- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
(*Contributed equally to this work, §Technical contact: weijw@webmail.hzau.edu.cn) Published: Vol 15, Iss 14, Jul 20, 2025 DOI: 10.21769/BioProtoc.5382 Views: 2159
Reviewed by: Alba BlesaLionel SchiavolinAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Thylakoid Membranes from the Cyanobacterium Synechocystis sp. PCC 6803 and Analysis of Their Photosynthetic Pigment-protein Complexes by Clear Native-PAGE
Josef Komenda [...] Tomas Zakar
Jan 5, 2019 8573 Views

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6191 Views
Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2080 Views
Abstract
The CRISPR-Cas system of Thermus thermophilus has emerged as a potent biotechnological tool, particularly its Cas6 endonuclease, which plays a crucial role in CRISPR RNA (crRNA) maturation. This protocol details a robust and reproducible method for the high-level expression and purification of recombinant T. thermophilus Cas6 proteins (Cas6-1 and Cas6-2) in E. coli. We describe a streamlined approach encompassing plasmid construction using seamless assembly, optimized bacterial heterologous expression, and multi-step purification leveraging affinity and size-exclusion chromatography. The protocol outlines the generation of both His-tagged and GST-tagged Cas6 variants, enabling flexibility in downstream applications. Key steps, including primer design, PCR optimization, competent cell transformation, and chromatography strategies, are meticulously detailed with critical parameters and troubleshooting guidance to ensure experimental success and high yields of highly pure and active T. thermophilus Cas6 proteins. This protocol is useful for researchers requiring purified T. thermophilus Cas6 for structural studies, biochemical characterization, and the development of CRISPR-based biotechnological tools.
Key features
• Robust method for expressing and purifying Thermus thermophilus Cas6 proteins in E. coli.
• Seamless assembly cloning and dual affinity tagging system: Offers options for both His-tag and GST-tag purification strategies for increased versatility.
• Applicable for diverse heterologous expression and purification of well-folding thermostable proteins in mesophilic host chassis cells [E. coli BL21(DE3)].
Keywords: CRISPR-Cas6Graphical overview
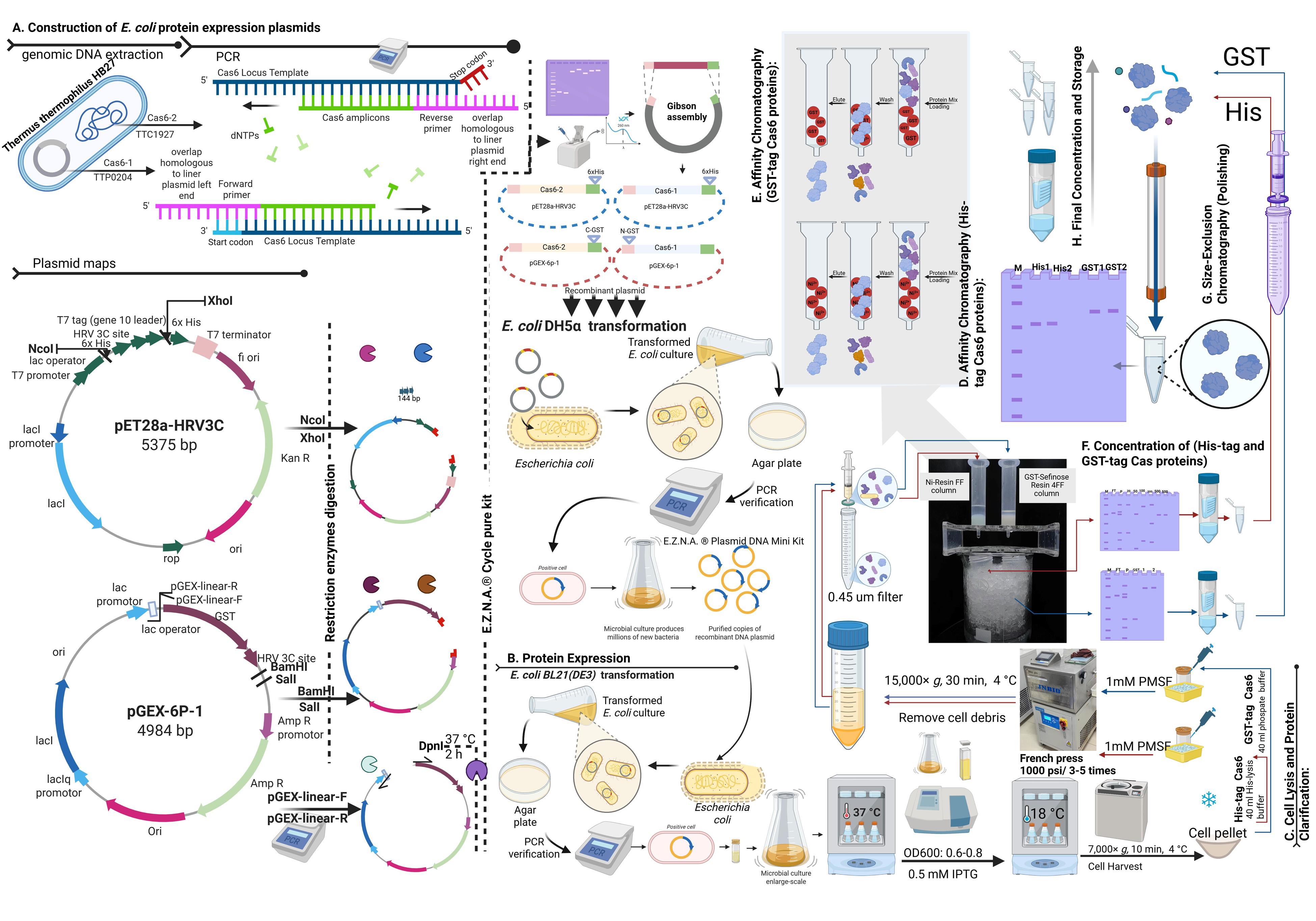
Flowchart outlining the steps in Cas6 heterologous expression and purification
Background
The CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats–CRISPR-associated proteins) system, a cornerstone of bacterial and archaeal adaptive immunity, relies on CRISPR RNAs (crRNAs) that guide effector complexes to target foreign nucleic acids, such as plasmids and bacteriophages, through sequence-specific nucleic acid targeting [1–3]. This system has enabled precise genome editing and innovative applications in therapeutics and biotechnology [3,4]. The biogenesis of mature crRNAs involves the precise cleavage of precursor CRISPR RNAs (pre-crRNAs) by specialized endonucleases, such as Cas6, which recognize and process repeat sequences within the CRISPR array [5,6].
Thermus thermophilus HB27, a thermophilic bacterium thriving in high-temperature environments, harbors a remarkably thermostable CRISPR-Cas system that has garnered significant interest for its biotechnological potential [3]. Thermophilic CRISPR-Cas systems, including those of T. thermophilus, are of particular interest due to their stability and functionality at elevated temperatures, which enhances their utility in industrial bioprocessing and high-temperature biochemical assays [7]. T. thermophilus is a model organism for studying thermozymes—enzymes adapted to extreme thermal conditions—owing to its rich repertoire of thermostable proteins, including DNA polymerases and ligases widely used in molecular biology [3]. Among its diverse CRISPR-Cas arsenal, T. thermophilus encodes multiple CRISPR-Cas subtypes, including Type I and Type III systems, which exhibit extraordinary diversity in their defensive mechanisms, making it a valuable system for studying CRISPR evolution and function [8,9].
Within this context, T. thermophilus Cas6 endonucleases, specifically Cas6-1 and Cas6-2, are critical for pre-crRNA processing, cleaving repeat sequences to generate mature crRNAs that integrate into CRISPR-Cas effector complexes for targeted nucleic acid recognition [3]. The thermostability of these enzymes, coupled with their precise RNA cleavage activity, positions them as promising tools for RNA manipulation, including in vitro RNA processing and diagnostic platforms requiring robust performance under stringent conditions [3]. However, structural and functional studies of T. thermophilus Cas6, as well as their biotechnological applications, have been hindered by the lack of efficient, high-yield recombinant production protocols.
This protocol addresses this gap by providing a comprehensive, optimized, and adaptable method for the expression and purification of recombinant T. thermophilus Cas6 proteins, including both His-tagged and GST-tagged variants. By enabling high-purity production, this procedure facilitates biochemical, structural, and applied research into these thermostable endonucleases, advancing their utility in CRISPR-based technologies and RNA manipulation tools.
Materials and reagents
Biological materials
1. Thermus thermophilus HB27 (ATCC, catalog number: BAA-163; extract gDNA according to Wei et al. [3])
2. Escherichia coli DH5α (Thermo Scientific, catalog number: 18265017, stored at -80 °C)
3. Escherichia coli BL21(DE3) (Thermo Scientific, catalog number: EC0114, stored at -80 °C)
Reagents
1. pET28a-HRV3C (Addgene ID: 241018, store at -20 °C)
2. pGEX-6p-1 (Addgene ID: 241019, store at -20 °C)
3. NcoI (Thermo Scientific, catalog number: FD0574, store at -20 °C)
4. XhoI (Thermo Scientific, catalog number: FD0694, store at -20 °C)
5. BamHI (Thermo Scientific, catalog number: FD0054, store at -20 °C)
6. SalI (Thermo Scientific, catalog number: FD0644, store at -20 °C)
7. Phanta Max Super-Fidelity DNA polymerase (Vazyme, catalog number: P505-d3-AA, store at -20 °C)
8. DpnI (Thermo Scientific, catalog number: FD1404, store at -20 °C)
9. ABclonal MultiF Seamless Assembly Mix (ABclonal, catalog number: RK21020, store at -20 °C)
10. Nuclease-free water (Sigma-Aldrich, CAS number: 7732-18-5)
11. 2× Phanta Max buffer (Vazyme, catalog number: P505-d3-Ab, store at -20 °C)
12. dNTP mix (10 mM each) (Vazyme, catalog number: P505-d3-Ac, store at -20 °C)
13. Tryptone (OXOID, catalog number: LP0042B)
14. Yeast extract (OXOID, catalog number: LP0021B)
15. Agar (BioFroxx, catalog number: 8211GR500)
16. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I6758-5G)
17. Sodium chloride (NaCl) (Sigma-Aldrich, CAS number: 7558-79-4)
18. HEPES (Sigma-Aldrich, catalog number: H3375)
19. Glycerol (Sigma-Aldrich, catalog number: G5516)
20. Imidazole (Sigma-Aldrich, catalog number: I2399)
21. Reduced glutathione (Sigma-Aldrich, catalog number: G4251)
22. Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541)
23. Dibasic sodium phosphate (Na2HPO4) (Sigma-Aldrich, catalog number: S3225)
24. Monobasic potassium phosphate (KH2PO4) (Sigma-Aldrich, catalog number: P9791)
25. Kanamycin (Sigma-Aldrich, catalog number: K1377)
26. Ampicillin (Sigma-Aldrich, catalog number: A5354-10ML)
27. Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626)
28. Sodium dodecyl sulfate (SDS) (BioFRoxx, catalog number: 3250GR500)
29. Tris base (2-Amino-2-(hydroxymethyl)-propane-1,3-diol,Tris base) (BioFRoxx, CAS number: 77-86-1)
30. Glycine (BioFRoxx, CAS number: 56-40-6)
31. Ammonium persulfate (APS) (SCR, CAS number: 7727-54-0)
32. Acryl/Bio 30% solution (37.5:1) (Sanggon Biotech, catalog number: B546018-0500, store at 2–8 °C)
33. TEMED (Sigma, CAS number: 110-12-9)
34. Disodium EDTA dihydrate (SCR, CAS number: 6381-92-6)
35. Sodium hydroxide (NaOH) (SCR, CAS number: 1310-73-2)
36. Glacial acetic acid (SCR, CAS number: 64-19-7)
37. Agarose (Singke, catalog number: TSJ001)
38. Goldview (GL Biotech, catalog number: H003-1)
Solutions
1. LB medium (LB) (see Recipes)
2. Kanamycin stock solution (30 mg/mL) (see Recipes)
3. IPTG stock solution (100 mM) (see Recipes)
4. His-lysis buffer (see Recipes)
5. 5 M Imidazole stock solution (see Recipes)
6. His-washing buffer (see Recipes)
7. 100 mM Imidazole His-elution buffer (see Recipes)
8. 200 mM Imidazole His-elution buffer (see Recipes)
9. 500 mM Imidazole His-elution buffer (see Recipes)
10. 1 M Imidazole His-elution buffer (see Recipes)
11. Phosphate buffer (see Recipes)
12. GST-elution buffer (see Recipes)
13. SEC buffer (see Recipes)
14. 10% SDS solution (see Recipes)
15. 1.5 M Tris-HCl (pH 8.8) stock solution (see Recipes)
16. 1.0 M Tris-HCl (pH 6.8) stock solution (see Recipes)
17. 2.5 M glycine stock solution (see Recipes)
18. 10% Ammonium persulfate (APS) (see Recipes)
19. SDS-PAGE running buffer (10× Tris-Glycine-SDS, pH 8.3) (see Recipes)
20. SDS-PAGE resolving gel (12% polyacrylamide) (see Recipes)
21. SDS-PAGE stacking gel (5% polyacrylamide) (see Recipes)
22. 0.5 M EDTA (pH 8.0) stock solution (see Recipes)
23. 50× TAE buffer (see Recipes)
24 1.5 % agarose gel (see Recipes)
Recipes
1. LB medium
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| NaCl | 10 g/L | 10 g |
| Water | n/a | to 1 L |
Dissolve reagents in approximately 900 mL of water. Adjust pH to 7.0 if necessary. Bring the volume to 1 L with water. For plate media, add agar to a final concentration of 1.5% (15 g/L). Sterilize by autoclaving at 121 °C for 20 min.
2. Kanamycin stock solution (30 mg/mL)
Dissolve 3 g of kanamycin in ddH2O to a final volume of 100 mL. Filter sterilize using a 0.22 μm filter. Store at -20 °C for long-term storage or 4 °C for short-term storage. This 1000-fold concentrated stock solution is designed to be added at 1 mL per 1 L of LB medium to achieve a final kanamycin concentration of 30 μg/mL.
Critical: Add kanamycin after the LB medium has cooled down to below 50 °C following autoclaving to prevent antibiotic degradation.
3. IPTG stock solution (100 mM)
Dissolve 2.383 g of IPTG in ddH2O to a final volume of 100 mL. Filter sterilize using a 0.22 μm filter. Store at -20 °C for long-term storage or 4 °C for short-term storage. This stock solution is designed to be added at 5 mL per 1 L of culture to achieve a final IPTG concentration of 0.5 mM.
4. His-lysis buffer (Base buffer)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 20 mM | 4.77 g |
| NaCl | 300 mM | 17.53 g |
| Glycerol (99%) | 5% (v/v) | 50 mL |
| Milli-Q water | n/a | to 1 L |
Dissolve HEPES in approximately 700 mL of Milli-Q water. HEPES dissolves best at neutral pH. Adjust the pH to 7.5 using a pH meter at room temperature. Then add the other components. Bring the final volume to 1 L with Milli-Q water. Filter using a 0.22 μm filter. Store at 4 °C for 1 week.
5. 5 M Imidazole stock solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Imidazole | 5 M | 34.02 g |
| Milli-Q water | n/a | to 100 mL |
Dissolve imidazole in Milli-Q water to a final volume of 100 mL. Adjust pH to 7.5 with HCl (imidazole dissolves best at neutral pH). Filter using a 0.22 μm filter. Store at -20 °C for long-term storage or 4 °C for short-term storage.
6. His-washing buffer
| Reagent | Final concentration | Quantity for 100 mL |
|---|---|---|
| His-lysis buffer | (Base buffer) | 99 mL |
| 5 M imidazole | 50 mM | 1 mL |
7. 100 mM Imidazole His-elution buffer
| Reagent | Final concentration | Quantity for 100 mL |
|---|---|---|
| His-lysis buffer | (Base buffer) | 98 mL |
| 5 M imidazole | 100 mM | 2 mL |
8. 200 mM Imidazole His-elution buffer
| Reagent | Final concentration | Quantity for 100 mL |
|---|---|---|
| His-lysis buffer | (Base buffer) | 96 mL |
| 5 M imidazole | 200 mM | 4 mL |
9. 500 mM Imidazole His-elution buffer
| Reagent | Final concentration | Quantity for 100 mL |
|---|---|---|
| His-lysis buffer | (Base buffer) | 90 mL |
| 5 M imidazole | 500 mM | 10 mL |
10. 1 M Imidazole His-elution buffer
| Reagent | Final concentration | Quantity for 100 mL |
|---|---|---|
| His-lysis buffer | (Base buffer) | 80 mL |
| 5 M imidazole | 1000 mM | 20 mL |
11. Phosphate buffer (pH 7.4)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 140 mM | 8.18 g |
| KCl | 2.7 mM | 0.20 g |
| Na2HPO4 | 10 mM | 1.42 g |
| KH2PO4 | 1.8 mM | 0.24 g |
| Milli-Q water | n/a | to 1 L |
Dissolve salts in approximately 800 mL of Milli-Q water and stir until fully dissolved. Adjust pH to 7.4. Bring the final volume to 1 L with Milli-Q water. Filter using a 0.22 μm filter. Store the buffer at 4 °C for up to 1 month.
Critical: Use anhydrous Na2HPO4 (141.96 g/mol) for accurate molarity. If using hydrated forms (Na2HPO4·7H2O, MW 268.07), adjust required mass = (10 mM × 268.07 g/mol) = 2.68 g/L.
12. GST-elution buffer (pH 8.0)
| Reagent | Final concentration | Quantity for 50 mL |
|---|---|---|
| Reduced glutathione | 10 mM | 153.66 mg |
| Phosphate buffer | (Base buffer) | to 50 mL |
Dissolve in 40 mL of phosphate buffer.
Critical: GSH dissolves slowly; stir gently at room temperature for 10–15 min. Adjust pH to 8.0 with 1 M NaOH (typically requires ~1–2 mL).
Critical: GST elution is optimal at pH 8.0–8.5. Bring the final volume to 50 mL. Filter using a 0.22 μm filter. Store the buffer at 4 °C for up to 1 week (GSH oxidizes over time).
13. SEC buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 20 mM | 4.77 g |
| NaCl | 150 mM | 8.77 g |
| Glycerol (99%) | 5% (v/v) | 50 mL |
| Milli-Q water | n/a | to 1 L |
14. 10% SDS Solution
Dissolve 50 g of sodium dodecyl sulfate (SDS) in Milli-Q water to a final volume of 500 mL with stirring. Store at room temperature for up to 6 months.
Caution: Wear a mask and gloves when handling SDS powder to avoid dust inhalation.
15. 1.5 M Tris-HCl (pH 8.8) (500 mL) stock solution
Dissolve 90.85 g of Tris base (MW 121.14 g/mol) in 300 mL of Milli-Q water with stirring. Adjust pH to 8.8 using concentrated HCl (add slowly, as Tris buffering capacity is high). Top up the volume to 500 mL. Store at 4 °C for up to 1 month.
Critical: Allow the solution to cool to room temperature before final pH adjustment, as Tris pH is temperature-sensitive.
16. 1.0 M Tris-HCl (pH 6.8) (50 mL) stock solution
Dissolve 6 g of Tris base (MW 121.14 g/mol) in 40 mL of Milli-Q water with stirring. Adjust pH to 6.8 using concentrated HCl. Top up the volume to 50 mL. Store at 4 °C for up to 1 month.
Critical: Recheck pH after mixing, as small deviations can affect gel stacking.
17. 2.5 M glycine (1 L) stock solution
Dissolve 187.7 g of glycine (MW 75.07 g/mol) in Milli-Q water to a final volume of 1 L with stirring. Store at room temperature for up to 6 months.
Critical: Ensure complete dissolution, as glycine can take time to dissolve fully.
18. 10% Ammonium persulfate (APS ) (1 mL)
Dissolve 0.1 g of APS in 1 mL of Milli-Q water immediately before use.
Critical: Do not store, as APS degrades rapidly in solution. Prepare fresh each day to ensure effective polymerization.
19. SDS-PAGE running buffer (10× Tris-Glycine-SDS, pH 8.3)
| Reagent | Final concentration (10×) | Quantity or Volume |
|---|---|---|
| 1.5 M Tris-HCl (pH 8.8) | 250 mM | 166.7 mL |
| 2.5 M glycine | 1.92 M | 768 mL |
| 10% SDS | 1% (w/v) | 100 mL |
| Milli-Q water | n/a | to 1 L |
Bring the volume to 1 L with Milli-Q water and mix gently by stirring. Check pH (should be ⁓8.3). Store at room temperature for up to 6 months. To prepare 1× working buffer, dilute 100 mL of 10× with 900 mL of Milli-Q water within 1 week for optimal performance.
Critical: Do not adjust pH with NaOH, as sodium ions can interfere with electrophoresis.
20. SDS-PAGE resolving gel (12% polyacrylamide)
| Reagent | Final concentration | Quantity for 10 mL |
|---|---|---|
| 30% Acrylamide/Bis (37.5:1) | 12% | 4.0 mL |
| 1.5 M Tris-HCl (pH 8.8) | 375 mM | 2.5 mL |
| 10% SDS | 0.1% | 100 μL |
| 10% APS | 0.1% | 100 μL |
| TEMED | 0.04% | 4 μL |
| Milli-Q water | n/a | to 10 mL (3.3 mL) |
In a 50 mL conical flask, combine 3.3 mL of Milli-Q water, 2.5 mL of 1.5 M Tris-HCl (pH 8.8), 4.0 mL of 30% acrylamide/Bis, and 0.1 mL of 10% SDS. Mix gently by inversion. Then, add 0.1 mL of 10% APS and 4 μL of TEMED immediately before pouring to initiate polymerization. Pour into the gel cassette with a 5 mL micropipette, leaving ~1 cm space for the stacking gel. Overlay with isopropanol or water to ensure a flat interface. Allow polymerization for 30–45 min at room temperature.
Caution/Critical: Acrylamide is neurotoxic; handle with gloves in a fume hood. Prepare APS fresh daily. Polymerization time may vary with ambient temperature.
21. SDS-PAGE stacking gel (5% polyacrylamide)
| Reagent | Final concentration (10×) | Quantity for 3 mL |
|---|---|---|
| 30% Acrylamide/Bis (37.5:1) | 5% | 500 μL |
| 1.0 M Tris-HCl (pH 6.8) | 125 mM | 375 μL |
| 10% SDS | 0.1% | 30 μL |
| 10% APS | 0.1% | 30 μL |
| TEMED | 0.1% | 3 μL |
| Milli-Q water | n/a | to 3 mL (2.062 mL) |
In a 15 mL centrifuge tube, combine 2,062 μL of Milli-Q water, 375 μL of 1.0 M Tris-HCl (pH 6.8), 500 μL of 30% acrylamide/bis, and 30 μL of 10% SDS. Mix gently by inversion. Then, add 30 μL of 10% APS and 3 μL of TEMED immediately before pouring to initiate polymerization. Remove the isopropanol/water overlay from the polymerized resolving gel. Pour the stacking gel mixture on top with a micropipette and insert the comb. Allow polymerization for 20–30 min at room temperature.
Critical: Ensure the resolving gel is fully polymerized before pouring the stacking gel. Use fresh APS for consistent polymerization. Measure volumes precisely using calibrated micropipettes.
22. 0.5 M EDTA (pH 8.0) stock solution
Dissolve 18.61 g of disodium EDTA dihydrate in 80 mL of Milli-Q water. Stir vigorously using a magnetic stir plate.
EDTA will not dissolve fully until the pH is ~8.0. Add NaOH (either pellets or a concentrated solution, e.g., 10 M) gradually while monitoring pH with a calibrated pH meter. Typically, ~0.5–1 g of NaOH pellets or ~0.5–1 mL of 10 M NaOH is sufficient. Continue stirring until the EDTA dissolves completely.
23. 50× TAE buffer
| Reagent | Final concentration (10×) | Quantity or Volume |
|---|---|---|
| Tris base | 2 M | 242 g |
| Glacial acetic acid | 1 M | 57.1 mL |
| 0.5 M EDTA (pH 8.0) | 50 mM | 100 mL |
| Milli-Q water | n/a | to 1 L |
Dissolve 242 g of Tris base in 700 mL of Milli-Q water with stirring until fully dissolved. Add 57.1 mL of glacial acetic acid and mix thoroughly. Add 100 mL of 0.5 M EDTA (pH 8.0) and mix. Bring the volume to 1 L with Milli-Q water. Stir gently. Verify the pH is ~8.0; adjust with acetic acid. Store at room temperature for up to 6 months. To prepare 1× TAE working buffer, dilute 20 mL of 10× with 980 mL of Milli-Q water.
Critical: Tris and EDTA solutions are pH-sensitive; ensure EDTA stock is at pH 8.0.
24. 1.5% agarose gel
Prepare a flask that can hold 2–4 times the volume of agarose you are preparing. Add 100 mL of the diluted buffer (1× TAE) to the flask, followed by 1.5 g of agarose. Heat the flask in the microwave in bursts of 30 s. Swirl after each burst. Cool it down to 40 °C. Add 10 μL of goldview. Pour in the tray. Add the combs.
Laboratory supplies
1. E.Z.N.A® Cycle Pure Kit (omega BIO-TEK, catalog number: D6492-02)
2. E.Z.N.A.® Plasmid DNA Mini Kit I (omega BIO-TEK, catalog number: D6943-02)
3. Bradford Protein Assay kit (Tiangen, catalog number: PA102)
4. Ni-charged Resin FF columns (GenScript, catalog number: L00666)
5. GST-Sefinose Resin 4FF columns (Sangon, catalog number: C600031)
6. Superdex 200 Increase 10/300 GL column (Cytiva, catalog number: 28990944)
7. Amicon Ultra centrifugal filter devices 10,000 MWCO (Millipore, catalog number: UFC901024)
8. Volumetric flasks, beakers, Erlenmeyer flasks
9. Pipettes (various volumes, calibrated) and pipette tips
10. Microcentrifuge tubes and centrifuge tubes (50 mL)
11. Petri dishes
12. Spreader for plating bacteria
13. Inoculation loops
14. Toothpicks (sterile, wooden)
15. Syringes
16. Syringe filters (0.22 μm, 0.45 μm)
Equipment
1. Thermocycler for PCR (Monad, model: Arhat 96)
2. Microcentrifuge (refrigerated) (Hunan Kecheng Instrument Equipment Co., model: H1-16KR)
3. Benchtop centrifuge (refrigerated) (Hunan Kecheng Instrument Equipment Co., model: H3-16KR)
4. Ultracentrifuge (refrigerated) (Beckman Coulter, model: optima xpn-100)
5. French Press (JNBIO, model: JNBIO-minipro)
6. NanoDrop (YoMin, model: Unano-1000)
7. Gel electrophoresis apparatus HET (Yeasen, catalog number: 80230ES01)
8. Power supply for agarose and SDS-PAGE EpPlus (Yeasen, catalog number: 80311ES05)
9. SDS-PAGE electrophoresis apparatus PET2 (Yeasen, catalog number: 80212ES05)
10. Gel documentation system (Clinx, model: GenoSens 2000)
11. pH meter (calibrated)
12. Magnetic stirrer and stir bars (Yeasen, model: MS400)
13. Rotary shaker/incubator (37 °C and 18 °C temperature control) (BLabotery, ZQPL-200)
14. Chromatography system (ÄKTA pure, catalog number: 21906560)
15. Autoclave (HefeiME, model: LX-B75L)
16. Static incubator (37 °C) (BLabotery, SPL-250)
17. Water bath
Software and datasets
1. SnapGene version 3.2.1 software was used for sequence analysis, primer design, PCR, and cloning stimulation
Procedure
A. Construction of E. coli protein expression plasmids
1. Design the Cas genes amplification primers as follows (Table 1):
Table 1. Optimized primer design for His- and GST-tagged Cas6 genes for cloning and expression
| Primer Name: Sequence (5' to 3') | Notes |
|---|---|
| His-Cas6-1-F: actttaagaaggagatataccatgcctcaggccgtggtcc | Underline = Cas6-1 gene sequence. Includes start codon (ATG), Locus (pTT27 megaplasmid): TTP0204. Italic = Overlap for pET28a-HRV3C of NcoI cut site. (Tm=65 °C) |
| His-Cas6-1-R: agtggtggtggtggtggtgctcgagggggactgccaggcc | Underline = Cas6-1 gene sequence, stop codon REMOVED. Italic = Overlap for pET28a-HRV3C of XhoI cut site + 6xHis. (Tm = 57 °C) |
| His-Cas6-2-F: actttaagaaggagatataccgtggtcctcgccgccttgg | Underline = Cas6-2 gene sequence. Includes start codon (ATG), Locus (chromosome): TTC1927. Italic = Overlap for pET28a-HRV3C of NcoI cut site. (Tm = 69 °C) |
| His-Cas6-2-R: agtggtggtggtggtggtgctcgaggcactttccgcccgggc | Underline = Cas6-2 gene sequence, stop codon REMOVED. Italic = Overlap for pET28a-HRV3C of XhoI cut site + 6xHis. (Tm = 65 °C) |
| N-GST-Cas6-1-F: tctgttccaggggcccctgggaatgcctcaggccgtggtcc | Underline = Cas6-1 gene sequence. Italic = Overlap for pGEX-6P-1 of BamHI cut site, in frame with HRV3C. (Tm = 61 °C) |
| N-GST-Cas6-1-R: cacgatgcggccgctcgagtcagagggggactgccaggcc | Underline = Cas6-1 gene sequence, Stop codon REMOVED. Italic = Overlap for pGEX-6P-1 of SalI cut site, in frame with HRV3C. Critical: Make sure both enzymes are in frame with HRV3C seq. (Tm = 57 °C) |
| C-GST-Cas6-2-F: tcacacaggaaacagtattcatgtggtcctcgccgccttgg | Underline = Cas6-2 gene sequence. Includes start codon (GTG). Italic = Overlap for pGEX-6P-1 for Gibson Assembly. (Tm = 64 °C) |
| C-GST-Cas6-2-R: taacctagtataggggacatggcactttccgcccgggc | Underline = Cas6-2 gene sequence, Stop codon REMOVED. Italic = Overlap for pGEX-6P-1 for Gibson Assembly. Critical: Ensure 20–40 bp overlaps with the linearized vector. (Tm = 64 °C) |
| pGEX-linear-F: atgtcccctatactaggttattgg | For linearizing pGEX-6P-1. (Tm = 55 °C) |
| pGEX-linear-R: gaatactgtttcctgtgtgaaattg | For linearizing pGEX-6P-1. (Tm = 56 °C) |
| pET28a-exam-F: gatggcgcccaacagtcc | To check and sequence (optional) the His tagged Cas6 gene insertion. Set the annealing temperature to 53 °C and the extension time to 1 min and 30 s. (Cas6-1~1,200 bp, Cas6-2~1,150 bp) |
| pET28a-exam-R: caaaaaacccctcaagacccg | |
| pGEX-exam-F: gggctggcaagccacgtttggtg | To check and sequence (optional) the N-GST tagged Cas6-1 gene insertion. Set the annealing temperature to 61 °C and the extension time to 1 min. (900 bp) |
| pGEX-exam-R: ccgggagctgcatgtgtcagagg | |
| pGEX-exam-C-F: ggcgtcaggcagccatcg | Use with pGEX-exam-R to check and sequence (optional) the C-GST tagged Cas6-2 gene insertion. Set the annealing temperature to 58 °C and the extension time to 2 min. (1,743 bp) |
2. Perform five separate PCR reactions:
a. PCR 1: Amplify the His-tag cas6-1 gene using the His-Cas6-1-F and His-Cas6-1-R primers.
b. PCR 2: Amplify the His-tag cas6-2 gene using the His-Cas6-2-F and His-Cas6-2-R primers.
c. PCR 3: Amplify the N-GST-tag cas6-1 gene using the N-GST-Cas6-1-F and N-GST-Cas6-1-R primers.
d. PCR 4: Amplify the C-GST-tag cas6-2 gene using the C-GST-Cas6-2-F and C-GST-Cas6-2-R primers.
e. PCR5: Linearize pGEX-6P-1 plasmid using the pGEX-linear-F and pGEX-linear-R primers.
3. Set up the PCR reactions as follows (Table 2):
Table 2. Streamlined PCR reaction setup for high-fidelity amplification
| Component | Volume per reaction |
|---|---|
| Nuclease-free water | To 50 µL |
| 2 × Phanta Max buffer | 25 μL |
| dNTP mix (10 mM each) | 1 μL |
| 10 µM F primer | 2 μL |
| 10 µM R primer | 2 μL |
| Template DNAa | x μL |
| Phanta Max Super-Fidelity DNA polymerase | 1 μL |
a.In a 50 μL system, the recommended template usage is 50–400 ng of genomic DNA of T. thermophilus HB27 and 30 ng for plasmid pGEX-6P-1.
4. Optimize thermocycler conditions as follows (Table 3):
Table 3. Optimized thermal cycling program for PCR amplification
| Step | Temperature (°C) | Time | Cycles |
| Initial denaturation | 95 | 5 min | 1 |
| Denaturation | 95 | 15 s | 35 |
| Primer annealing | Tm-5 | 15 s | |
| Extension | 72 | 1 min per kb | |
| Final extension | 72 | 5 min | 1 |
| Hold | 4 | ∞ |
Critical: The annealing temperature is critical and MUST be optimized for each primer pair. Start with a temperature of 5 °C below the lowest calculated Tm of the primer pair.
Note: To maximize PCR product yields, perform three programs and use the following conditions for the primer annealing and extension steps:
PCR1 and PCR3: Set the annealing temperature to 52 °C and the extension time to 1 min.
PCR2 and PCR4: Set the annealing temperature to 60 °C and the extension time to 1 min.
PCR5: Set the annealing temperature to 50 °C and the extension time to 5 min.
5. Digest pET28a-HRV3C with NcoI and XhoI (for His-tag Cas6-1,2).
6. Digest pGEX-6P-1with BamHI and SalI (for N-GST-tag cas6-1).
7. Prepare the restriction enzyme digestion mixture as follows: 600 ng of plasmid, 5 μL of buffer, 1 μL of enzyme + 1 μL of enzyme, top up the volume to 50 µL with nuclease-free water. Gently mix and briefly centrifuge. Reaction conditions: incubate at 37 °C overnight.
8. Linearize the circular pGEX-6P-1 backbone using a PCR reaction mixture based on pGEX-linear-F and pGEX-linear-R primers (PCR5) as in step A3 and A4 for C-GST-Cas6-2 Gibson assembly.
9. Add 1 μL of DpnI enzyme to the PCR product of step A8 to digest and remove the methylated circular plasmid. Incubate at 37 °C for 2 h.
10. Quantify the product of steps A4, A5, A6, and A9 using 1.5% agarose gel to check the right products [linear plasmid, cas amplicons, negative control (without restriction enzyme digestion)].
Note: PCR1 amplicon length is 795 bp. PCR2 amplicon length is 720 bp. PCR3 amplicon length is 795 bp. PCR4 amplicon length is 720 bp. PCR5 amplicon length is 4,984 bp. Store the DNA at 4 °C until the whole process is completed.
11. Purify the DNA of PCR Cas amplicons (step A4) and digested plasmids (steps A5, A6, and A9) using the E.Z.N.A.® Cycle Pure kit. Follow the manufacturer's instructions.
Critical: Elute the purified DNA in ddH2O or other low EDTA buffers. Accurate DNA quantification is essential for optimal ligation/assembly ratios (measure the quantity of the purified DNA using Nanodrop).
12. Set up seamless assembly to the purified PCR Cas genes (step A4) with linear purified plasmid (step A11) using ABclonal MultiF Seamless Assembly as follows (Table 4):
Table 4. Gibson assembly reaction setup for seamless Cas6 genes cloning
| Component | Final concentration |
|---|---|
| Liner plasmid | 1× (e.g., 100 ng 1 µL) |
| Cas 6 amplicon | 5× (e.g., 100 ng 5 µL) |
| 2× MultiF Seamless Assembly Mix | 10 µL |
| Nuclease-free water | To 20 μL |
Critical: Refer to Table 1 to verify the Cas-purified amplicon and its corresponding plasmid.
13. Incubate the Gibson assembly mixture (step A12) at 50 °C in a dry thermal bath for 20 min.
14. Thaw 50 μL of competent E. coli DH5α cells on ice for 5 min (four 1.5 mL microcentrifuge tubes, one for each gene construction).
Critical: Handle competent cells very gently. Do not vortex.
15. Add 10 µL of the assembly reaction (step A13) to competent cells.
16. Mix gently by pipetting and incubate on ice for 25–30 min.
Critical: Avoid touching the bottom of the tube with warm fingers.
17. Heat-shock the cells at 42 °C for 45 s.
18. Immediately place the cells back on ice for 2 min.
19. Add 950 µL of LB broth (without antibiotics) to the cells.
20. Incubate at 37 °C with shaking (200–250 rpm) for 1 h.
21. Centrifuge at 4,000× g for 1 min at room temperature.
22. Discard 700 μL of the supernatant and suspend the pellet in the residual supernatant (200 μL).
23. Plate 50–100 µL of the transformation mixture onto selection LB agar.
Critical: The antibiotic selectable marker for pET28a-HRV3C is 30 μg/mL kanamycin, while pGEX-6P-1 is 100 µg/mL ampicillin.
24. Incubate the plates upside down at 37 °C overnight (16–18 h).
25. Prepare a PCR master mix as described in step A3, using the exam primer pairs mentioned in Table 1
26. Mark several (8–16) colonies from the transformation plates.
27. Using sterile toothpicks, pick colonies and resuspend each in the PCR mixture.
Critical: Do not mix the colonies during sampling and keep sterilized conditions, since you will be back to pick up the PCR-positive colony. Do not pick the whole colony for PCR.
28. Run the PCR as illustrated in step A4 with annealing temperature and extension time mentioned in Table 1 (negative control: water or the original plasmid).
29. Analyze the PCR products by agarose gel electrophoresis (refer to Table 1 for band size).
30. Isolate colonies that show a band of the expected size.
31. Inoculate 5 mL of selection LB broth with the PCR-verified colony and incubate for 8 h at 37 °C with shaking (200–250 rpm).
Critical: Do not forget to submit samples to sequencing using the exam primers mentioned in Table 1. Save a copy at -80 °C.
32. Analyze the sequencing results carefully. Verify that the cas6-1 or cas6-2 gene is in-frame with the affinity tag and there are no mutations or frameshifts. Any strains with errors MUST be discarded.
33. Extract the plasmid from the verified cell culture using the E.Z.N.A.® Plasmid DNA Mini Kit I. Follow the manufacturer's instructions.
34. Quantify and qualify the plasmid concentration using NanoDrop and 1.5% agarose gel.
35. Repeat steps A14–31 using E. coli BL21(DE3). No need for resequencing.
B. Protein expression
1. Use the verified 5 mL culture from step A35 as a starter culture to scale up the culture into 1 L of selection LB broth at a 1:200 dilution.
2. Incubate the culture overnight at 37 °C with shaking (220 rpm).
3. Monitor the culture OD. When the OD600 reaches 0.6–0.8 (mid-log phase), add IPTG to a final concentration of 0.5 mM.
Critical: Do not overgrow the culture before induction, as this can lead to inclusion body formation.
4. Reduce the incubation temperature to 18 °C and continue shaking (220 rpm) for 18 h.
Critical: Lowering the temperature often improves the solubility of recombinant proteins.
5. Harvest the cells by centrifugation at 7,000× g for 10 min at 4 °C. Discard the supernatant.
Critical: Keep the cell pellet on ice from this point forward to minimize protein degradation.
Pause point: Store the cell pellet at -80 °C or proceed directly to cell lysis.
C. Cell lysis and protein clarification
1. Resuspend the cell pellet in 40 mL of ice-cold lysis buffer (use His-lysis buffer for His-tag Cas6 and phosphate buffer for GST-tag Cas6).
Critical: Ensure complete resuspension of the cell pellet by pipetting, since incomplete resuspension will lead to reduced protein yield. Do not use sonication to resuspend the cells.
2. Add PMSF to the cell suspension to a final concentration of 1 mM to inhibit protease activity.
3. Lyse the cells using a French press at 1,000 psi. Pass the lysate through the French press 3–5 times or until the lysate becomes significantly less viscous and more translucent. Keep the lysate on ice during the entire process.
Critical: Monitor the pressure and keep the French press cell cold at 4 °C; overheating can denature the protein.
4. Centrifuge the lysate at 15,000× g for 30 min at 4 °C to remove cell debris.
5. Collect the supernatant using a syringe without disturbing the pellet.
6. Filter the collected supernatant through a 0.45 µm filter.
Critical: If the supernatant is still cloudy, repeat the centrifugation step.
D. Affinity chromatography (His-tag Cas6 proteins)
1. Pack Ni-Resin FF into a gravity-flow chromatography column (e.g., 5 mL column, 1 cm diameter). Allow the resin to settle by gravity (typically, 1 mL of bed volume per 1 L of culture). Ensure no air bubbles are trapped.
2. Wash with 5 mL of nuclease-free water (5 bed volumes) at 1 mL/min to remove ethanol.
Critical: Use 20% ethanol to wash or store the Ni-Resin after use (after completing steps D3–7 for His-tagged Cas 6-1 and before repeating the same steps for His-tagged Cas 6-2).
3. Flow 10–20 mL of His-washing buffer (10–20 bed volumes) through the column at 1 mL/min using gravity.
4. Load the crude protein extract (the filtrate supernatant from step C6) onto the equilibrated Ni-Resin column at a flow rate of 1–2 mL/min.
Critical: Monitor the flow-through with the Bradford Protein Assay kit and repeat loading of the collected flow-through of the Ni-Resin column until the flow-through sample reaction remains reddish-brown (baseline color).
5. Wash the column with His-washing buffer to remove weakly bound contaminants.
Critical: Monitor the wash fractions with the Bradford Protein Assay kit and continue washing until it returns to baseline.
6. Elute the His-tagged Cas6 proteins with a stepwise gradient of imidazole (100 mM, 200 mM, 500 mM, and 1,000 mM).
Critical: Monitor each elute fraction with the Bradford Protein Assay kit and continue eluting until it returns to baseline.
7. Collect fractions (e.g., 1–2 mL each).
Critical: A stepwise gradient is generally preferred over a linear gradient for initial purification, as it helps to identify the optimal imidazole concentration for elution and minimizes co-elution of contaminants.
8. Analyze the fractions by SDS-PAGE to identify those containing the His-tagged Cas6 protein (Figure 1A, B).
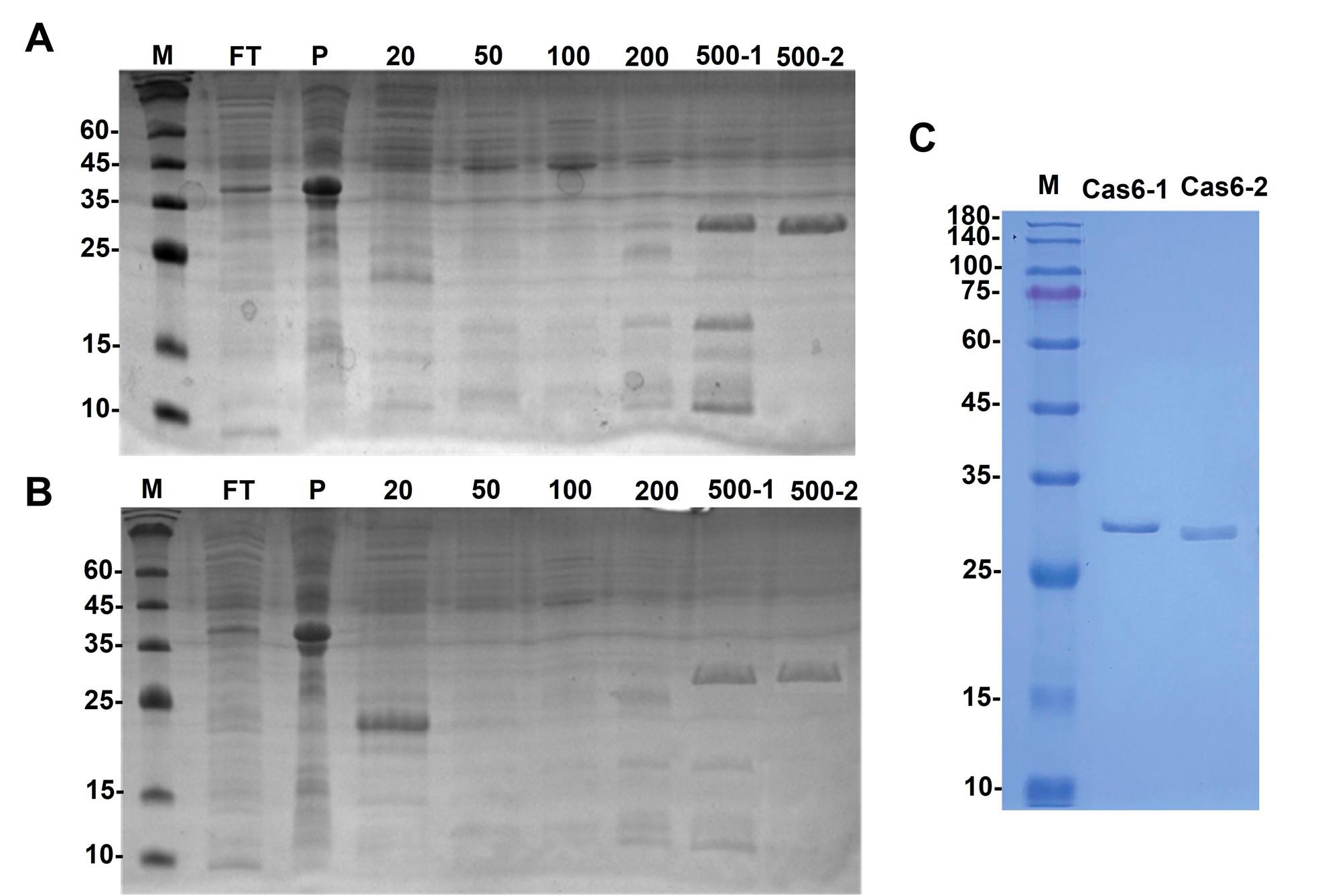
Figure 1. Coomassie-stained SDS-PAGE gel of the expressed His-tagged Cas6 proteins. (A) His-tagged Cas6-1 protein with MW: 29.9 kDa shows a clear band in the 500-2 elution fraction of Ni-Resin FF affinity chromatography. (B) His-tagged Cas6-2 protein with MW: 27.3 kDa shows a clear band in the 500-1, 500-2 elution fraction of Ni-Resin FF affinity chromatography. (C) Polished His-tagged Cas6 proteins. Lane M: 12% SDS-PAGE (Tri-Gly) marker.
E. Affinity chromatography (GST-tag Cas6 proteins)
1. Pack GST-Sefinose Resin 4FF into a gravity-flow chromatography column (e.g., 5 mL column, 1 cm diameter). Allow resin to settle by gravity (typically, 1 mL of bed volume per 1 L of culture). Ensure no air bubbles are trapped.
2. Wash with 5 mL of nuclease-free water (5 bed volumes) at 1 mL/min to remove ethanol.
Critical: Use 20% ethanol to wash or store the GST-Sefinose Resin after use (after completing steps E3–6 for N-GST tagged Cas 6-1 and prior to repeating the same steps for C-GST tagged Cas 6-2).
3. Flow 10–20 mL of phosphate buffer (10–20 bed volumes) through the column at 1 mL/min using gravity.
4. Load the crude protein extract (step C6) onto the equilibrated column at a flow rate of 1–2 mL/min.
Critical: Monitor the flow-through with Bradford protein assay kit. Repeatedly sample the collected flow-through of GST-Resin column until the reaction color of the flow-through samples remains reddish-brown (baseline color).
5. Wash the column with phosphate buffer.
Critical: Monitor the wash fractions with the Bradford Protein Assay kit and continue washing until it returns to baseline.
6. Elute the GST-tagged protein with GST-elution buffer. Collect fractions (e.g., 1–2 mL each).
Critical: Monitor elute fractions with the Bradford Protein Assay kit and continue eluting until it returns to baseline.
Critical: Freshly prepared glutathione is crucial for efficient elusion, because oxidized glutathione will not elute the protein effectively.
7. Analyze the fractions by SDS-PAGE to identify those containing the GST-tagged protein (Figure 5b, c in [3]).
F. Concentration of His-tag and GST-tag Cas proteins
1. Pool the fractions containing the Cas6 proteins from steps D7 and E6 (as determined by SDS-PAGE, steps D8 and E7) and filter through a 0.45 μm syringe filter to avoid filter membrane polarization.
Critical: Do not pool different Cas proteins together. Keep each type in a separate pool.
2. Concentrate them using an Amicon Ultra-15 centrifugal filter with 10,000 MWCO, and ensure the total loaded volume does not exceed 15 mL (maximum capacity).
Critical: Select a molecular weight cut-off (MWCO) at least two-fold smaller than the target protein’s molecular weight to prevent protein loss (Cas6 proteins are ~30 kDa). Monitor the concentration factor using volume gradations to avoid over-concentration, which may cause the Cas6 protein solution to become viscous and lead to precipitation. Visually inspect the sample against a light source during centrifuge pauses; slight cloudiness or turbidity may indicate impending precipitation. Cease concentration immediately if turbidity appears or if filtration slows significantly. Do not allow the filter membrane to dry and load the rest of the protein into the filter cup. To prevent filter membrane polarization, pre-filter the sample using a 0.45 μm membrane before loading.
Optional: Use a Bradford assay to track protein concentration.
3. Centrifuge at 4,000× g for 10–15 min intervals at 4 °C.
Critical: Place the device into the centrifuge with one membrane panel parallel to the top of the centrifuge, and the other membrane toward the center of the centrifuge. The panels should not be perpendicular to the outer edge of the centrifuge. Keep the filtrate on ice until the end of the assay to avoid any loss of the molecule of interest.
4. Pause every 10–15 min to check the remaining volume (using tube gradations) and inspect for viscosity or cloudiness.
Critical: Gently mix the retentate by pipetting to resuspend proteins and prevent membrane clogging.
5. Continue steps F1–4 until the volume is reduced to ~1–2 mL (5–10-fold concentration, targeting ~0.5–5 mg/mL).
6. Collect the concentrated Cas6 proteins from the membrane filter cup using the pipette tip.
G. Size-exclusion chromatography (polishing)
1. Connect the SuperdexTM 200 Increase 10/300 GL column to an FPLC system (ÄKTA pure).
2. Prime the system with nuclease-free water or SEC buffer to remove air bubbles from the tubing.
3. Pump the SEC buffer (see Recipe 13) through the column at a flow rate of 0.5 mL/min.
Critical: Use 50–60 mL of SEC buffer (2–2.5 column volumes, as the column bed volume is ~24 mL) to equilibrate.
4. Monitor UV absorbance at 280 nm (for protein detection) and conductivity (for salt content, ~15–20 mS/cm for 150 mM NaCl) using the FPLC system’s detectors.
Critical: Equilibration is complete when the UV baseline is stable (flat, <10 mAU variation) and conductivity matches the SEC buffer. If the column was stored in 20% ethanol, first flush with 2 column volumes (48 mL) of nuclease-free water at 0.5 mL/min to remove ethanol before equilibrating with SEC buffer.
5. Filter the concentrated Cas6 samples (0.5–1 mL, ~1–5 mg/mL from step F6) through a 0.45 μm syringe filter to prevent column clogging.
6. Inject the concentrated Cas6 protein obtained in step G5 into the column at 0.5 mL/min (maximum volume: 2% of the column volume).
Critical: Ensure the protein sample is clear and free of aggregates before loading by filtering the sample through a 0.45 µm filter (step F5).
7. Elute the protein with 1–1.5 column volumes (24–36 mL) of SEC buffer at a flow rate of 0.5 mL/min. Collect fractions (e.g., 0.5–1 mL each) and monitor the A280.
8. Monitor UV absorbance at 280 nm to identify Cas6 protein peaks (~30 kDa His-tag elutes ~14–16 mL; ~56 kDa GST-tag elutes ~12–14 mL, based on column calibration).
9. Analyze the fractions by SDS-PAGE to identify fractions containing the pure Cas6 protein (Figure 1C).
H. Final concentration and storage
1. Repeat steps F1–6 with the fractions containing purified protein (as determined by SDS-PAGE) to concentrate the protein to the desired final concentration.
2. Aliquot the concentrated pure Cas6 protein into small volumes to avoid repeated freeze-thaw cycles. Flash-freeze the aliquots in liquid nitrogen and store at -80 °C.
Critical: Avoid repeated freeze-thaw cycles, as this can denature the protein.
Validation of protocol
This protocol has been used and validated in the following research article:
Wei et al. [3]. A type III-associated Cas6 functions as a negative regulator of type I-B CRISPR-Cas system in Thermus thermophilus. Commun Biol (Figure 4b, c; Figure 5c, d).
General notes and troubleshooting
Troubleshooting
Problem 1: Plasmid linearization error.
Possible cause: Inappropriate incubation time of restriction enzyme digestion. Inactive restriction enzymes due to storage issues.
Solution: Optimize the enzyme digestion system by increasing the amount of restriction endonuclease (e.g., 2 µL per 600 ng plasmid) or extending the incubation time to the whole day to reduce background from uncut plasmid. In a 50 μL PCR reaction, adjust the circular plasmid template to ≤30 ng to minimize the false positive rate.
Problem 2: No colonies or bacterial lawn on transformation plates.
Possible cause: The antibiotic concentration is not correct or expired. Transformation failure.
Solution: Verify antibiotic concentration (e.g., 50 µg/mL kanamycin for pET28). If expired, antibiotics may fail, leading to a bacterial lawn due to the growth of non-transformed cells. Prepare fresh antibiotic stocks. Test competent cell viability and transformation efficiency with a control plasmid (e.g., pUC19, 1 ng, expecting ~108 CFU/µg). Repeat transformation if necessary, ensuring proper heat shock (42 °C, 30 s) and recovery (1 h, 37 °C).
Problem 3: False-positive colonies.
Possible cause: Incomplete vector linearization. Assembly failure. Inappropriate PCR setting.
Solution: Purify the linearized vector and PCR product prior to the assembly reaction (gel recovery for digested vector). Optimize the molar ratio between insert and vector as suggested in the manufacturer's instructions. If both the target and control bands are missing, it is recommended to optimize the PCR reaction program.
Problem 4: PCR amplification failure.
Possible cause: High GC content in the T. thermophilus template. Suboptimal annealing temperature or reaction components.
Solution: For T. thermophilus HB27 DNA (~69% GC), add 5%–10% DMSO or 1 M betaine to the PCR mix to relax secondary structures. Optimize the annealing temperature as mentioned in the procedure. Use another high-fidelity polymerase (e.g., Phusion). Run a gradient PCR if bands are absent.
Problem 5: Low protein expression.
Possible cause: Inefficient codon usage. Low inducer concentration. Premature stop codons in the gene sequence.
Solution: Check the codon sequence used for E. coli. Increase IPTG concentration. Try different induction temperatures (e.g., 16 °C) or verify the incubator temperature.
Problem 6: French press blockage.
Possible cause: Cell clumps were not removed prior to processing, and clogging of the valve occurs.
Solution: The unit must be cleaned thoroughly before processing can continue. Make sure that the cell pellet is completely resuspended in the lysis buffer. Pay attention to the equipment's sound tone.
Problem 7: Protein is in inclusion bodies.
Possible cause: High expression rate. protein misfolding.
Solution: Reduce induction temperature. Lower IPTG concentration. Use the recommended E. coli strain in this protocol.
Problem 8: Protein does not bind to the affinity resin.
Possible cause: Incorrect buffer pH. Resin is degraded.
Solution: Check buffer pH and composition. Use fresh resin. Ensure the tag is present by western blot.
Problem 9: Protein elutes in the washing step.
Possible cause: Wash buffer contains too high imidazole concentration.
Solution: Reduce imidazole concentration in wash buffer.
Problem 10: Protein aggregates during purification.
Possible cause: High protein concentration. Buffer conditions.
Solution: Lower protein concentration prior to size-exclusion chromatography (SEC). Adjust buffer pH and ionic strength, or include additives [e.g., TCEP (to prevent oxidation) or DDM (to enhance membrane protein solubility)] to minimize aggregation.
Problem 11: Low protein purity after SEC.
Possible cause: Overloading the column. Insufficient resolution.
Solution: Reduce the sample volume loaded onto the SEC column. Use a longer column.
Acknowledgments
Resources, Methodology, Investigation, Writing–original draft, Reviewing & Editing, J.W.W. & M.M.; Conceptualization, Supervision, Proofreading, Funding acquisition, Project administration, Resources, Reviewing, Y.J.L. This research was supported by the National Key Research and Development Program of China (2022YFA0912200), National Natural Science Foundation of China (32170096), and Key Research and Development Project of Hubei Province (2023BBB025). This protocol has been used and validated in Wei et al. [3]. This protocol is adapted from Han et al., Li et al., and Liu et al. [7,11,12]. The graphical abstract was created in BioRender. MOHAMED, M. (2025) https://BioRender.com/7f251wj
Competing interests
The authors declare no conflicts of interest.
References
- Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D. A. and Horvath, P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315(5819): 1709–1712. https://doi.org/10.1126/science.1138140
- Marraffini, L. A. (2015). CRISPR-Cas immunity in prokaryotes. Nature. 526(7571): 55–61. https://doi.org/10.1038/nature15386
- Wei, J., Shao, Y., Liang, Y., Bu, X., Zhou, W., Liang, Y. and Li, Y. (2025). A type III-associated Cas6 functions as a negative regulator of type I-B CRISPR-Cas system in Thermus thermophilus. Commun Biol. 8(1): 793. https://doi.org/10.1038/s42003-025-08223-4
- Doudna, J. A. and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science. 346(6213): e1258096. https://doi.org/10.1126/science.1258096
- Carte, J., Wang, R., Li, H., Terns, R. M. and Terns, M. P. (2008). Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22(24): 3489–3496. https://doi.org/10.1101/gad.1742908
- Haurwitz, R. E., Jinek, M., Wiedenheft, B., Zhou, K. and Doudna, J. A. (2010). Sequence- and Structure-Specific RNA Processing by a CRISPR Endonuclease. Science. 329(5997): 1355–1358. https://doi.org/10.1126/science.1192272
- Li, Y., Pan, S., Zhang, Y., Ren, M., Feng, M., Peng, N., Chen, L., Liang, Y. X. and She, Q. (2015). Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 44(4): e34–e34. https://doi.org/10.1093/nar/gkv1044
- Agari, Y., Sakamoto, K., Tamakoshi, M., Oshima, T., Kuramitsu, S. and Shinkai, A. (2010). Transcription Profile of Thermus thermophilus CRISPR Systems after Phage Infection. J Mol Biol. 395(2): 270–281. https://doi.org/10.1016/j.jmb.2009.10.057
- Staals, R. H. J., Agari, Y., Maki-Yonekura, S., Zhu, Y., Taylor, D. W., van Duijn, E., Barendregt, A., Vlot, M., Koehorst, J. J., Sakamoto, K., et al. (2013). Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 52(1): 135–145. https://doi.org/10.1016/j.molcel.2013.09.013
- Han, W., Li, Y., Deng, L., Feng, M., Peng, W., Hallstrøm, S., Zhang, J., Peng, N., Liang, Y. X., White, M. F., et al. (2016). A type III-B CRISPR-Cas effector complex mediating massive target DNA destruction. Nucleic Acids Res. 45(4): 1983–1993. https://doi.org/10.1093/nar/gkw1274
- Liu, T., Li, Y., Wang, X., Ye, Q., Li, H., Liang, Y., She, Q. and Peng, N. (2015). Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 43(2): 1044–1055. https://doi.org/10.1093/nar/gku1383
Article Information
Publication history
Received: Apr 14, 2025
Accepted: Jun 16, 2025
Available online: Jul 9, 2025
Published: Jul 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Wei, J., Motawaa, M. and Li, Y. (2025). Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification. Bio-protocol 15(14): e5382. DOI: 10.21769/BioProtoc.5382.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








