- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rapid and Multiplex Diagnosis of Malaria Using Chelex-100 Extraction and LAMP-MS Assay
Published: Vol 15, Iss 13, Jul 5, 2025 DOI: 10.21769/BioProtoc.5375 Views: 1829
Reviewed by: Ivonne SehringManousos E. KambourisAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Rapid FRET Real-Time PCR Protocol for Simultaneous Quantitative Detection and Discrimination of Human Plasmodium Parasites
Renate Schneider [...] Julia Walochnik
Apr 5, 2022 2362 Views
Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking
Yinwei Gu [...] Haixing Li
Dec 5, 2025 933 Views
Abstract
Malaria remains a major public health threat, especially in tropical and subtropical regions. Accurate and rapid diagnosis is essential for effective disease management and control, yet conventional malaria diagnostics, including blood smear microscopy using Giemsa staining, PCR, and rapid diagnostic tests (RDTs), are limited by the need for trained personnel, reliance on laboratory infrastructure, and reduced sensitivity at low parasite densities, respectively. This protocol details an innovative, rapid, and economical diagnostic platform combining a simplified Chelex-100 resin-based nucleic acid extraction method with a multiplex loop-mediated isothermal amplification microscanner (LAMP-MS) assay. The malaria diagnostic platform enables simultaneous detection of Plasmodium falciparum (Pf), Plasmodium vivax (Pv), pan-malaria (Pan), and an internal control (IC) within 40 min, from DNA extraction to result interpretation. It demonstrates sensitivity and specificity comparable to traditional PCR-based diagnostics, making it a practical and scalable solution for use in resource-constrained environments.
Key features
• Low-cost and simple DNA extraction using Chelex-100 resin.
• Multiplex detection of Pan malaria, Pf, Pv, and IC using microchip chambers.
• Visual endpoint detection using a microscope or a microscanner.
• Field-deployable with minimal equipment.
Keywords: MalariaGraphical overview
Schematic illustration of the malaria Pan/Pf/Pv/IC loop-mediated isothermal amplification microscanner (LAMP-MS) assay
Background
Malaria continues to be a major global public health challenge, particularly in tropical and subtropical regions [1]. In 2020 alone, an estimated 241 million cases and over 627,000 deaths were reported, with the majority of fatalities occurring among children under five in sub-Saharan Africa [2,3]. Early and accurate diagnosis is critical not only for initiating timely treatment but also for controlling disease transmission and reducing associated mortality and morbidity [4].
Conventional malaria diagnostic methods, such as blood smear microscopy using Giemsa staining, rapid diagnostic tests (RDTs), and polymerase chain reaction (PCR), each present distinct strengths and limitations. Blood smear microscopy is cost-effective and widely accessible but requires trained personnel and is subject to inter-operator variability [5]. PCR offers high sensitivity and specificity but depends on advanced laboratory infrastructure and skilled technicians, making it impractical in many low-resource settings [6,7]. RDTs, though widely adopted for their simplicity and speed, may exhibit reduced sensitivity in cases of low parasite density [8]. To overcome these limitations, loop-mediated isothermal amplification (LAMP) has emerged as a promising alternative. LAMP employs three pairs of primers: inner primers (FIP/BIP), outer primers (F3/B3), and loop primers (LoopF/LoopB). The inner and outer primers initiate strand displacement amplification, while the loop primers enhance the reaction speed by promoting faster cycling. LAMP offers sensitivity and specificity comparable to PCR while requiring only minimal equipment, making it highly suitable for field-based applications in endemic areas [9–11].
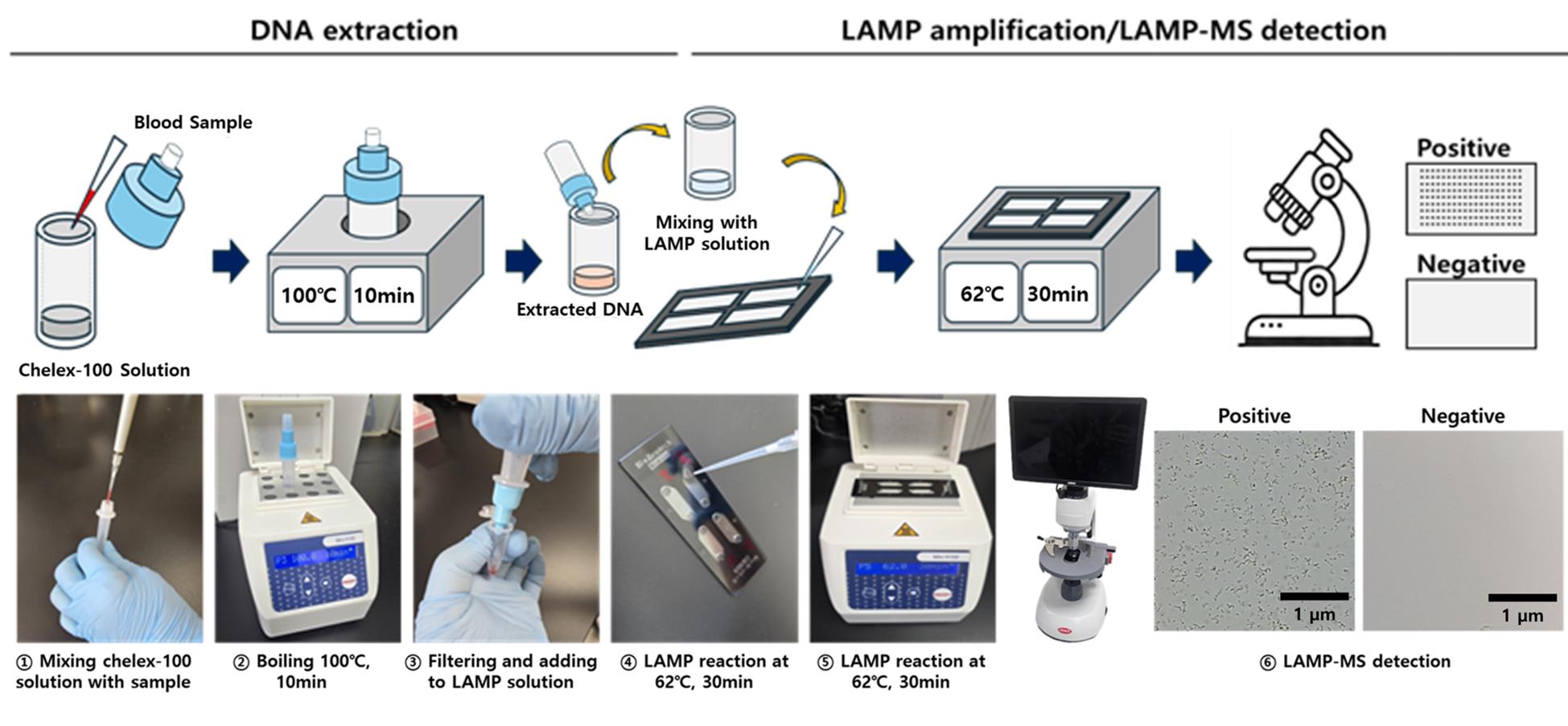
Building on this potential, our research group has developed a novel diagnostic platform that integrates a Chelex-100/boiling-based DNA extraction method with a microchip-based LAMP-MS (microscanner) assay [12]. As illustrated in the Graphical overview, the LAMP-MS workflow begins with heat-based DNA extraction using Chelex-100, followed by multiplexed LAMP reaction in a microchip, and visual endpoint detection under a microscope. This system allows for rapid, multiplex detection of Plasmodium falciparum, P. vivax, and pan-malarial targets through visually interpretable results without the need for complex instrumentation.
This protocol presents a practical and scalable approach that combines Chelex-100 resin-based nucleic acid extraction with a microchip-integrated LAMP-MS assay, enabling simultaneous detection of multiple malaria species. Its simplicity, cost-effectiveness, and rapid visual readout make this platform especially well-suited for resource-limited settings [12].
Materials and reagents
Biological materials
Malaria residual blood samples were collected from patients suspected of malaria infection at Korea University Guro Hospital between 2013 and 2022. Whole blood samples were collected from patients using standard venipuncture techniques into EDTA-containing tubes, gently mixed, and stored at 4 °C until further processing. After routine diagnostic testing, they were stored at -80 °C. Malaria infection was confirmed through various methods, including Giemsa staining, blood smear microscopy, rapid antigen tests, and PCR tests. Procedures involving clinical samples, including handling and nucleic acid extraction, were conducted under Biosafety Level 2 (BSL-2) conditions. This study was approved by the Medical Ethics Committee of Korea University’s Guro Hospital (2021GR0337).
1. 80 P. falciparum clinical samples
2. 80 P. vivax clinical samples
3. 100 negative clinical samples
4. Zika virus, Chikungunya virus, and Japanese encephalitis virus, provided by the Korea Disease Control and Prevention Agency (KDCA)
Reagents
1. Chelex-100 resin (Bio-Rad, catalog number: 142-1253)
2. Tris-EDTA buffer solution (Sigma-Aldrich, catalog number: T9285)
3. Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 10010-023)
Solutions
1. LAMP primer mix (see Recipes)
Recipes
1. LAMP primer mix
a. Prepare primer solutions separately for each target (Pan, Pf, Pv, IC) at the following final concentrations:
i. 4 μM of each outer primer (F3 and B3)
ii. 32 μM of each inner primer (FIP and BIP)
iii. 10 μM of each loop primer (FLP and BLP)
b. Each primer set solution (Pan, Pf, Pv, IC) is prepared from 100 μM stock solutions by mixing 4 μL each of F3 and B3, 32 μL each of FIP and BIP, and 10 μL each of FLP and BLP, followed by the addition of 18 μL of nuclease-free water (Table 1).
Table 1. Multi-malaria pan/Pf/Pv/IC LAMP-MS primer sets used in this study
| Target | Name | Sequence (5′-3′) | Length (mer) |
| Malaria pan (18S rRNA gene) | Malaria pan F3 | GTA TCA ATC GAG TTT CTG ACC | 21 |
| Malaria pan B3 | CTT GTC ACT ACC TCT CTT CT | 20 | |
| Malaria pan FIP | TCG AAC TCT AAT TCC CCG TTA CCT ATC AGC TTT TGA TGT TAG GGT | 45 | |
| Malaria pan BIP | CGG AGA GGG AGC CTG AGA AAT AGA ATT GGG TAA TTT ACG CG | 41 | |
| Malaria pan FLP | CGT CAT AGC CAT GTT AGG CC | 20 | |
| Malaria pan BLP | AGC TAC CAC ATC TAA GGA AGG CAG | 24 | |
| Plasmodium falciparum (LDH gene) | P. falciparum F3 | AAT TGT AAA CTT ACA TGC ATC AC | 23 |
| P. falciparum B3 | TCA AAT TTA GCT TTT TCC TCA CT | 23 | |
| P. falciparum FIP | GCC CTT CTA ACA AGG TTG AGC AGC TGC TGC TAT TAT CGA AAT | 42 | |
| P. falciparum BIP | TAT GGA CAC TCC GAT ATA TTC GGT GTA ATT CGA TAA CTT GTT CAA CAC | 48 | |
| P. falciparum FLP | CAA ATC TTT AAG TAG GAT TCA GCC | 24 | |
| P. falciparum BLP | GGT ACA CCT GTT GTT TTA GGT GCT | 24 | |
| Plasmodium vivax (16S rRNA gene) | P. vivax F3 | GGA ATG ATG GGA ATT TAA AAC CT | 23 |
| P. vivax B3 | ACG AAG TAT CAG TTA TGT GGA T | 22 | |
| P. vivax FIP | CTA TTG GAG CTG GAA TTA CCG CTC CCA AAA CTC AAT TGG AGG | 42 | |
| P. vivax BIP | AAT TGT TGC AGT TAA AAC GCT CGT AAG CTA GAA GCG TTG CT | 41 | |
| P. vivax FLP | GCT GCT GGC ACC AGA CTT | 18 | |
| P. vivax BLP | AGT TGA ATT TCA AAG AAT CG | 20 | |
| Human (actin gene) | IC F3 | AGT ACC CCA TCG AGC ACG | 18 |
| IC B3 | AGC CTG GAT AGC AAC GTA CA | 20 | |
| IC FIP | GAG CCA CAC GCA GCT CAT TGT ATC ACC AAC TGG GAC GAC A | 40 | |
| IC BIP | CTG AAC CCC AAG GCC AAC CGG CTG GGG TGT TGA AGG TC | 38 | |
| IC FLP | TGT GGT GCC AGA TTT TCT CCA | 21 | |
| IC BLP | CGA GAA GAT GAC CCA GAT CAT GT | 23 |
Laboratory supplies
1. 1.5 mL microcentrifuge tubes (Axygen, catalog number: MCT-150-C)
2. Mmiso® DNA Amplification kit master mix (Mmonitor, catalog number: 22202)
3. DNA extraction tube set (Biozentech, catalog number: NAE-03)
4. 4-chambers grid microchip (Biozentech, catalog number: BZC-04G)
5. Sealing tape (Bio-Rad, catalog number: MSB1001)
6. Aluminum foil (Taeshin Bioscience, catalog number: TS-TF3050)
Equipment
1. Micropipettes 0.1–2.5 μL, 2–20 μL, 20–200 μL (Eppendorf, catalog numbers: 3123000012, 3123000098, 3123000039)
2. Aptical RV400TMCL microscope (OMAX OMB microscope, Gyeonggi-do, Korea; field-deployable with built-in battery and monitor)
3. Microscanner (OMAX, model: RV400TMCL)
4. Vortex-Genie® 2 (Scientific Industries, catalog number: SI-0246A)
5. Heat block (BioFree, catalog number: BF-40HB)
6. Complete blood count (CBC) analyzer (Beckman Coulter, catalog number: B23858)
7. Incubator (L&T Co., catalog number: LT-201S)
8. Refrigerator (LG Electronics, catalog number: LG B607W)
Software and datasets
1. Diagnostic test calculator: https://www.medcalc.org/calc/diagnostic_test.php
Procedure
A. Preparation of 10% Chelex-100 solution
1. Weigh 1 g of Chelex-100 resin using an analytical balance and transfer to a sterile 15 mL conical tube.
2. Add 10 mL of TE buffer (see Recipes) to the tube containing the resin.
3. Vortex vigorously for 1 min until the resin is uniformly suspended.
4. Dispense 195 μL aliquots into DNA extraction tubes (Figure 1) and store at 4 °C until use.
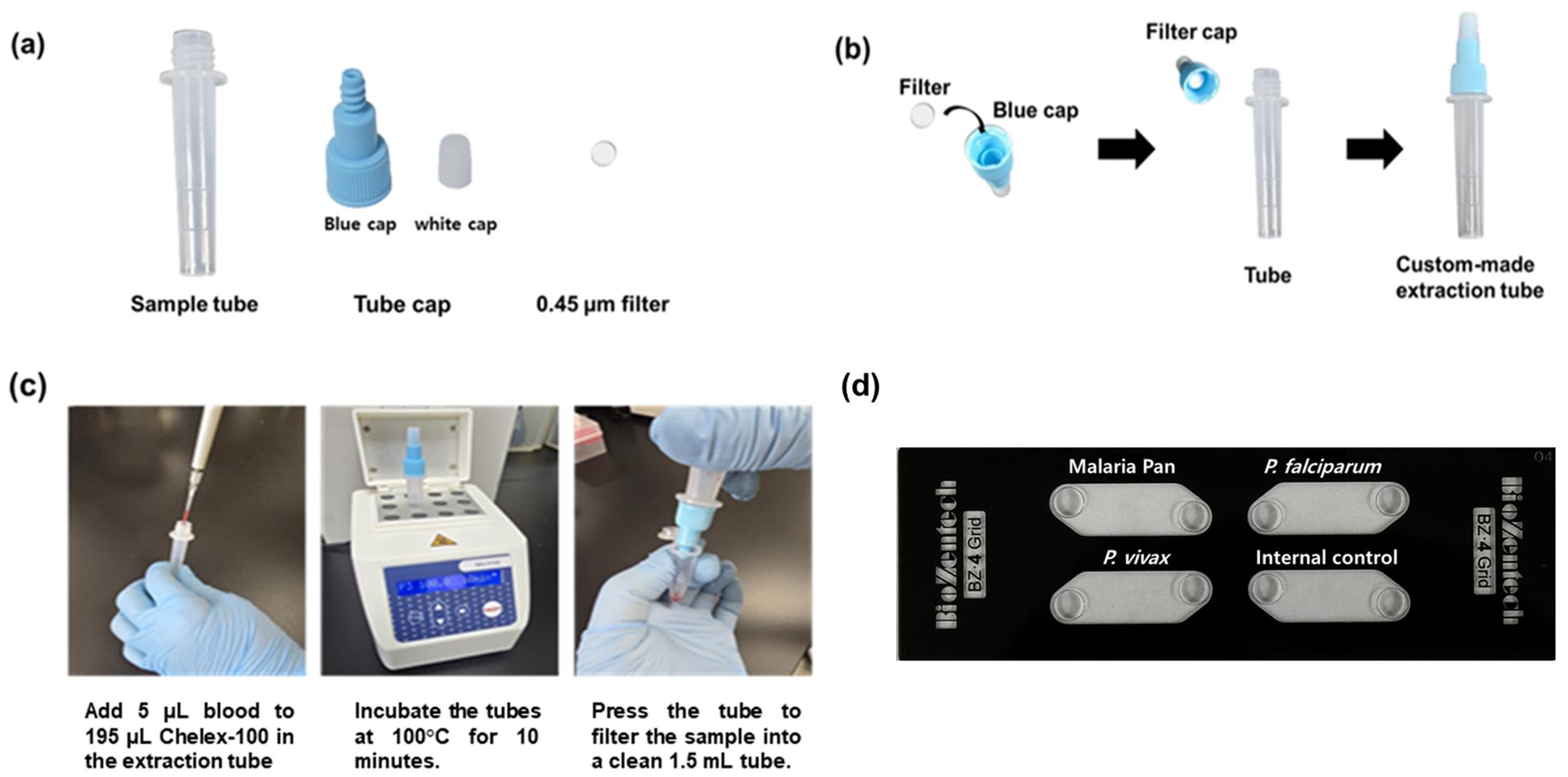
Figure 1. Workflow of Chelex-100/boiling extraction using a custom tube and its application to a four-chamber LAMP-MS chip. (A) Components of the Chelex-100/boiling nucleic acid extraction tube. (B) Assembly steps of the tube. (C) Step-by-step illustration of the nucleic acid extraction procedure using the assembled tube. (D) Layout of the four-chamber microchip used in the LAMP-MS assay, consisting of detection chambers for malaria pan, P. falciparum, P. vivax, and an internal control.
B. DNA extraction by Chelex-100/boiling method
1. Thaw frozen blood samples gently at room temperature and vortex briefly to mix.
2. Add exactly 5 μL of blood sample into the pre-filled extraction tube containing 195 μL of Chelex-100 suspension (Figure 1c)
3. Cap tubes tightly and vortex briefly to mix thoroughly.
4. Incubate tubes in a heating block set to 100 °C for exactly 10 min.
5. Immediately after incubation, gently press the tube to filter the heated sample through the built-in filter into a clean 1.5 mL microcentrifuge tube.
6. Collect approximately 100 μL of filtered DNA-containing solution and store immediately at -20 °C until further analysis.
Pause point: Extracted DNA can be stored at -20 °C for up to one month before the LAMP assay.
C. Microchip preparation
1. Pipette exactly 0.5 μL of each primer solution into each respective chamber of the four-chamber microchip (Figure 1d).
2. Dry loaded microchips at 60 °C in an incubator for precisely 1 h.
3. After drying, label the microchips with the preparation date and primer targets. Wrap them in aluminum foil and store at room temperature until use.
D. LAMP reaction setup
1. Prepare LAMP reaction mix for each sample as follows:
a. 25 μL of Mmiso master mix.
b. 4 μL of Mmiso enzyme.
c. 15 μL of distilled water.
d. 6 μL of extracted DNA (prepared and stored as described in step B7).
2. Vortex gently to homogenize the reaction mix.
3. Load exactly 10 μL of the prepared reaction mix into each chamber of the dried microchip, taking care to avoid bubble formation.
4. Seal microchip chambers with sealing tape carefully to prevent evaporation during incubation.
5. Place the microchip onto a heating block preheated and maintained accurately at 62 °C. Incubate for exactly 30 min.
E. Result interpretation
1. After incubation, immediately remove the microchip from the heating block.
2. Examine each chamber visually under a microscanner or optical microscope.
3. Positive reactions show clearly visible white granules, representing magnesium pyrophosphate, indicating successful amplification.
4. Negative reactions appear clear without visible granules.
5. Record results promptly and dispose of used microchips safely according to laboratory biosafety guidelines.
Data analysis
Visual results; statistical performance (sensitivity/specificity) analyzed via MedCalc (https://www.medcalc.org/calc/diagnostic_test.php).
Validation of protocol
A. Limit of detection (LoD)
1. Clinical whole blood samples confirmed to be positive for Plasmodium falciparum (Pf) and Plasmodium vivax (Pv) were selected. Parasite densities in the original samples were estimated based on hospital patient records and microscopic examination of Giemsa-stained blood smears, in conjunction with white blood cell (WBC) counts.
2. Microscopic examination
a. Parasites (ring, trophozoite, and schizont) are counted against 100 WBCs, and the result is expressed as the number of parasites per microliter of blood, based on the actual WBC count obtained from a complete blood count (CBC) analyzer.
b. Calculation formula:
The actual WBC count was obtained from hospital records or measured using a CBC analyzer.
3. Based on this analysis, the estimated parasite densities were determined to be:
a. P. falciparum: 114,511 parasites/μL
b. P. vivax: 20,832 parasites/μL
c. Each clinical sample was serially diluted 10-fold in PBS to generate a range of parasite concentrations. For P. falciparum, dilutions ranged from 1.1 × 105 to 1.1 × 10-1 parasites/μL, and for P. vivax, from 2 × 104 to 2 × 10-2 parasites/μL.
d. Genomic DNA was extracted from each dilution using the Chelex-100/boiling method.
4. Extracted DNA samples were tested using the malaria pan/Pf/Pv/IC LAMP-MS assay.
5. The limit of detection (LoD) test of the malaria pan/Pf/Pv/IC LAMP-MS assay was repeated in three independent experiments.
6. The LoD was defined as the lowest parasite concentration that yielded consistent positive results in all three replicates.
7. The LOD of the malaria pan/Pf/Pv/IC LAMP-MS assay was determined to be 1.1 × 102 parasites/μL for Plasmodium falciparum and 2 × 102 parasites/μL for Plasmodium vivax (Table 2, Figure 2).
Table 2. Limit of detection (LOD) tests of malaria pan/Pf/Pv/IC LAMP-MS assay using 10-fold serial diluted P. falciparum and P. vivax samples
| Assay | Samples | Target | Parasites/μL | |||||||
| 1.1×105 | 1.1×104 | 1.1×103 | 1.1×102 | 1.1×101 | 1.1×100 | 1.1×10-1 | DW | |||
| Malaria pan LAMP-MS (P/N) | P. falciparum | Pan | P | P | P | P | N | N | N | N |
| Pf | P | P | P | P | N | N | N | N | ||
| Pv | N | N | N | N | N | N | N | N | ||
| IC | P | P | P | N | N | N | N | N | ||
| 2×104 | 2×103 | 2×102 | 2×101 | 2×100 | 2×10-1 | 2×10-2 | DW | |||
| P. vivax | Pan | P | P | P | N | N | N | N | N | |
| Pf | N | N | N | N | N | N | N | N | ||
| Pv | P | P | P | N | N | N | N | N | ||
| IC | P | P | P | N | N | N | N | N | ||
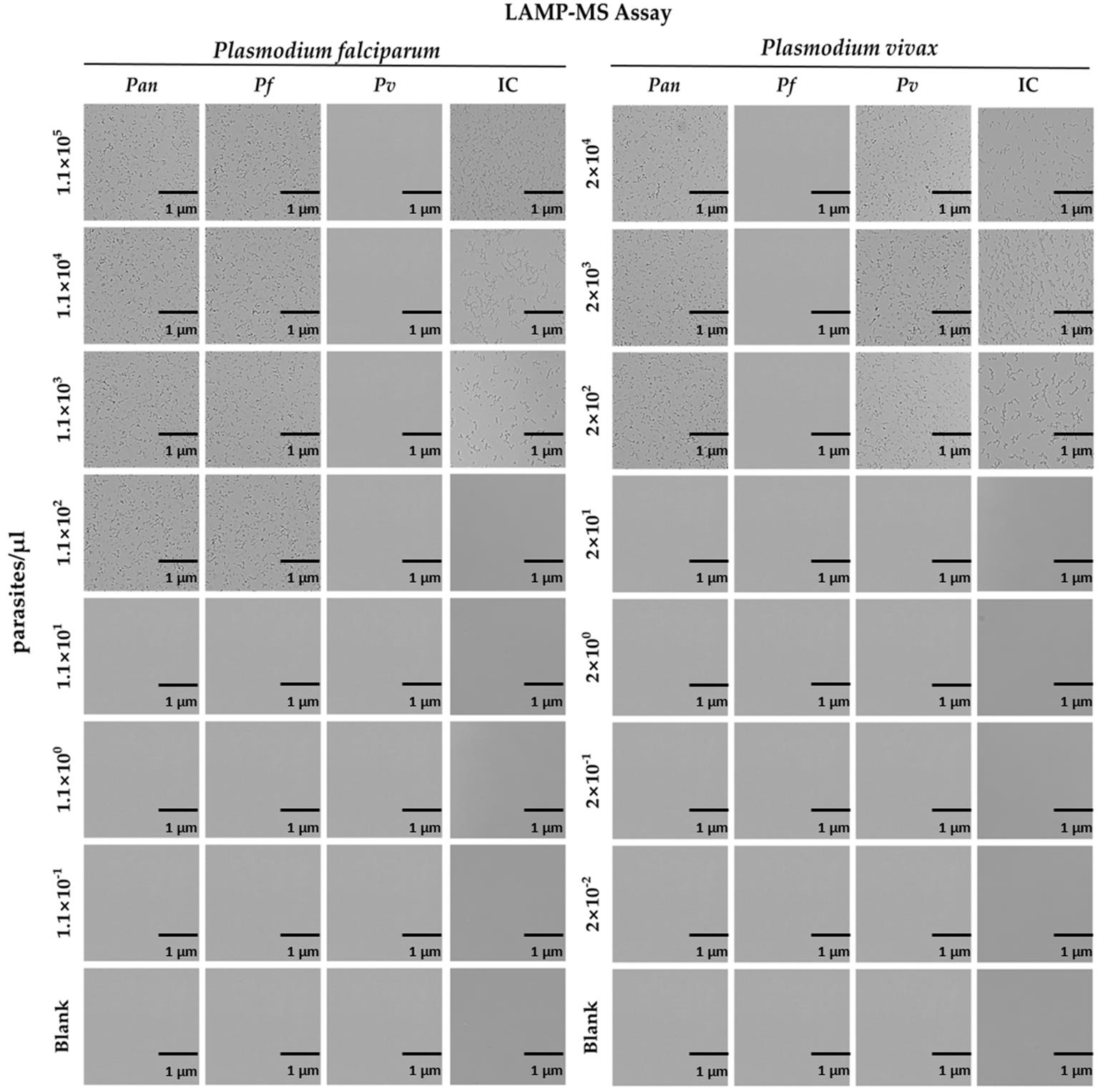
Figure 2. Limit of detection (LOD) tests for the malaria pan/Pf/Pv/IC LAMP-MS assay using 10-fold serially diluted P. falciparum (left panel) and P. vivax (right panel) DNA samples extracted by the Chelex-100/boiling method
B. Clinical test
1. The clinical performance of the malaria pan/Pf/Pv/IC LAMP-MS assay was evaluated using a total of 260 clinical whole blood samples (Table 3, Figure 3), composed of:
a. 80 samples positive for Plasmodium falciparum (Pf)
b. 80 samples positive for Plasmodium vivax (Pv)
c. 100 non-infected (negative control) samples
Table 3. Sensitivity and specificity of malaria pan/Pf/Pv/IC LAMP-MS assay against P. falciparum, P. vivax, and non-infection clinical samples
| Clinical samples | Malaria pan/Pf/Pv/IC LAMP-MS assay | ||||
| Pan | Pf | Pv | IC | ||
| P. falciparum (n = 80) | P/N | 78/80 | 80/80 | 0/80 | 80/80 |
| Sensitivity [95% CI] | 97.5% [91.3–99.7] | 100% [95.5–100.0] | - | 100% [95.5–100.0] | |
| Specificity [95% CI] | - | - | 100% [95.5–100.0] | - | |
| P. vivax (n = 80) | P/N | 76/80 | 0/80 | 75/80 | 80/80 |
| Sensitivity [95% CI] | 95% [87.7–98.2] | - | 94% [86.0–97.9] | 100% [95.5–100.0] | |
| Specificity [95% CI] | - | 100% [95.5–100.0] | - | - | |
| Non-infection (n = 100) | P/N | 0/100 | 0/100 | 0/100 | 100/0 |
| Sensitivity [95% CI] | - | - | - | 100% [96.4–100.0] | |
| Specificity [95% CI] | 100% [96.4–100.0] | 100% [96.4–100.0] | 100% [96.4–100.0] | - | |
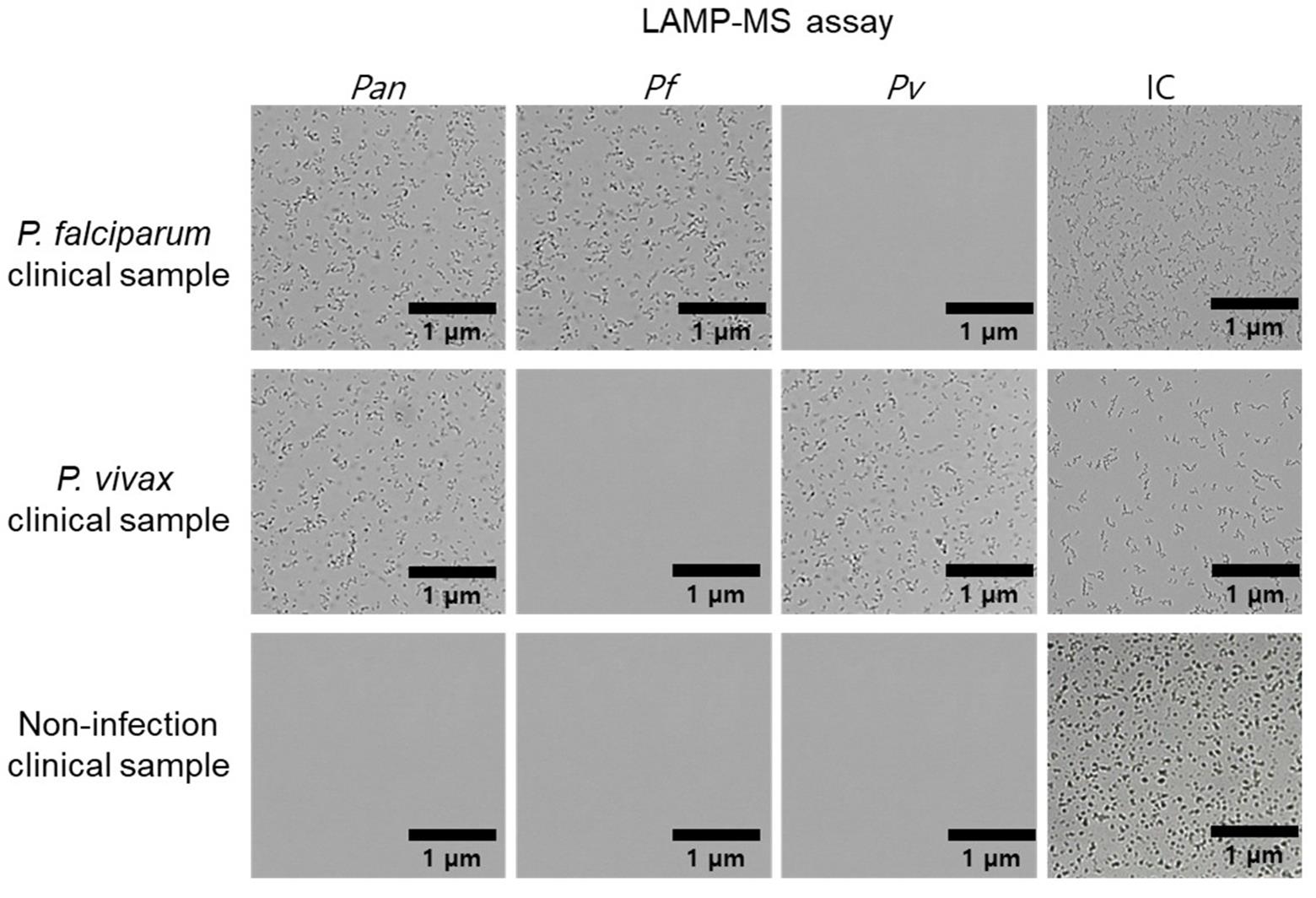
Figure 3. Representative malaria pan/Pf/Pv/IC LAMP-MS assay results for clinical malaria samples. Microscopic images show the results of the LAMP-MS assay performed on clinical samples infected with Plasmodium falciparum and Plasmodium vivax and non-infected clinical samples. Four detection channels are shown: pan-malaria (Pan), P. falciparum-specific (Pf), P. vivax-specific (Pv), and internal control (IC). Amplified products appear as dense clustered signals, while negative results show clear backgrounds. Scale bars: 1 μm.
2. DNA was extracted from all samples using the Chelex-100/boiling DNA extraction method and subsequently analyzed using the LAMP-MS assay.
3. For P. falciparum samples:
a. Malaria pan/Pf/Pv/IC LAMP-MS assay showed a sensitivity of 97.5% for the pan target (95% CI: 91.3–99.7) and 100% for the Pf-specific target (95% CI: 95.5–100.0).
b. Malaria pan/Pf/Pv/IC LAMP-MS assay exhibited 100% specificity, with no false positives.
4. For P. vivax samples:
a. Malaria pan/Pf/Pv/IC LAMP-MS assay showed a sensitivity of 95.0% for the pan target (95% CI: 87.7–98.2) and 94.0% for the Pv-specific target (95% CI: 86.0–97.9).
b. Malaria pan/Pf/Pv/IC LAMP-MS assay demonstrated 100% specificity, with no false positives.
5. For the 100 non-infected samples:
• Malaria pan/Pf/Pv/IC LAMP-MS assay achieved 100% specificity across all diagnostic targets (pan, Pf, Pv) with no false positives.
• Internal control (IC) was detected in 100% of the samples, confirming sample integrity and reaction validity.
C. Cross-reactivity
1. To evaluate the specificity of the malaria pan/Pf/Pv/IC LAMP-MS assay, potential cross-reactivity was assessed using RNA or DNA from clinical samples infected with non-malarial pathogens.
2. The assay was tested against the following viruses: Zika virus (ZIKV), Chikungunya virus (CHIKV), Japanese encephalitis virus (JEV), and Dengue virus (DENV), including all four serotypes (DENV-1, DENV-2, DENV-3, DENV-4).
3. Additional negative control tests were conducted using human whole blood genomic DNA (malaria-negative donors) and distilled water (no-template control).
4. Each sample was subjected to the standard LAMP-MS protocol, and amplification was evaluated across all diagnostic targets: Pan, Plasmodium falciparum (Pf), Plasmodium vivax (Pv), and internal control (IC).
5. The LAMP-MS assay produced negative results for all malaria targets (Pan, Pf, Pv) and the internal control (IC) in all non-malarial viral samples. In contrast, the assay yielded positive results only for the internal control (IC) when tested with human whole blood DNA, confirming assay validity and high specificity (Table 4).
Table 4. Cross-reactivity of the malaria pan/Pf/Pv/IC LAMP-MS assay against other virus infection samples
| Samples | Malaria pan/Pf/Pv/IC LAMP-MS assay | |||
| Pan | Pf | Pv | IC | |
| Zika virus | N | N | N | N |
| Dengue 1 virus | N | N | N | N |
| Dengue 2 virus | N | N | N | N |
| Dengue 3 virus | N | N | N | N |
| Dengue 4virus | N | N | N | N |
| Chikungunya virus | N | N | N | N |
| Japanese encephalitis | N | N | N | N |
| Human whole blood DNA | N | N | N | P |
| Distilled water | N | N | N | N |
This protocol or parts of it has been used and validated in the following research article(s):
• Jang et al. [12]. Development of a rapid malaria LAMP-MS assay for diagnosis of malaria infections. Scientific Reports (Figure 3B).
General notes and troubleshooting
1. To prevent cross-contamination during chip loading, use filtered pipette tips and change tips between samples. Cross-contamination can lead to false-positive results, especially in isothermal amplification assays [13].
2. Air bubbles introduced during pipetting can inhibit LAMP reactions by disrupting uniform thermal distribution and reagent mixing, potentially leading to false-negative or inconsistent results. To minimize bubble formation, we recommend pipetting slowly using low-retention tips and taking care to avoid introducing air into the reaction mixture during reagent loading. In addition, Mahdavi et al. [14] reported that centrifugal microfluidic chips effectively eliminate bubbles and residual volumes, contributing to more stable amplification.
3. This method is recommended for use in resource-limited or field settings due to its simplicity and minimal equipment requirements. However, full integration into automated high-throughput systems may be limited. To enable full field applicability of the assay, development of a battery-operated heat block would be necessary.
Acknowledgments
This work was supported by a grant from the Korea Health Technology R&D Project, through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HR20C0021); a grant from the National Research Foundation of Korea (NRF), funded by the Korean Government, MSIP (RS-2023-00207833); and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant number: 2022R1I1A1A01063976). This protocol was used in [12].
Competing interests
The authors declare no competing interests.
Ethical considerations
This study was approved by the Medical Ethics Committee of Korea University’s Guro Hospital (2021GR0337).
References
- Paton, D. G., Childs, L. M., Itoe, M. A., Holmdahl, I. E., Buckee, C. O. and Catteruccia, F. (2019). Exposing Anopheles mosquitoes to antimalarials blocks Plasmodium parasite transmission. Nature. 567(7747): 239–243. https://doi.org/10.1038/s41586-019-0973-1
- Rosenthal, P. J. (2022). Malaria in 2022: Challenges and Progress. Am J Trop Med Hyg. 106(6): 1565–1567. https://doi.org/10.4269/ajtmh.22-0128
- Roberts, D. and Matthews, G. (2016). Risk factors of malaria in children under the age of five years old in Uganda. Malar J. 15: 246. https://doi.org/10.1186/s12936-016-1290-x
- Landier, J., Parker, D. M., Thu, A. M., Carrara, V. I., Lwin, K. M., Bonnington, C. A., Pukrittayakamee, S., Delmas, G. and Nosten, F. H. (2016). The role of early detection and treatment in malaria elimination. Malar J. 15: 363. https://doi.org/10.1186/s12936-016-1399-y
- Yoon, J., Jang, W. S., Nam, J., Mihn, D. C. and Lim, C. S. (2021). An Automated Microscopic Malaria Parasite Detection System Using Digital Image Analysis. Diagnostics. 11(3): 527. https://doi.org/10.3390/diagnostics11030527
- Mouatcho, J. C. and Goldring, J. P. D. (2013). Malaria rapid diagnostic tests: challenges and prospects. J Med Microbiol. 62(Pt 10): 1491–1505. https://doi.org/10.1099/jmm.0.052506-0
- Britton, S., Cheng, Q. and McCarthy, J. S. (2016). Novel molecular diagnostic tools for malaria elimination: a review of options from the point of view of high-throughput and applicability in resource limited settings. Malar J. 15: 88. https://doi.org/10.1186/s12936-016-1158-0
- McMorrow, M. L., Aidoo, M. and Kachur, S. P. (2011). Malaria rapid diagnostic tests in elimination settings--can they find the last parasite? Clin Microbiol Infect. 17(11): 1624–1631. https://doi.org/10.1111/j.1469-0691.2011.03639.x
- Lucchi, N. W., Ndiaye, D., Britton, S. and Udhayakumar, V. (2018). Expanding the malaria molecular diagnostic options: opportunities and challenges for loop-mediated isothermal amplification tests for malaria control and elimination. Expert Rev Mol Diagn. 18(2): 195–203. https://doi.org/10.1080/14737159.2018.1431529
- Abdul-Ghani, R., Al-Mekhlafi, A. M. and Karanis, P. (2012). Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: would it come to clinical reality as a point-of-care test? Acta Trop. 122(3): 233–240. https://doi.org/10.1016/j.actatropica.2012.02.004
- Cuadros, J., Pérez-Tanoira, R., Prieto-Pérez, L., Martin-Martin, I., Berzosa, P., González, V., Tisiano, G., Balcha, S., Ramos, J. M. and Górgolas, M. (2015). Field Evaluation of Malaria Microscopy, Rapid Malaria Tests and Loop-Mediated Isothermal Amplification in a Rural Hospital in South Western Ethiopia. PLoS One. 10(11): e0142842. https://doi.org/10.1371/journal.pone.0142842
- Jang, W. S., Choi, M. K., Choe, Y. L. and Lim, C. S. (2025). Development of a rapid malaria LAMP-MS assay for diagnosis of malaria infections. Sci Rep. 15(1): 8547. https://doi.org/10.1038/s41598-025-92935-4
- Shakeri, A., Yousefi, H., Abu Jarad, N., Kullab, S., Al-Mfarej, D., Rottman, M. and Didar, T. F. (2022). Contamination- and carryover-free handling of complex fluids using lubricant-infused pipette tips. Sci Rep. 12: 15002. https://doi.org/10.1038/s41598-022-18756-x
- Mahdavi, F., Niyyati, M., Shamloo, A., Dezhkam, R., Hoseinian, S. M., Seyyedtabaei, S. J. and Mirjalali, H. (2025). Design and fabrication of multiplex microfluidic LAMP PCR for simultaneous detection of opportunistic protozoa. Sens Bio-Sens Res. 48: 100787. https://doi.org/10.1016/j.sbsr.2025.100787
Article Information
Publication history
Received: Apr 17, 2025
Accepted: Jun 5, 2025
Available online: Jun 19, 2025
Published: Jul 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Lim, M. S., Choe, Y. L. and Jang, W. S. (2025). Rapid and Multiplex Diagnosis of Malaria Using Chelex-100 Extraction and LAMP-MS Assay. Bio-protocol 15(13): e5375. DOI: 10.21769/BioProtoc.5375.
Category
Microbiology > Pathogen detection > RT-LAMP
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link