R., Busetto, L., Canty, M., Forrest, D., Ghosh, A., Golicher, D., Gray, J., Greenberg, J. A., Hiemstra, P., Hingee, K., Geosciences, I. for M. A., Karney, C., Mattiuzzi, M., Mosher, S., Wueest, R. (2020). raster: Geographic Data Analysis and Modeling (3.1-5) [Computer software]. Website https://CRAN.R-project.org/package=raster
Hortal, J., & Lobo, J. M. (2005). An ED-based Protocol for Optimal Sampling of Biodiversity. Biodiversity and Conservation, 14(12): 2913–2947. https://doi.org/10.1007/s10531-004-0224-z
Hortal, J., Lobo, J. M., & Jiménez-Valverde, A. (2007). Limitations of Biodiversity Databases: Case Study on Seed-Plant Diversity in Tenerife, Canary Islands. Conservation Biology, 21(3): 853–863. https://doi.org/10.1111/j.1523-1739.2007.00686.x
iDigBio. (2021). Integrated digitized biocollections. IDigBio. Website https://www.idigbio.org/home [accessed 7 October 2021]
iNaturalist. (2021). iNaturalist. Website https://www.inaturalist.org/ [accessed 15 July 2021]
IPNI. (2020). International Plant Names Index. Website https://www.ipni.org/ [accessed 11 September 2020]
Jin, J., & Yang, J. (2020). BDcleaner: A workflow for cleaning taxonomic and geographic errors in occurrence data archived in biodiversity databases. Global Ecology and Conservation, 21: e00852. https://doi.org/10.1016/j.gecco.2019.e00852
Lepage, D., Vaidya, G., & Guralnick, R. (2014). Avibase – a database system for managing and organizing taxonomic concepts. ZooKeys, 420: 117–135. https://doi.org/10.3897/zookeys.420.7089
Lima, R. A. F., Sánchez‐Tapia, A., Mortara, S. R., Steege, H., & Siqueira, M. F. (2021). plantR: An R package and workflow for managing species records from biological collections. Methods in Ecology and Evolution, 2041-210X.13779. https://doi.org/10.1111/2041-210X.13779
Maitner, B. (2020). BIEN: Tools for Accessing the Botanical Information and Ecology Network Database (1.2.4) [Computer software]. Website https://CRAN.R-project.org/package=BIEN
Maitner, B. S., Boyle, B., Casler, N., Condit, R., Donoghue, J., Durán, S. M., Guaderrama, D., Hinchliff, C. E., Jørgensen, P. M., Kraft, N. J. B., McGill, B., Merow, C., Morueta‐Holme, N., Peet, R. K., Sandel, B., Schildhauer, M., Smith, S. A., Svenning, J.-C., Thiers, B., … Enquist, B. J. (2018). The bien r package: A tool to access the Botanical Information and Ecology Network (BIEN) database. Methods in Ecology and Evolution, 9(2): 373–379. https://doi.org/10.1111/2041-210X.12861
Mammal Diversity Database. (2020). Mammal Diversity Database (1.2) [Data set]. Zenodo. https://doi.org/10.5281/ZENODO.4139818
R Core Team. (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Website https://www.R-project.org/.
Robertson, M. P., Visser, V., & Hui, C. (2016). Biogeo: An R package for assessing and improving data quality of occurrence record datasets. Ecography, 39(4): 394–401. https://doi.org/10.1111/ecog.02118
Soberón, J., & Peterson, T. (2004). Biodiversity informatics: Managing and applying primary biodiversity data. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 359(1444): 689–698. https://doi.org/10.1098/rstb.2003.1439
Sullivan, B. L., Wood, C. L., Iliff, M. J., Bonney, R. E., Fink, D., & Kelling, S. (2009). eBird: A citizen-based bird observation network in the biological sciences. Biological Conservation, 142(10): 2282–2292. https://doi.org/10.1016/j.biocon.2009.05.006
Töpel, M., Zizka, A., Calió, M. F., Scharn, R., Silvestro, D., & Antonelli, A. (2016). SpeciesGeoCoder: Fast Categorization of Species Occurrences for Analyses of Biodiversity, Biogeography, Ecology, and Evolution. Systematic Biology, syw064. https://doi.org/10.1093/sysbio/syw064
Uetz, P., Freed, P., Aguilar, R., & Hošek, J. (2021). The Reptile Database. Website http://www.reptile-database.org
USDA, NRCS. 2022. The PLANTS Database. National Plant Data Team, Greensboro, NC USA. Website http://plants.usda.gov [accessed 2 October 2022
VertNet. (2021). VertNet. Website http://vertnet.org/ [accessed 7 October 2021]
Wickham, H. (2020). plyr: Tools for Splitting, Applying and Combining Data (1.8.6) [Computer software]. Website https://CRAN.R-project.org/package=plyr
Wickham, H., François, R., Henry, L., Müller, K., & RStudio. (2020). dplyr: A Grammar of Data Manipulation (1.0.0) [Computer software]. Website https://CRAN.R-project.org/package=dplyr
Wickham, H., Girlich, M., & RStudio. (2022). tidyr: Tidy Messy Data (1.2.0) [Computer software]. Website https://CRAN.R-project.org/package=tidyr
Wickham, H., Hester, J., Chang, W., & RStudio. (2021). devtools: Tools to Make Developing R Packages Easier (2.4.2) [Computer software]. Website https://CRAN.R-project.org/package=devtools
Wickham, H., Hester, J., Francois, R., R, R. C. T. , J. J. , M. J. (2018). readr: Read Rectangular Text Data (1.3.1) [Computer software]. Website https://CRAN.R-project.org/package=readr
Zizka, A., Silvestro, D., Andermann, T., Azevedo, J., Duarte Ritter, C., Edler, D., Farooq, H., Herdean, A., Ariza, M., Scharn, R., Svantesson, S., Wengström, N., Zizka, V., & Antonelli, A. (2019). CoordinateCleaner: Standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution, 10(5): 744–751. https://doi.org/10.1111/2041-210X.13152
- Coca-de-la-Iglesia, M, Valcárcel, V and Medina, N G(2022). Biodiversity online databases: An applied R protocol to get and curate spatial and climatic data. Bio-protocol Preprint. bio-protocol.org/prep1573.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link
