- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Real-Time Imaging of Specific Genomic Loci With CRISPR/dCas9 in Human Cells Using CRISPRainbow
Published: Vol 15, Iss 17, Sep 5, 2025 DOI: 10.21769/BioProtoc.5432 Views: 3650
Reviewed by: David PaulAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring of Sperm-Independent Calcium Oscillations in Immature Oocytes of Mice
Sae Horiike [...] Hidehiko Ogawa
Feb 5, 2026 215 Views

Time-Lapse Into Immunofluorescence Imaging Using a Gridded Dish
Nick Lang [...] Andrew D. Stephens
Feb 20, 2026 189 Views

How to Train Custom Cell Segmentation Models Using Cell-APP
Anish J. Virdi and Ajit P. Joglekar
Feb 20, 2026 266 Views
Abstract
Proper genome organization is essential for genome function and stability. Disruptions to this organization can lead to detrimental effects and the transformation of cells into diseased states. Individual chromosomes and their subregions can move or rearrange during transcriptional activation, in response to DNA damage, and during terminal differentiation. Techniques such as fluorescence in situ hybridization (FISH) and chromosome conformation capture (e.g., 3C and Hi-C) have provided valuable insights into genome architecture. However, these techniques require cell fixation, limiting studies of the temporal evolution of chromatin organization in detail. Our understanding of the heterogeneity and dynamics of chromatin organization at the single-cell level is still emerging. To address this, clustered regularly interspaced short palindromic repeats (CRISPR)/dead Cas9 (dCas9) systems have been repurposed for precise live-cell imaging of genome dynamics. This protocol uses a system called CRISPRainbow, a powerful tool that allows simultaneous targeting of up to seven genomic loci and tracks their locations over time using spectrally distinct fluorescent markers to study real-time chromatin organization. Multiple single-guide RNA (sgRNA), carrying specific RNA aptamers for labeling, can be cloned into a single vector to improve transfection efficiency in human cells. The precise targeting of CRISPRainbow offers distinct advantages over previous techniques while also complementing them by validating findings in live cells.
Key features
• Simultaneous imaging of up to seven specific genomic loci in living cells.
• Multicolor imaging using a single CRISPR system from Streptococcus pyogenes.
• Signal amplification through targeting repetitive sequences.
• Targeting endogenous DNA without the need for foreign DNA insertion.
Keywords: CRISPRainbowGraphical overview
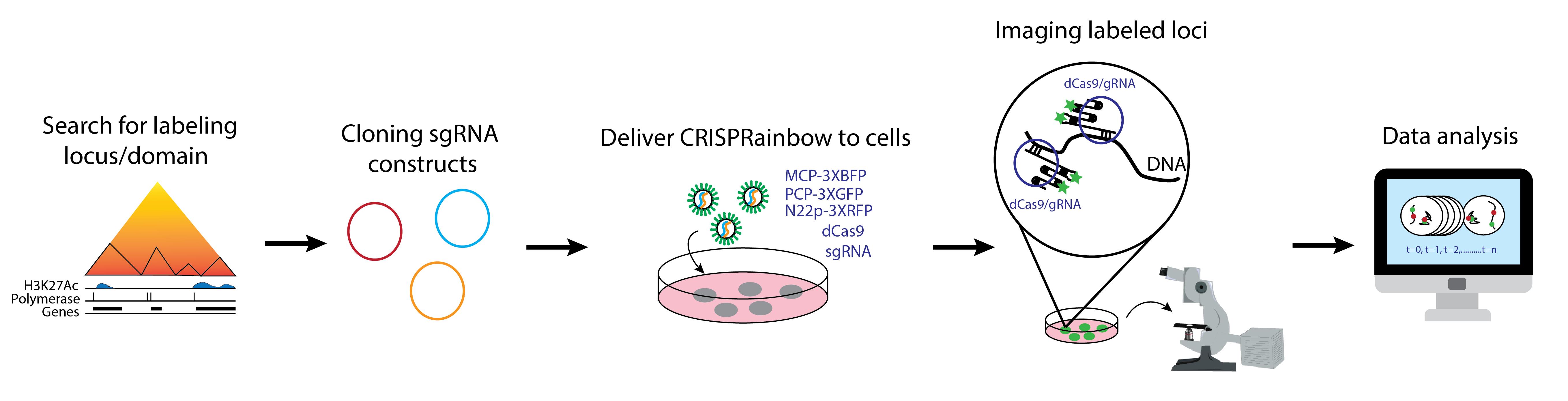
Background
CRISPR/Cas systems from Streptococcus pyogenes have been widely utilized in gene editing and silencing across various organisms owing to their ability to precisely target specific DNA sequences [1,2,3]. A particular adaptation has been the use of catalytically inactive Cas9 (dCas9), which binds DNA without introducing double-strand breaks. This enabled the repurposing of CRISPR/dCas9 into a powerful imaging platform for visualizing specific genomic loci and probing dynamic chromatin architecture in living cells [4,5,6]. Prior to the development of the CRISPR/dCas9-based imaging, studies of nuclear organization relied heavily on labor-intensive tools such as fluorescent transcription activator-like effector nucleases (TALENs) or endpoint techniques like fluorescence in situ hybridization (FISH), which are limited to fixed cells and provide only static snapshots of chromatin structure [7,8]. Although live-cell imaging using fluorescently tagged histone molecules (e.g., H2B-GFP) has provided valuable insights into global chromatin dynamics, their inability to resolve individual loci or local specific transcriptional states limits their utility in fine-scale studies of genome function. In contrast, systems such as the lac operon, which require insertion of foreign DNA elements, pose risks of artificial perturbation to the native chromatin environment. CRISPR-based imaging methods overcome many of these limitations. They allow for endogenous locus visualization without genomic modification, require minimal prior knowledge of genomic sequence context, and enable simultaneous tracking of multiple loci at the single-cell level. The ability to image chromatin structure in three dimensions and in real time opens the door to transformative insights into genome organization, transcriptional regulation, and epigenetic control—areas of immense relevance for understanding development, disease, and genome plasticity.
Labeling of genomic loci with the CRISPR/dCas9 system was initially achieved by fusing a fluorescent protein, such as EGFP, to the dCas9, allowing only one color per CRISPR system [4,5,6]. In our protocol, we engineered single-guide RNAs (sgRNAs) to carry RNA scaffolds that recruit fluorescent protein-fused RNA coat proteins for locus-specific labeling [3,9]. This method, known as CRISPRainbow, which uses spectrally separated fluorophores, facilitates multiplexed, multicolor imaging of distinct genomic regions in single cells by a single CRISPR system [9]. Such advancements greatly improve the resolution and scalability of CRISPR imaging and offer powerful complementary data when integrated with genome-wide mapping techniques like Hi-C. Altogether, CRISPR-based imaging offers unprecedented opportunities to investigate the spatial and temporal dynamics of the genome with high precision and minimal perturbation.
Materials and reagents
Biological materials
1. NEB® Stable Competent E. coli (high efficiency) (New England Biolabs, catalog number: C3040H) or One ShotTM TOP10 Chemically Competent E. coli (Thermo Fisher Scientific, catalog number: C404010), storage: -80 °C
2. One ShotTM ccdB SurvivalTM 2 T1R Competent Cells (Thermo Fisher Scientific, catalog number: A10460), storage: -80 °C
3. Human U2OS cells (ATCC, catalog number: HTB-96), storage: liquid nitrogen tank
Reagents
A. Annealing oligonucleotides
1. Custom forward and reverse oligonucleotides with standard desalting purification (Integrated DNA Technologies or similar vendors; see Section A for details), storage: -20 °C as a 100 μM stock solution. If oligos are ordered as powder, briefly spin the tubes in a minicentrifuge and dissolve the pellet in molecular biology–grade water to prepare a 100 μM solution
2. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: room temperature (RT)
3. Tris-HCl, 1 M, pH = 8.0, molecular biology grade, ultrapure (Thermo Fisher Scientific, catalog number: J22638-AP), storage: RT
4. Sodium chloride (NaCl) (Fisher Chemical, catalog number: S271-1), storage: RT
5. Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884), storage RT
B. Cloning annealed oligos into vectors
1. CRISPR sgRNA vectors with stem loops: pLH-sgRNA1-2XMS2 (Addgene, ID: 75389), pLH-sgRNA1-2XPP7 (Addgene, ID: 75390), pLH-sgRNA1-2XboxB (Addgene, ID: 75391), pLH-sgRNA1-MS2-PP7 (Addgene, ID: 75392), pLH-sgRNA1-PP7-boxB (Addgene, ID: 75393), pLH-sgRNA1-boxB-MS2 (Addgene, ID: 75396), and pLH-sgRNA1-boxB-MS2-PP7 (Addgene, ID: 75397), storage: -20 °C
2. ATP, 100 mM (New England Biolabs, catalog number: P0756S), storage: -20 °C
3. BbsI-HF® (New England Biolabs, catalog number: R3539L), storage: -20 °C
4. T7 DNA Ligase (New England Biolabs, catalog number: M0318L), storage: -20 °C
5. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
6. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
7. Zyppy® Plasmid Miniprep kit (Zymo Research, catalog number: D4019), storage: RT (except for the neutralization buffer, storage: 4 °C)
C. LB-carbenicillin plates
1. LB broth with agar (Lennox) (Sigma-Aldrich, catalog number: L2897), storage: RT
2. Carbenicillin solution, 100 mg/mL (Teknova, catalog number: C2199), storage: -20 °C
D. LB-carbenicillin liquid media
1. LB broth (Lennox) (Sigma-Aldrich, catalog number: L7658), storage: RT
E. Restriction digest of individual sgRNA
1. NotI-HF®, 20,000 U/mL (New England Biolabs, catalog number: R3189S), storage: -20 °C
2. EcoRI-HF®, 20,000 U/mL (New England Biolabs, catalog number: R3101S), storage: -20 °C
3. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
4. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
F. 1% agarose gel
1. TAE buffer, tris-acetate-EDTA, 50× (Fisher BioreagentsTM, catalog number: BP13321), storage: RT
2. Agarose (Sigma-Aldrich, catalog number: A9539), storage: RT
3. Ethidium bromide solution, 10 mg/mL (Sigma-Aldrich, catalog number: E1510), storage: RT
4. Gel loading dye, purple 6× (New England Biolabs, catalog number: B7024S), storage: -20 °C
5. 1 kb Plus DNA ladder, 500 μg/mL (New England Biolabs, catalog number: N3232L), storage: -20 °C
6. Monarch® PCR & DNA Cleanup kit (5 μg) (New England Biolabs, catalog number: T1030L), storage: RT
G. PCR amplification of individual CRISPRainbow sgRNA inserts
1. PCR primers (Integrated DNA Technologies or other vendors that synthesize single-stranded DNA oligos at 25 nmol with standard desalting purification). Dissolve the primers using molecular biology–grade water into a 100 μM stock solution, storage: -20 °C
a. For template pLH-sgRNA1-2XMS2, the forward and reverse primers are:
Forward primer sgRNA-Ex-F1: AATAATggtctcaTGACccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R1: AATAATggtctcATTGCTGCCATTTGTCTCGAGGTCGAGA
b. For template pLH-sgRNA1-2XPP7, the forward and reverse primers are:
Forward primer sgRNA-In-F2: AATAATggtctcaGCAAccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R2: AATAATggtctcAGTTCTGCCATTTGTCTCGAGGTCGAGA
c. For template pLH-sgRNA1-2XboxB, the forward and reverse primers are:
Forward primer sgRNA-In-F3: AATAATggtctcaGAACccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R3: AATAATggtctcAACTGTGCCATTTGTCTCGAGGTCGAGA
d. For template pLH-sgRNA1-MS2-PP7, the forward and reverse primers are:
Forward primer sgRNA-In-F4: AATAATggtctcaCAGTccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R4: AATAATggtctcAGTGTTGCCATTTGTCTCGAGGTCGAGA
e. For template pLH-sgRNA1-PP7-boxB, the forward and reverse primers are:
Forward primer sgRNA-In-F5: AATAATggtctcaACACccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R5: AATAATggtctcAGACATGCCATTTGTCTCGAGGTCGAGA
f. For template pLH-sgRNA1-boxB-MS2, the forward and reverse primers are:
Forward primer sgRNA-In-F6: AATAATggtctcaTGTCccttcaccgagggcctatttcc
Reverse primer sgRNA-Ex-R1: AATAATggtctcAGAGTTGCCATTTGTCTCGAGGTCGAGA
2. dNTP, 10 mM (New England Biolabs, catalog number: N0447S), storage: -20 °C
3. Q5® Reaction Buffer pack, 5× (New England Biolabs, catalog number: B9027S), storage: -20 °C
4. Q5® high-fidelity DNA polymerase, 2,000 U/mL (New England Biolabs, catalog number: M0491L), storage: -20 °C
5. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
H. Assembling CRISPRainbow for the targets of interest
1. pCRISPRainbow-DONOR1 (Addgene, ID: 75398), storage: -20 °C
2. BsaI-HF® v2, 20,000 U/mL (New England Biolabs, catalog number: R3733L), storage: -20 °C
3. T7 DNA ligase, 3,000,000 U/mL (New England Biolabs, catalog number: M0318L), storage: -20 °C
4. ATP, 100 mM (New England Biolabs, catalog number: P0756S), storage: -20 °C
5. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
6. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
I. Restriction digest of assembled CRISPRainbow
1. ApaLI, 10,000 U/mL (New England Biolabs, catalog number: R0507S), storage: -20 °C
2. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
3. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
J. Assembled CRISPRainbow amplification
1. ZymoPURETM II Plasmid Midiprep kit (Zymo Research, catalog number D4200), storage: RT (except for ZymoPURETM P1, storage: 4 °C)
2. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
K. U2OS cell growth
1. Dulbecco’s modified Eagle medium (DMEM), 1× (Corning, catalog number: 10-013-CV), storage: 4 °C
2. Fetal bovine serum (FBS) (Gibco, catalog number: A5256701), storage: -20 °C
3. Penicillin/streptomycin (Pen/Strep, 100×), 10,000 U/mL (Gibco, catalog number: 15140122), storage: -20 °C
4. Dulbecco’s phosphate buffered saline (DPBS), 1× (Corning, catalog number: 21-030-CV), storage: 4 °C
5. TrypLETM Express enzyme, no phenol red, 1× (Gibco, catalog number: 12604013), storage: RT
L. Transfection
1. Opti-MEMTM I reduced serum medium, 1× (Gibco, catalog number: 31985062), storage: 4 °C
2. Lipofectamine® 2000 (Life Technologies, catalog number: 11668027), storage: 4 °C
3. Trypan Blue solution, 0.4% (Thermo Fisher Scientific, catalog number: 15250061), storage: RT
4. pHAGE-TO-dCas9 (Addgene, ID: 75381), storage: -20 °C
5. pHAGE-EFS-MCP-3XBFPnls (Addgene, ID: 75384), pHAGE-EFS-PCP-3XGFPnls (Addgene, ID: 75385), pHAGE-EFS-N22p-3XRFPnls (Addgene, ID: 75387), storage: -20 °C
M. Imaging
1. Olympus Type F immersion oil (Thorlabs, catalog number: MOIL-30), storage: RT
2. TetraSpeckTM microspheres, 0.1 μm, fluorescent blue/green/orange/dark red (Life Technologies, catalog number: T7279), storage: 4 °C
Solutions
1. Annealing buffer (see Recipes)
2. 1× TAE buffer (see Recipes)
3. Medium for U2OS cells (see Recipes)
Recipes
1. Annealing buffer (25 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M Tris-HCl, pH = 8.0 | 100 mM (v/v) | 0.25 mL |
| 5 M NaCl | 50 mM (v/v) | 0.25 mL |
| 0.5 M EDTA, pH = 8.0 | 1 mM (v/v) | 0.05 mL |
| ddH2O | - | 24.5 mL |
2. 1× TAE buffer (1L)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 50× TAE buffer | 2% (v/v) | 20.0 mL |
| ddH2O | 98% (v/v) | 980.0 mL |
3. Medium for U2OS cells (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DMEM, 1× | 89.1% (v/v) | 450 mL |
| FBS | 9.9% (v/v) | 50 mL |
| Pen/Strep, 100× | 1.0% (v/v) | 5 mL |
Laboratory supplies
1. TIPONE® pipette tips in racks, 10 μL (USA Scientific, catalog number: 1111-3200)
2. TIPONE® pipette tips in racks, 200 μL (USA Scientific, catalog number: 1110-1200)
3. TIPONE® pipette tips in racks, 1,000 μL (USA Scientific, catalog number: 1111-2721)
4. Corning® 100 mm × 20 mm style dish (Corning, catalog number: 430293)
5. 0.2 mL 8-strip PCR tube with flat cap, clear (GeneMate, catalog number: 490003-710)
6. DNA LoBind® tubes, 1.5 mL (Eppendorf, catalog number: 022431021)
7. FisherbrandTM sterile polystyrene disposable serological pipettes with magnifier stripe, 5 mL (Thermo Fisher Scientific, catalog number: 13-678-11D)
8. FisherbrandTM sterile polystyrene disposable serological pipettes with magnifier stripe, 10 mL (Thermo Fisher Scientific, catalog number: 13-678-11E)
9. FisherbrandTM sterile polystyrene disposable serological pipettes with magnifier stripe, 50 mL (Thermo Fisher Scientific, catalog number: 13-678-11F)
10. VWR® centrifuge tube, 15 mL (Avantor, catalog number: 525-1068)
11. VWR® centrifuge tube, 50 mL (Avantor, catalog number: 525-1075)
12. Falcon® 25 cm2 rectangular canted neck cell culture flask with plug seal screw cap (Corning, catalog number: 353082)
13. 35 mm glass bottom dishes, No. 1.5 coverslips (MatTek, catalog number: P35G-1.5-10-C)
14. L-shaped cell spreader (Thermo Fisher Scientific, catalog number: 14665230)
15. Corning Falcon round-bottom polypropylene culture tubes (Thermo Fisher Scientific, catalog number: 352059)
Equipment
1. Eppendorf Research® plus, 6-pack with Pipette Carousel 2 (Eppendorf, model: 3123000942)
2. Easypet 3 Motorized Pipette Controller (Eppendorf, model: 4430000018)
3. Mastercycler® nexus GX2 (Eppendorf, model: 6336000023)
4. EnduroTM UV Transilluminator (Labnet Internation, Inc., model: U1002-230V)
5. EnduroTM Gel XL Electrophoresis System (Labnet International, Inc., model: E0160)
6. NewClassic MF Analytical Balance (Mettler Toledo, model: MS303S)
7. Synergy® UV with integrated remote dispenser (Merck, model: 171-0774)
8. MaxQTM 6000 Incubated/Refrigerated Stackable Shakers (Thermo Fisher Scientific, model: SHKE6000-7)
9. NanoDropTM 2000c Spectrophotometer (Thermo Fisher Scientific, model: ND-2000C)
10. myECL Imager (Thermo Fisher Scientific, model: 62236X)
11. myFUGETM 12 Mini Centrifuge 100-240V (Benchmark, model: C1012)
12. Mini Vortexer (Thermo Fisher Scientific, model: 02215365)
13. CO2 Incubator (Sanyo, model: MCO-18AIC)
14. Hermle Refrigerated Centrifuge (Labnet, model: Z 400 K)
15. Class II A2 Biological Safety Cabinet (Thermo Forma, model: 1284)
16. Centrifuge (Eppendorf, model: 5424 R)
17. Large centrifuge (Eppendorf, model: 5810 R)
18. QIAvac 24 Plus (Qiagen, model: 19413)
19. Phase Contrast Microscope (Nikon, model: Diaphot 200/300, for cell culture)
20. 4 °C refrigerator
21. -20 °C refrigerator
22. -80 °C freezer
23. LocatorTM Plus Rack and Box Systems (Thermo Fisher Scientific, model: Locator 8 Plus)
24. Countess® II FL Automated Cell Counter (Invitrogen, discontinued) or Countess® 3 FL Automated Cell Counter (Invitrogen, model: A49866)
25. Epifluorescence microscope (Leica, model: DMIRB, or similar) equipped with an EMCCD camera (Andor model: iXon-897D, or similar) and 100× apochromatic objective lens (oil immersion, Olympus, model: UPLXAPO100XO, NA 1.4 or higher)
Software and datasets
1. A plasmid Editor (ApE) (M. Wayne Davis, v3.1.7) (https://jorgensen.biology.utah.edu/wayned/ape/), free to use
2. Fiji (NIH, version 2.14.0 or newer) (https://imagej.net/software/fiji/downloads), free to use
3. MetaMorph (Molecular Devices) (https://www.moleculardevices.com/products/cellular-imaging-systems/high-content-analysis/metamorph-microscopy), requires a license (Leica Microsystems)
4. Camera registration codes are available upon request to Dr. David Grunwald at the University of Massachusetts Chan Medical School.
5. OriginLab Pro (OriginLab) (https://www.originlab.com/), requires a license
Procedure
A. Search and anneal sgRNA oligos
1. CRISPR sgRNA targeting sites can be searched on CRISPRbar (http://genome.ucf.edu/CRISPRbar/). In the general settings, select the chromosome where your genomic region of interest is located and set the number of repetitive sequences (# Copies) on the chromosome. To search the entire chromosome, check the “entire chromosome” box. Design forward and reverse protospacer oligos by removing the PAM sequences (5′-NGG-3′) at the 3′ end for a total length of 12 nucleotides. A minimum crRNA length for effective locus labeling is 11 nucleotides. Shorter lengths will significantly reduce signal brightness or result in no signal. Then, add ACCG to the 5′ end of the forward oligo, and AAAC to the 5′ end of the reverse oligo for vector annealing. The forward and reverse oligos must be reverse complements of each other, excluding the overhang sequences (ACCG and AAAC) (see Table 1 for examples). Order both forward and reverse oligos from Integrated DNA Technologies (standard desalting, 25 nmol).
Note: CRISPRbar is provided as an example website for identifying CRISPR targeting sites. Other tools or websites designed for mining CRISPR target sequences can also be used to identify labeling sites for this protocol.
Table 1. Oligonucleotide sequences for annealing
| Oligo | Sequence (5′-3′) | PAM | Target chromosome | Vector |
|---|---|---|---|---|
| C3-Forward | ACCGTGATATCACAG | TGG | Chromosome 3 | pLH-sgRNA1-boxB-MS2 |
| C3-Reverse | AAACCTGTGATATCA | Chromosome 3 | ||
| C13-Forward | ACCGACCATTCCTTC | AGG | Chromosome 13 | pLH-sgRNA1-PP7-boxB |
| C13-Reverse | AAACGAAGGAATGGT | Chromosome 13 | ||
| C1-Forward | ACCGAGATGCTCACC | TGG | Chromosome 1 | pLH-sgRNA1-MS2-PP7 |
| C1-Reverse | AAACGGTGAGCATCT | Chromosome 1 | ||
| C7-Forward | ACCGATGGTGAGAGTG | TGG | Chromosome 7 | pLH-sgRNA1-2XboxB |
| C7-Reverse | AAACCACTCTCACCAT | Chromosome 7 | ||
| C14-Forward | ACCGCTCAGGAGAGAG | AGG | Chromosome 14 | pLH-sgRNA1-2XPP7 |
| C14-Reverse | AAACCTCTCTCCTGAG | Chromosome 14 | ||
| Cx-Forward | ACCGTAGAGTCTGCAT | TGG | Chromosome X | pLH-sgRNA1-2XMS2 |
| Cx-Reverse | AAACATGCAGACTCTA | Chromosome X |
Notes:
1. ApE can be used to generate reverse-complement oligo sequences.
2. For targeting the seventh color, insert the additional targeting sequence into the vector pLH-sgRNA1-boxB-MS2-PP7 (Addgene ID: 75397) using the same 5′ and 3′ annealing sequences.
2. In a PCR tube, mix the forward and reverse oligonucleotides for each protospacer in a 1:1 ratio as follows: 5 μL of forward protospacer oligo 100 μM stock, 5 μL of reverse protospacer oligo 100 μM stock, and 40 μL of annealing buffer.
Caution: Work in a DNase-free space with proper equipment to avoid degrading the DNA!
3. Anneal the oligonucleotides using the following program in a PCR machine (Figure 1):
95 °C for 3 min
37 °C for 30 min
95 °C to 25 °C at -0.1 °C/s
4 °C pause
4. Dilute annealed oligonucleotides with molecular biology–grade water to a final concentration of 200 nM (about 3 μg/μL) in a 1.5 mL tube (i.e., mix 5 μL of annealed oligos in 245 μL of ddH2O).
Pause point: You can store the dilutions at -20 °C and continue the next day.
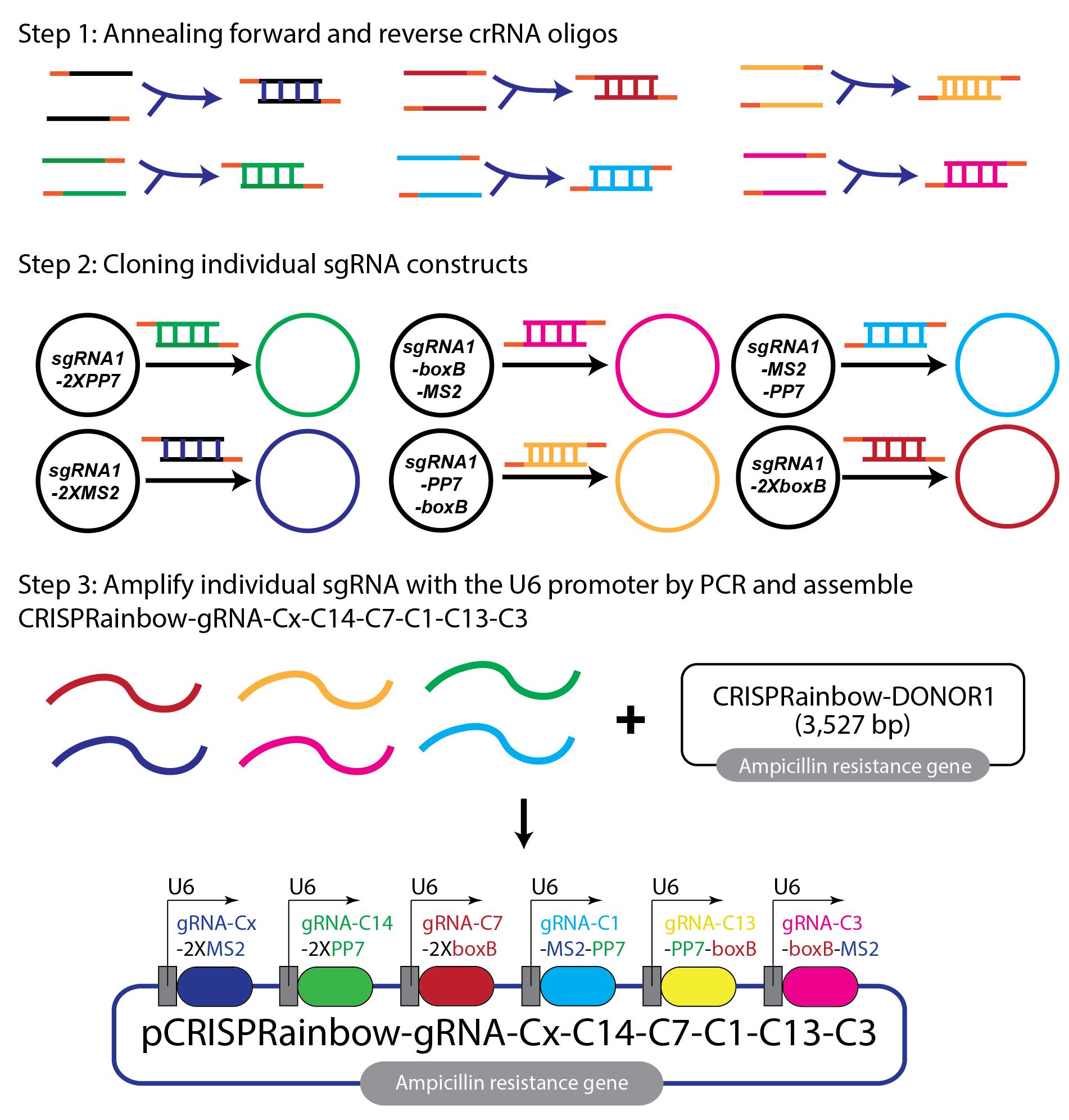
Figure 1. Assembly of the CRISPRainbow-6xsgRNA construct. The schematic illustrates the steps for constructing the CRISPRainbow-6xsgRNA plasmid. First, individual crRNA oligos targeting different chromosomes are annealed into double-stranded DNA. Second, each double-stranded crRNA is cloned into a vector containing a unique combination of stem loops. Note that secondary colors, cyan, yellow, and magenta, are defined from combinations of primary colors (blue, green, and red). Third, individual sgRNA cassettes (with the U6 promoter) are amplified by PCR and assembled into CRISPRainbow-gRNA-Cx-C14-C7-C1-C13-C3 construct using a one-step Gibson reaction. Primary colors are used on chromosome X (Cx, blue, 2XMS2), chromosome 14 (C14, green, 2XPP7), and chromosome 7 (C7, red, 2XboxB). Secondary colors are used on chromosome 1 (C1, cyan, a combination of blue and green, MS2-PP7), chromosome 13 (C13, yellow, a combination of green and red, PP7-boxB), and chromosome 3 (C3, magenta, a combination of red and blue, boxB-MS2). A final assembled CRISPRainbow-gRNA-Cx-C14-C7-C1-C13-C3 construct is 4,627 bp in size.
B. Clone annealed oligos into vectors and transform into NEB stable competent E. coli
1. In a PCR tube, prepare the cloning reactions (10 μL) as follows:
1 μL of annealed oligo 200 nM
1 μL of sgRNA vector (e.g., pLH-sgRNA1-2XMS2) 100 ng/μL
1 μL of CutSmart BufferTM 10×
1 μL of ATP 10 mM
0.5 μL of BbsI-HF® 20,000 U/mL
0.3 μL of T7 DNA Ligase 3,000,000 U/mL
5.2 μL of molecular biology grade water
Caution: All tubes, except for molecular biology–grade water, should be kept on ice to prevent reagents from degradation or loss of activity.
2. Incubate in a PCR machine or water bath at 37 °C for 15 min.
3. Thaw NEB® Stable Competent E. coli (high efficiency) on ice (about 5 min) and immediately add 5 μL of the cloning reaction mix.
4. Mix gently by tapping the tube and let sit on ice for 4–5 min.
Caution: Do not pipette up and down or vortex!
5. Spread the whole volume (55 μL) on an LB plate with carbenicillin (100 μg/mL) and incubate overnight at 37 °C.
Note: Successful cloning will remove the ccdB gene for proper selection of transformed E. coli.
6. After 16–19 h, select 1–3 colonies from each plate (for each protospacer), culture in 5 mL of LB broth containing carbenicillin (100 μg/mL) in a 14 mL polypropylene round-bottom culture tube, and incubate overnight at 37 °C with shaking (225–250 rpm) in the MaxQTM 6000 Incubated/Refrigerated Stackable Shakers.
Note: It is recommended to perform steps B1–6 in a sterile biosafety cabinet.
7. After ~16–29 h, isolate the cloned vectors using the Zyppy® Plasmid Miniprep kit following the manufacturer’s instructions.
Note: Alternative miniprep kits from other vendors can be used.
8. Measure eluted sgRNA insert plasmid using the NanoDropTM 2000c Spectrophotometer in ng/μL.
Pause point: You can store the plasmid at -20 °C and continue the next day.
9. Confirm the successful insertion of protospacer sequences by Sanger sequencing using the sequencing primer: 5′-CTATTCTTTCCCCTGCACTGTACCC-3′. Alternatively, perform a diagnostic digestion reaction (steps B9–14, optional) on isolated plasmids and control (vector without the crRNA insertion) in PCR tubes as follows:
5 μL of isolated plasmid 100 ng/μL
0.5 μL of NotI-HF® 20,000 U/mL stock
0.5 μL of EcoRI-HF® 20,000 U/mL stock
1 μL of CutSmart BufferTM 10× stock
3 μL of molecular biology–grade water
10. Incubate in a PCR machine or water bath at 37 °C for 15–30 min.
11. Prepare a 1% agarose gel: Mix 1% agarose in 1× TAE buffer and microwave for 40–60 s, swirling the flask after microwaving. Then, add the ethidium bromide solution (5 μL per 60 mL of agarose gel) and swirl the flask again to mix. Then, pour the gel mixture into a gel tray with the desired gel comb.
12. Once incubation is complete, add 2 μL of gel loading dye, purple 6× stock to each tube.
13. Load 1 μL of 1 kb Plus DNA ladder 500 μg/mL stock and samples on the gel and run at 100 V for 30 min.
14. Image gel in myECL Imager and ensure bands are the correct sizes (6,353 bp, 898 bp, and 423 bp) relative to the control (6,353 bp, 1,076 bp, and 898 bp) (Figure 2).
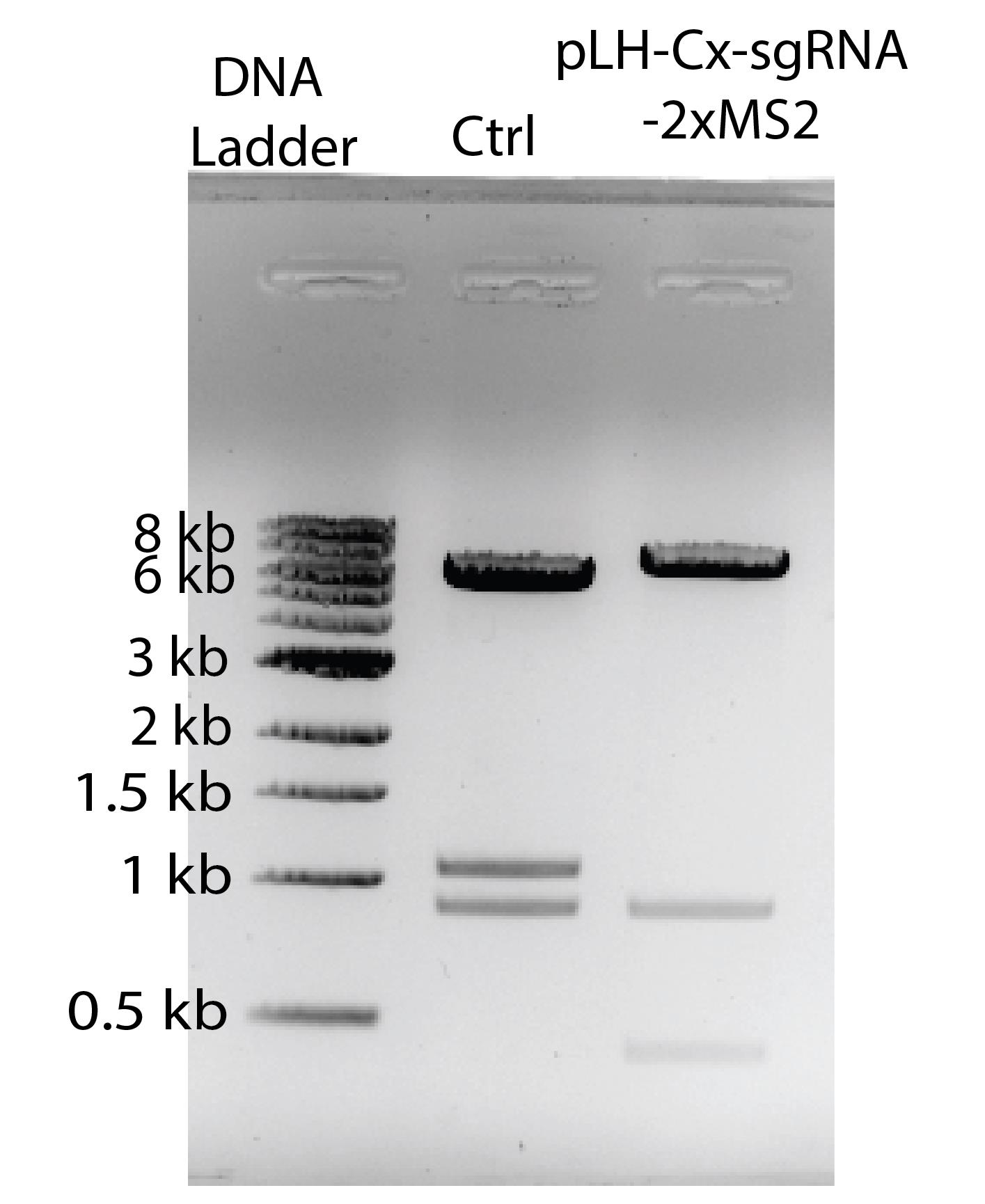
Figure 2. Digestion test of the construct with successful sgRNA insertion. DNA ladder, NEB 1 kb DNA ladder. Ctrl, unmodified pLH-sgRNA1-2xMS2 vector digested with NotI and EcoRI, resulting in three bands at 6,353 bp, 1,076 bp, and 898 bp. Digestion of the sgRNA-inserted pLH-Cx-sgRNA1-2xMS2 construct produced three DNA fragments at 6,353 bp, 898 bp, and 423 bp. The reduction in fragment size indicates the loss of ccdB gene. Image is inverted for enhanced visibility.
C. PCR amplify pLH-sgRNA1 with the U6 promoter and desired crRNA sequence
1. Dilute the isolated plasmids to 10 ng/μL using molecular biology–grade water.
2. In a PCR tube, mix the reaction as follows:
1 μL of isolated plasmid 10 ng/μL
2.5 μL of forward primer 10 μM (see section G in the Reagent list)
2.5 μL of reverse primer 10 μM (see section G in the Reagent list)
1 μL of dNTP 10 μM
10 μL of Q5® reaction buffer 5×
0.5 μL of Q5® high-fidelity DNA polymerase 2,000 U/mL
32.5 μL of molecular biology–grade water
3. Amplify pLH-sgRNA1 with the desired crRNA sequence using the following program in a PCR machine:
98 °C for 30 s
98 °C for 10 s (35 cycles)
55 °C for 30 s (35 cycles)
72 °C for 30 s (35 cycles)
72 °C for 10 min
4 °C pause
Pause point: You can store the amplified pLH-sgRNA1 at -20 °C and continue the next day.
4. Prepare a 1% agarose gel: Mix 1% agarose in 1× TAE buffer and microwave for 40–60 s, swirling the flask after microwaving. Then, add the ethidium bromide solution (5 μL per 60 mL of agarose gel) and swirl the flask again to mix. Then, pour the gel mixture into a gel tray with the desired gel comb.
5. Add 2 μL of gel loading dye, purple 6×stock to each tube containing 10 μL of the PCR product.
6. Load 1 μL of 1 kb Plus DNA ladder 500 μg/mL stock and samples on the gel and run at 100 V for 30 min.
7. Isolate amplified pLH-sgRNA1 using Monarch® PCR & DNA Cleanup kit following the manufacturer’s instructions.
8. Measure isolated pLH-sgRNA1 plasmid concentration using the NanoDropTM 2000c spectrophotometer in ng/μL.
Pause point: You can store the isolated pLH-sgRNA1 at -20 °C and continue the protocol the next day.
D. Assembling CRISPRainbow for the gene of interest
1. Dilute isolated pLH-sgRNA1 so that they are 10 ng/μL.
2. In a PCR tube, mix the digestion reaction as follows:
1 μL of CRISPRainbow-DONOR1 100 ng/μL
1 μL of 10× CutSmart BufferTM
1 μL of ATP 10 mM
0.6 μL of BsaI-HF® v2 20,000 U/mL
0.4 μL of T7 DNA ligase 3,000,000 U/mL
1 μL of each isolated pLH-sgRNA1 (total of 6 μL) 10 ng/μL stock
Note: CRISPRainbow-DONOR1 plasmid must be amplified by One ShotTM ccdB SurvivalTM 2 T1R Competent Cells.
3. Incubate in a PCR machine or water bath at 37 °C for 1 h.
4. Thaw NEB® Stable Competent E. coli (high efficiency) on ice (~5 min) and immediately add 5 μL of the cloning reaction mix.
5. Mix gently by tapping the tube and let sit on ice for 4–5 min.
Caution: Do not pipette up and down or vortex!
6. Spread the whole volume (55 μL) on an LB plate containing carbenicillin (100 μg/mL) and incubate overnight at 37 °C.
7. After 16–19 h, select 4–6 colonies from the plate, culture in 5 mL of LB broth containing carbenicillin (100 μg/mL) in a 14 mL polypropylene round-bottom culture tube, and incubate overnight at 37 °C with shaking (225–250 rpm) in the MaxQTM 6000 Incubated/Refrigerated Shaker.
8. After 16–19 h, isolate the cloned vectors using the Zyppy® Plasmid Miniprep kit and follow the protocol provided. The assembled CRISPRainbow-sgRNA-Cx-C14-C7-C1-C13-C3 sequence is shown in Figure 3.
Note: If pausing the experiment for more than 12 h, 0.5 mL of bacterial culture can be stored in a -80 °C freezer with 15% glycerol (final concentration).

Figure 3. Assembled CRISPRainbow-sgRNA-Cx-C14-C7-C1-C13-C3 sequence visualized in ApE. The U6 promoter is highlighted in light grey. Inserted sgRNA sequences are highlighted in orange. The MS2, PP7, and boxB stem loops are highlighted in blue, green, and red, respectively. Overhanging annealing sequences are marked in violet.
9. Measure assembled CRISPRainbow construct (Figure 1) using the NanoDropTM 2000c spectrophotometer in ng/μL.
Note: A minimal concentration of 20 ng/μL is needed for whole plasmid sequencing.
Pause point: You can store the isolated assembled CRISPRainbow at -20 °C and continue the next day.
10. (Steps D10–15 are optional) Perform restriction digest on CRISPRainbow plasmids in PCR tubes as follows:
3 μL of isolated plasmid 100 ng/μL
0.5 μL of ApaLI 10,000 U/mL
1 μL of 10× CutSmart BufferTM
5.5 μL of molecular biology–grade water
11. Incubate in a PCR machine or water bath at 37 °C for 15–30 min.
12. Prepare a 1% agarose gel: Mix 1% agarose in 1× TAE buffer and microwave for 40–60 s, swirling the flask after microwaving. Then, add the ethidium bromide solution (5 μL per 60 mL of agarose gel) and swirl the flask again to mix. Then, pour the gel mixture into a gel tray with the desired gel comb.
13. Once incubation is complete, add 2 μL of gel loading dye, purple 6× stock to each tube.
14. Load 1 μL of 1 kb Plus DNA ladder 500 μg/mL and samples on the gel and run at 100 V for 30 min.
15. Image the gel in myECL Imager and ensure bands are in correct sizes (3,226 bp, 1,246 bp, and 288 bp) for the successful insertion of all six pLH-sgRNA1 PCR products (Figure 4).
Note: ApaLI will cut in three positions in plasmids that have successful insertions, but will cut four positions in the empty vector. The size of the largest band should reflect the number of inserted sgRNA.
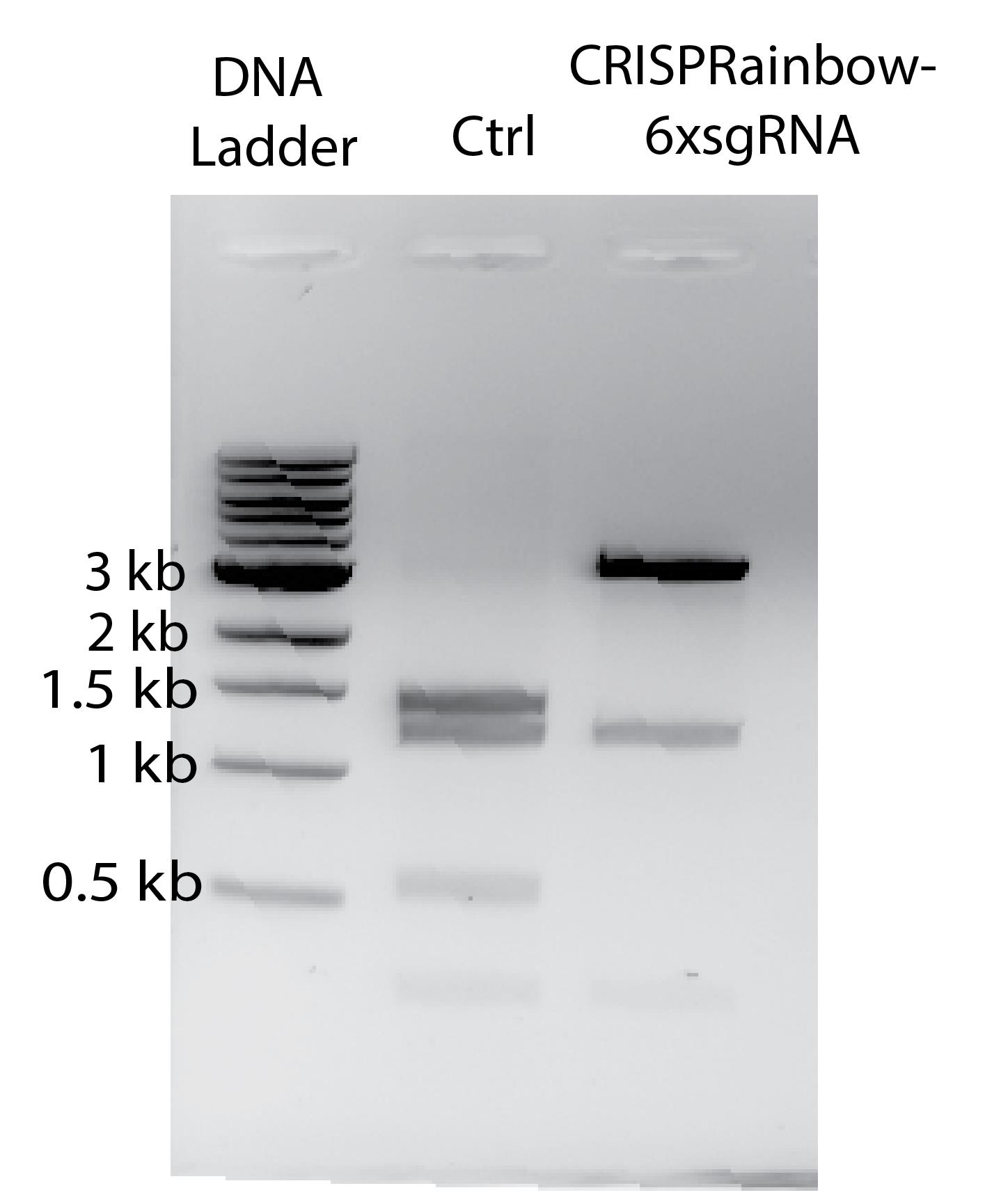
Figure 4. Digestion test of the assembled CRISPRainbow-6xsgRNA construct. DNA ladder, NEB 1 kb DNA ladder. Ctrl, uninserted CRISPRainbow vector digested with ApLI, resulting in four bands at 1,434 bp, 1,246 bp, 559 bp, and 288 bp. ApLI digestion of the assembled CRISPRainbow-6xsgRNA construct yielded three DNA fragments at ~3,226 bp, 1,246 bp, and 288 bp. The image is inverted for better visibility.
16. Order whole plasmid sequencing for samples using PLASMIDSAURUS (https://plasmidsaurus.com/).
Notes:
1. Prepare samples following the instructions on the website.
2. Alternatively, use two sequencing primers to confirm the assembled CRISPRainbow plasmid:
DONOR1-Forward, 5′-GCTACAACAAGGCAAGGCTTGACCGA-3′
DONOR1-Reverse, 5′-AACGCCAGCAACGCGGCCTT-3′
17. Once sequencing results come back, analyze and make sure the assembled CRISPRainbow plasmid has all pLH-sgRNA1 inserts and check for random mutations using ApE software.
Caution: Mutations within the pLH-sgRNA1 inserts can significantly impact targeting the gene of interest and may result in no labeling.
E. Amplification of assembled CRISPRainbow
1. After confirming the sequence, use a sterile tip to scratch a small piece of ice from the frozen bacterial stock (step D8) of the colony with the correct assembled CRISPRainbow and add 5 mL of LB broth containing carbenicillin (100 μg/mL) in a 14 mL polypropylene round-bottom culture tube. Incubate for 16–19 h at 37 °C with shaking (225–250 rpm) in the MaxQTM 6000 Incubated/Refrigerated Shaker.
Notes:
1. It is recommended to perform step E1 in a sterile biosafety cabinet.
2. If the transformed bacteria were not stored at -80 °C, retransformation of the correct plasmid is necessary for the amplification.
2. Add 0.5 mL of the bacterial solution to 49.5 mL of LB broth containing carbenicillin (100 μg/mL) in a 250 mL flask and incubate at 37 °C with shaking (180–200 rpm) overnight.
3. After 16–19 h, isolate the assembled CRISPRainbow using the ZymoPURETM II Plasmid Midiprep kit following the manufacturer’s instructions.
4. Measure eluted CRISPRainbow plasmid using the NanoDropTM 2000c spectrophotometer in ng/μL.
Pause point: You can store the isolated assembled CRISPRainbow at -20 °C and continue the next day.
F. Preparing U2OS cells for transfection
1. Warm U2OS culture medium (see Recipes) in a water bath for 20 min.
2. Take the U2OS cells out from the liquid nitrogen (LocatorTM Plus Rack and Box Systems) and thaw for 30 s in the water bath until it is almost melted.
3. Prepare a 15 mL VWR® centrifuge tube with 9 mL of the U2OS culture medium and add 1 mL of the U2OS stock solution.
Note: This step is important in clearing DMSO and cell debris from the stock.
4. Centrifuge the tube at 300× g for 3 min to form a small pellet and aspirate the supernatant.
5. Resuspend the U2OS pellet in 5 mL of fresh media (for a 25 cm2 flask) by gently pipetting up and down.
Caution: Do not vortex!
6. Transfer resuspended cells to a 25 cm2 cell culture flask and incubate at 37 °C in a CO2 incubator.
7. The next day, check for cell growth and confluency using a phase contrast microscope.
8. Split cells when the confluency reaches approximately 75%. After 72 h, prepare cells for transfection.
Note: To maintain cells, when confluency reaches near 75%, remove media and add 1 mL of DPBS 1× and rock the flask back and forth three to four times. Then, remove the DPBS and add 1 mL of TrypLETM Express Enzyme, no phenol red, 1×. Rock back and forth three to four times and then remove 800 μL and incubate at 37 °C in a CO2 incubator for 2 min. Add 5 mL of media for U2OS cells and give the flask a hard smack to the side to release the cells into the media. Then, split the cells as desired.
9. Add the 5 mL of suspended cells to a 15 mL VWR® centrifuge tube and centrifuge at 300× g for 3 min at RT.
10. Aspirate the media and resuspend the pellet in 500 μL of fresh media by gently pipetting up and down.
11. Mix 10 μL of 0.4% Trypan Blue dye with 10 μL of cell solution by pipetting up and down a few times.
12. Add 10 μL of cell/dye mixture to the cell counter slide, cover with a coverslip, and wait for 40 s.
13. Place the cell counter slide into the white plastic holder, then count the cells using the Countess® II FL Automated Cell Counter (or similar cell counting instruments).
Note: Cells can be counted manually or using cell counters from other vendors.
14. Seed 1 × 105 cells in 1.5 mL culture media in a 35 mm imaging dish a day before transfection.
Note: Optimal cell seeding number can vary depending on the cell counter used. In general, cells should reach about 50%-70% confluency before transfection.
15. Save the rest of the cells in a new 25 cm2 flask in a 37 °C CO2 incubator for the next experiment.
G. U2OS transfection with assembled CRISPRainbow
1. Bring Opti-MEMTM I reduced serum medium and Lipofectamine® 2000 to room temperature before transfection.
2. Vortex and briefly centrifuge down all plasmid solutions (1,000× g for 30 s on a minicentrifuge machine), the assembled CRISPRainbow plasmid, pHAGE-EFS-MCP-3XBFPnls, pHAGE-EFS-PCP-3XGFPnls, pHAGE-EFS-N22p-3XRFPnls, and the pHAGE-TO-dCas9 plasmid. Then, prepare the tubes according to Table 2.
Table 2. Preparing transfection tubes
| Tube 1 | Tube 2 | ||
| Opti-MEMTM I reduced serum medium, 1× | 150 μL | Opti-MEMTM I reduced serum medium, 1× | 100 μL |
| Lipofectamine® 2000 | 5 μL | Assembled CRISPRainbow-6XsgRNA plasmid | 750 ng |
| pHAGE-TO-dCas9 | 150 ng | ||
| pHAGE-EFS-MCP-3XBFPnls | 20 ng | ||
| pHAGE-EFS-PCP-3XGFPnls | 20 ng | ||
| pHAGE-EFS-N22p-3XRFPnls | 20 ng | ||
3. Combine the contents of the two tubes by adding Tube 2 to Tube 1 and mixing by pipetting up and down 8–10 times. Incubate at room temperature for 5 min.
4. After incubation, add 250 μL of the DNA-lipid complex to the imaging dish containing cells and incubate in a 37 °C CO2 incubator.
5. After 24–48 h, record the fluorescence signals from brightfield with enhanced contrast, such as differential interference contrast (DIC), blue, red, and green, under a microscope (Figure 5). For tracking genomic locus movement, time-lapse videos must be recorded. For short-time dynamics, a video of 50–200 frames with 50 ms exposure time can be recorded and analyzed. For long-time dynamics, a time interval from minutes to hours can be implemented. To register images from distinct channels, capture coverslip-immobilized microsphere (0.1 μm) images in all channels before imaging cells.
Notes:
1. When recording long-time dynamics, corrections for cell movements and rotations are recommended for accurate data interpretation.
2. U2OS seeding density can be modified to maximize the transfection efficiency.
3. A real-time video can be found in the supplementary information of the original publication in Nature Biotechnology [9].
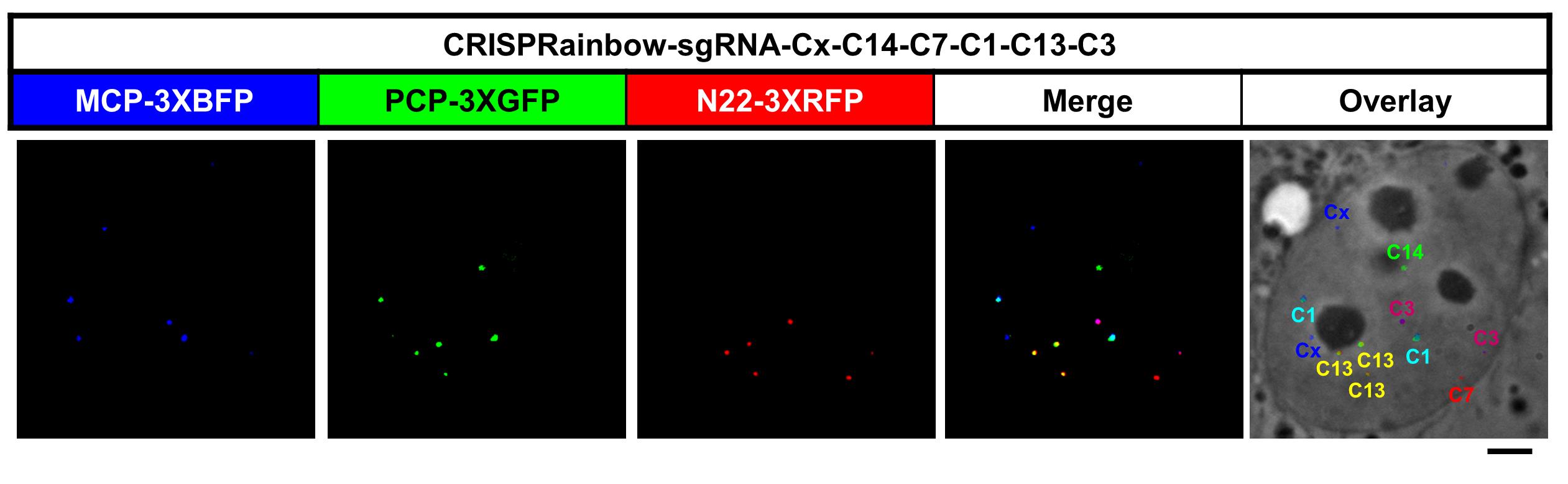
Figure 5. Localization of multiple chromosome-specific loci in a U2OS cell. Transfection of pCRISPRainbow-sgRNA-Cx-C14-C7-C1-C13-C3, MCP-3XBFP, PCP-3XmNeonGreen, and N22-3XRFP into U2OS cells. Scale bar, 5 μm. False colors were added to enhance visualization.
Data analysis
A. Image overlay and single-particle detection
Generally, two fluorescent signals are observed per genomic locus in diploid cell lines. Fiji can be used to merge channels (Image > Color > Merge Channels), and fluorescent particles can be automatically counted by the function Analyze Particles (Analyze > Analyze Particles). Loci showing only in one channel are labeled with primary colors: blue, green, and red. In this protocol, the crRNA sequence (5′-TAGAGTCTGCAT-3′) cloned into the vector pLH-sgRNA1-2xMS2 targeting the locus on chromosome X is labeled blue, the crRNA sequence (5′-ATGGTGAGAGTG-3′) cloned into the vector pLH-sgRNA1-2xboxB targeting the locus on chromosome 7 is labeled red, and the crRNA sequence (5′-ATGGTGAGAGTG-3′) cloned into the vector pLH-sgRNA1-2xPP7 targeting the locus on chromosome 14 is labeled green (Figure 5, see Table 1 for details). Loci labeled with secondary colors, cyan, magenta, and yellow, appear as overlapping signals from two channels. For example, the crRNA sequence (5′-TGATATCACAG-3′) cloned into the vector pLH-sgRNA1-boxB-MS2 (magenta) targeting the locus on chromosome 3 is detected in both blue and red channels, the crRNA sequence (5′-AGATGCTCACC-3′) cloned into the vector pLH-sgRNA1-MS2-PP7 (cyan) targeting the locus on chromosome 1 is detected in both green and blue channels, and the crRNA sequence (5′-ACCATTCCTTC-3′) cloned into the vector pLH-sgRNA1-PP7-boxB (cyan) targeting the locus on chromosome 13 is detected in both green and red channels.
Note: When cells enter late S phase, more than two signals can be observed. In such cases, signals from sister chromatids are observed in close proximity to each other.
B. Tracking movement of single loci over time
If time-lapse videos are recorded, tracking of individual loci can be performed on Fiji, using the plugin TrackMate [10]. Instructions for the user interface can be found on https://imagej.net/plugins/trackmate/. Output the trajectory coordinates as .csv files.
Note: Do not crop images before performing locus tracking because cropping will result in the loss of original coordinate information.
C. Plotting locus trajectories using OriginPro
We used OriginPro to plot locus trajectories, but other software, such as Microsoft Excel, can also be used for this purpose. The output .csv files from TrackMate can be directly imported into OriginPro to create plots using the x and y coordinates, which are given in camera pixel units. To generate plots in real-world units, convert the x and y coordinates to micrometers (μm) or nanometers (nm) by multiplying the pixel size of your imaging system. For example, if the camera pixel size is 16 μm × 16 μm and the sample is imaged using a 100× objective lens, the pixel size is approximately 160 nm × 160 nm in both x and y directions.
Validation of protocol
This protocol has been used and validated in the following research article [9]:
• Ma, H., Tu, L.-C., Naseri, A., Huisman, M., Zhang, S., Grunwald, D. & Pederson, T. (2016). Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 34: 528–530. https://doi.org/10.1038/nbt.3526
A modified protocol for HEK293, hTERT-RPE1, HFF1, TIG1, and MRC5 cells has been used and validated in the following research article [11]:
• Takata H., Masuda Y. & Ohmido N. (2023). CRISPR imaging reveals chromatin fluctuation at the centromere region related to cellular senescence. Sci. Rep. 13: 14609. https://doi.org/10.1038/s41598-023-41770-6
A modified protocol for plants has been used and validated in the following research article [12]:
• Khosravi S, Schindele P, Gladilin E, Dunemann F, Rutten T, Puchta H. & Houben A. (2020). Application of aptamers improves CRISPR-based live imaging of plant telomeres. Front. Plant Sci. 11: 1254. https://doi.org/10.3389/fpls.2020.01254
General notes and troubleshooting
Troubleshooting
| No. | Section | Problem | Possible cause | Suggestion/solution |
|---|---|---|---|---|
| 1 | B, D | Incorrect bands on diagnostic digestion. | 1) Reduced restriction enzyme activity. 2) Unsuccessful insertion of sgRNA sequence. 3) Expiration of reaction buffers. | 1) Adjust the amounts of restriction enzyme/DNA used. 2) Adjust incubation time. 3) Reattempt insertion of sgRNA sequence. 4) Pick new colonies. |
| 2 | D | No colonies on the plate. | 1) Unsuccessful assembly of CRISPRainbow, 2) The antibiotic selection is too strong or incorrect. 3) The competent cells have lost viability or competence. | 1) Reattempt insertion of sgRNA sequence. 2) Remake bacterial plates with the correct antibiotic concentration. 3) Reattempt transformation with a new tube of competent cells. |
| 3 | Data analysis A | No fluorescent signal. | 1) Plasmid ratio imbalance. 2) CRISPR target sites blocked by DNA binding proteins. 3) Unsuccessful delivery of plasmids. | 1) Increase sgRNA amount. 2) Change a sgRNA targeting sequence. 3) Reattempt transfection with a new tube of the transfection reagent. 4) Optimize cell numbers vs. the amount of the transfection reagent. |
| 4 | Data analysis A | Signal variability. | 1) Off-target binding by sgRNAs. 2) The cell line is aneuploid. | 1) Change a sgRNA targeting sequence. 2) Use a diploid cell line. |
Acknowledgments
Conceptualization, L.C.T.; Investigation, T.V.; Writing—Original Draft, T.V.; Figures and Tables, L.C.T. and T.V.; Writing—Review & Editing, D.N., L.C.T., and T.V.; Funding acquisition, L.C.T.; Supervision, L.C.T. This work was funded by the NIH grant R35 GR132998 to L.C.T. and the OSU Undergraduate Research Scholarship to D.N. This protocol was adapted from previous work [9].
Competing interests
The authors declare no competing financial interests.
References
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. and Charpentier, E. (2012). A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337(6096): 816–821. https://doi.org/10.1126/science.1225829
- Hsu, P. D., Lander, E. S. and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157(6): 1262–1278. https://doi.org/10.1016/j.cell.2014.05.010
- Zalatan, J.G., Lee, M.E., Almeida, R., Gilbert, L.A., Whitehead, E.H., La Russa, M., Tsai, J.C., Weissman, J.S., Dueber, J. E., Qi, L. S. and Lim, W. A. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 160(1–2): 339–350. https://doi.org/10.1016/j.cell.2014.11.052
- Chen, B., Gilbert, L. A., Cimini, B. A., Schnitzbauer, J., Zhang, W., Li, G.-W., Park, J., Blackburn, E. H., Weissman, J. S., Qi, L. S. and Huang, B. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 155(7): 1479–1491. https://doi.org/10.1016/j.cell.2013.12.001
- Anton, T., Bultmann, S., Leonhardt, H. and Markaki, Y. (2014). Visualization of specific DNA sequences in living mouse embryonic stem cells with programmable fluorescent CRISPR/Cas system. Nucleus. 5(2): 163–172. https://doi.org/10.4161/nucl.28488
- Ma, H., Naseri, A., Reyes-Gutierrez, P., Wolfe, S. A., Zhang, S. and Pederson, T. (2015). Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA. 112(10): 3002–3007. https://doi.org/10.1073/pnas.1420024112
- Ren, R., Deng, L., Xue, Y., Suzuki, K., Zhang, W., Yu, Y., Wu, J., Sun, L., Gong, X., Luan, H., et al. (2017). Visualization of aging-associated chromatin alterations with an engineered TALE system. Cell Res. 27: 483–504. https://doi.org/10.1038/cr.2017.18
- Bey, T. D., Koini, M. and Fransz, P. (2018). Fluorescence In Situ Hybridization and Immunolabeling on 3D Preserved Nuclei. Methods Mol Biol. 1675: 467–480. https://doi.org/10.1007/978-1-4939-7318-7_27
- Ma, H., Tu, L.-C., Naseri, A., Huisman, M., Zhang, S., Grunwald, D. and Pederson, T. (2016). Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol. 34(5): 528–530. https://doi.org/10.1038/nbt.3526
- Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., Bednarek S.Y., Shorte S. L. and Eliceiri, K. W. (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods. 115: 80–90. https://doi.org/10.1016/j.ymeth.2016.09.016
- Takata H., Masuda Y. and Ohmido N. (2023). CRISPR imaging reveals chromatin fluctuation at the centromere region related to cellular senescence. Sci Rep. 13: 14609. https://doi.org/10.1038/s41598-023-41770-6
- Khosravi, S., Schindele, P., Gladilin, E., Dunemann, F., Rutten, T., Puchta, H. and Houben, A. (2020). Application of aptamers improves CRISPR-based live imaging of plant telomeres. Front Plant Sci. 11: 1254. https://doi.org/10.3389/fpls.2020.01254
Article Information
Publication history
Received: May 20, 2025
Accepted: Jul 29, 2025
Available online: Aug 11, 2025
Published: Sep 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Versosky, T. J., Nishonov, D. U. and Tu, L. C. (2025). Real-Time Imaging of Specific Genomic Loci With CRISPR/dCas9 in Human Cells Using CRISPRainbow. Bio-protocol 15(17): e5432. DOI: 10.21769/BioProtoc.5432.
Category
Biophysics > Microscopy > Single-molecule localization microscopy
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








