- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA PolyA Tailing Assay to Qualitatively Analyze Circular RNA Manufacturing
Published: Vol 15, Iss 10, May 20, 2025 DOI: 10.21769/BioProtoc.5310 Views: 1647
Reviewed by: Joyce ChiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Spin Labeling of RNA Using “Click” Chemistry for Coarse-grained Structure Determination via Pulsed Electron-electron Double Resonance Spectroscopy
Maria F. Vicino [...] Olav Schiemann
May 5, 2021 5987 Views
An Improved m6A-ELISA for Quantifying N6-methyladenosine in Poly(A)-purified mRNAs
Wei Yee Chan [...] Folkert J. Van Werven
Jun 20, 2025 2989 Views
Examining the Roles of m6A Sites in mRNA Using the Luciferase Gene Fused With Mutated RRACH Motifs
Nobuko Katoku-Kikyo and Nobuaki Kikyo
Nov 5, 2025 1934 Views
Abstract
A covalently closed loop structure provides circular RNA (circRNA) with more stability than conventional RNAs in linear form, making circRNA an emerging tool in RNA therapeutics. The qualification and quantification of circRNA after production is critical for its design and effectiveness assessments, particularly when the following applications could be affected by byproduct RNAs. Despite PCR-based methods effectively detecting low-abundance circRNA, they are unsuitable for assessing uncircularized RNA in a mass production fraction to maintain quality control. Here, we present a straightforward protocol for evaluating uncircularized byproduct RNAs from circRNA production. This method enrolls the template-independent RNA polymerase activity to add adenine tails (polyA) to the 3' ends of a linear RNA, making it easy to distinguish trace byproducts or uncircularized RNA from a pool of mass circRNA products. With conventional linear RNA and RNase R-treated circRNA as the positive and negative controls, the purity of a circRNA preparation could be readily resolved. Regardless of circRNA production strategies, this protocol provides a reliable and practical way to ensure the consistent quality of homemade circRNAs or to recheck circRNA quality from commercial manufacturing.
Key features
• The RNA circularization assessment relies on detecting a free 3' end OH group of the non-circRNA in the circRNA products.
• An RNA denaturing electrophoresis is required to detect nucleotide length changes of the non-circRNA post-3' end tailing.
• Rather than quantitatively, the molecular weight changes qualitatively highlight the trace non-circRNA in a circRNA preparation.
• While there are different techniques for producing circRNA, the tailing method is effective for most, detecting leftover linear RNA contaminants.
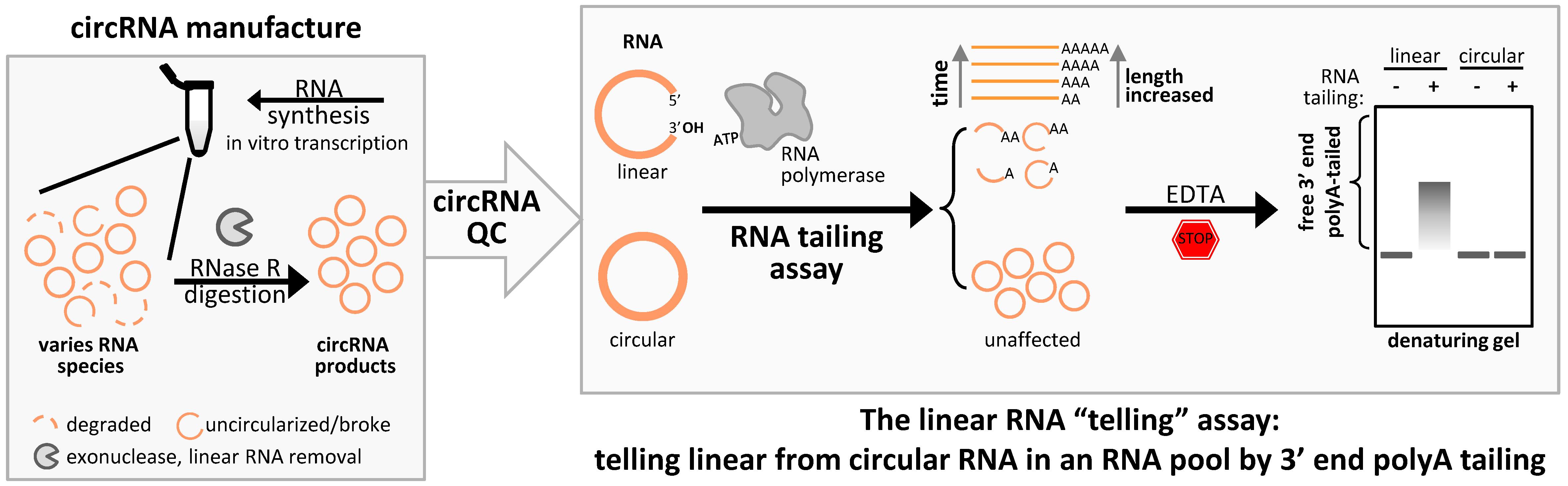
Keywords: RNA tailing/telling assayGraphical overview
Background
Circular RNAs (circRNAs) are unique RNAs characterized by their covalently closed-loop structures. These structures confer increased stability compared to their linear counterparts by rendering them resistant to exonuclease-mediated degradation [1]. This natural stability makes circRNAs promising candidates for RNA-based therapeutics, including gene regulation, biomarker development, and targeted drug delivery [2]. As the diversity of circRNA design strategies and applications continues to expand, accurately assessing circRNA yield and purity has become an emerging and critical need.
Current circRNA detection methods [3] mainly detect and quantify a relatively small amount of circRNAs in a specific context. High-throughput RNA sequencing (RNA-Seq), combined with bioinformatics, analyzes various types of genome-wide circRNAs present in low abundance [4]. By targeting the junction sequences of RNA back-splicing/ligation, quantitative reverse transcription PCR (qRT-PCR) offers sensitive and specific quantification for trace circRNA from an RNA pool [5]. Even though these approaches effectively detect circRNA, none directly assess the accompanying linear RNAs in the same pool. This is especially crucial for quality control and downstream applications of circRNA after mass production.
Linear RNA contaminants can be found in a circRNA preparation for several reasons, such as yet-to-be circularized RNA, somehow disconnected circRNAs, or any byproduct RNAs with free 3' ends in circRNA production processes. Here, we present a streamlining assay that leverages the activity of template-independent RNA polymerase to append adenine tails (polyA) to the 3' ends of linear RNAs [6]. As a consequence, the prerequisite for the success of this polyA tailing assay significantly relies on optimizing denaturing formaldehyde gel conditions and testing appropriate tailing assay parameters in-house. Denaturing gel electrophoresis offers better resolution and prevents RNA degradation, reducing misleading band shifts compared to non-denaturing TBE gels. This strategy clearly tells that any linear RNA remains in the circRNA products, allowing for a reliable assessment of circRNA purity, predominantly when the sample is mainly composed of circRNA.
Exonuclease resistance is a circRNA signature, distinguishing it from linear RNA [7]. However, RNase R can be used for linear RNA removal in circRNA preparation rather than for detecting circRNA. To evaluate linear RNA in the bulk of circRNA preparation more straightforwardly and intuitively, we repurposed the polyA tailing assay from its primary application for in vitro mRNA preparation [8]. This method provides an effective and practical approach to assessing circRNA integrity.
Materials and reagents
Reagents
1. Agarose biomax (Hispanagar, CAS number: 9012-36-6)
2. Diethyl pyrocarbonate (DEPC) (VWR, catalog number: E174-25G)
3. E. coli PolyA polymerase (NEB, catalog number: M0276S)
4. EDTA, disodium salt, dihydrate, crystal (J. T. Baker, catalog number: 8993-01)
5. Ethyl alcohol, 99%, HPLC/ACS (Everdine Technology, CAS number: 64-17-5)
6. Formaldehyde solution (Sigma, catalog number: F1635)
7. GelRed nucleic acid gel stain (Biotium, catalog number: 41003)
8. MEGAclear Transcription Clean-Up kit (Invitrogen, catalog number: AM1908)
9. MOPS buffer (10×) (ZGENEBIO, catalog number: GB33)
10. RNA loading dye (2×) (NEB, catalog number: B0363S)
11. RNase R (Abcam, catalog number: ab286929)
12. ssRNA ladder (NEB, catalog number: N0362S)
13. TBE buffer (5×) (VWR, catalog number: J885-5L)
14. Adenosine 5'-triphosphate (ATP) (NEB, catalog number: P0756S)
Solutions
1. DEPC-treated water (see Recipes)
2. MOPS buffer (1×) (see Recipes)
3. Denaturing formaldehyde agarose gel (1%) (see Recipes)
4. TBE buffer (0.5×) (see Recipes)
5. Non-denaturing TBE agarose gel (1%) (see Recipes)
6. EDTA solution (0.5 M) (see Recipes)
7. RNA polyA tailing reaction mixture (see Recipes)
8. EDTA stop solution (see Recipes)
9. RNase R digestion (see Recipes)
Recipes
1. DEPC-treated water
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DEPC | 0.1% | 1 mL |
| Deionized water | 999 mL | |
| Total | 1 L |
Stir overnight. To remove DEPC, autoclave the solution for 15–45 min at 15 psi.
2. MOPS buffer (1×)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MOPS buffer (10×) | 1× | 50 mL |
| DEPC-treated water | 450 mL | |
| Total | 500 mL |
3. Denaturing formaldehyde agarose gel (1%)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1% | 0.5 g |
| MOPS buffer (1×) | 42 mL | |
| Formaldehyde | 2 M | 8 mL |
| Total | 50 mL |
4. TBE buffer (0.5×)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| TBE buffer (5×) | 0.5× | 100 mL |
| DEPC-treated water | 900 mL | |
| Total | 1 L |
5. Non-denaturing TBE agarose gel (1%)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1% | 0.5 g |
| TBE (1×) | 50 mL | |
| GelRed nucleic acid gel stain (10,000×) | 1× | 5 μL |
| Total | 50 mL |
6. EDTA solution (0.5 M)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EDTA | 0.5 M | 18.61 g |
| Deionized water | 80 mL | |
| DEPC | 0.1% | 100 μL |
| Total | 100 mL |
Adjust the final volume to 100 mL using deionized water and stir overnight. To remove DEPC, autoclave the solution for 15–45 min at 15 psi.
7. RNA polyA tailing reaction mixture
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| RNA sample | 1.25 μg | 15 μL |
| E. coli PolyA polymerase reaction buffer (10×) | 1× | 2 μL |
| ATP (10 mM) | 1 mM | 2 μL |
| E. coli PolyA Polymerase | 5 U | 1 μL |
| Total | 20 μL |
8. EDTA stop solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| RNA polyA tailing reaction mixture | 20 μL | |
| EDTA (50 mM) | 10 mM | 5 μL |
| Total | 25 μL |
9. RNase R digestion
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| RNA sample | 100 μg | 88 μL |
| RNase R reaction buffer (10×) | 1× | 10 μL |
| RNase R | 20 U | 2 μL |
| Total | 100 μL |
Adjust the final volume to 100 μL using DEPC-treated water.
Laboratory supplies
1. Eppendorf DNA LoBind tubes (Eppendorf, catalog number: 022431021)
2. 0.1–10 μL sterilized filter tips (GILSON, catalog number: F171203)
3. 10–100 μL sterilized filter tips (GILSON, catalog number: F171403)
4. 2–200 μL sterilized filter tips (GILSON, catalog number: F171503)
5. 50–1,200 μL sterilized filter tips (GILSON, catalog number: F171803)
Equipment
1. 37 °C incubator (Astec, catalog number: SCA-165DS)
2. Block incubator (Astec, catalog number: BI-525)
3. Gel electrophoresis system (Major Science, catalog number: MJ-105-S)
4. Electrophoresis power supply (Thermo, catalog number: EC250-90)
5. Orbital shaker (TKS, catalog number: OS701)
6. Microcentrifuge (Eppendorf, catalog number: 5424)
Software and datasets
1. Quantum CX5 Gel Imaging System (VILBER, BIO VISION)
2. ImageJ (NIH, 1.54p)
Procedure
A. Visualize RNA samples by denaturing gel electrophoresis
A1. Preparation of 1% denaturing formaldehyde agarose gel
1. Prepare MOPS buffer (1×) (see Recipe 2).
2. Weigh 0.5 g of agarose in 42 mL of 1× MOPS buffer and heat until dissolved (see Recipe 3).
3. Thoroughly mix 8 mL of 37% formaldehyde (2 M) into the melted agarose.
Caution: Carry out all procedures involving formaldehyde in the chemical fume hood.
4. Pour the formaldehyde agarose into the gel casting tray and allow the gel to set at room temperature.
Note: Clean the gel tray and gel tank before use and cover with plastic wrap to prevent contamination.
5. Place the solidified agarose in the electrophoresis tank and cover with 1× MOPS buffer.
6. Prepare 100–200 ng of RNA samples and 200 ng of markers with 2× RNA loading dye in equal volumes.
7. Heat RNA samples at 70 °C for 5 min, then load the samples into the wells.
8. Run the gel at 100 V for 30 min.
9. Soak the gel in deionized water for 15 min to remove residual formaldehyde and allow RNA renaturation.Note: This step aims to eliminate formaldehyde, enabling RNA to renature for staining.
10. Stain the gel with 15 μL of 3× GelRed nucleic acid gel stain in 50 mL deionized water, shaking at 50 rpm for 30 min for even staining.
11. Soak in deionized water for an additional 10 min to reduce background staining.
12. Capture images with the Quantum CX5 gel imaging system.
Note: Adjust exposure to 4 s with a 6× zoom for better image clarity.
A2. Preparation of 1% non-denaturing TBE agarose gel
1. Prepare 0.5× TBE running buffer (see Recipe 4).
2. Weigh 0.5 g of agarose in 50 mL of 0.5× TBE buffer and heat until dissolved (see Recipe 5).
3. Add 5 μL of 1× GelRed nucleic acid gel stain to 50 mL of melted agarose and mix thoroughly.
4. Pour the agarose into the gel casting tray and allow the agarose to solidify completely.
5. Immerse the solidified gel in 0.5× TBE running buffer within the electrophoresis tank.
Note: Clean the gel tray and gel tank before use and cover with plastic wrap to prevent contamination.
6. Prepare 100–200 ng of RNA samples and 200 ng of markers with 2× RNA loading dye in equal volumes.
7. Heat RNA samples at 70 °C for 5 min and then load the samples into the wells.
8. Run the gel at 100 V for 30 min.
9. Capture images using the Quantum CX5 gel imaging system.
Note: Set exposure to 4 s with 6× zoom for optimal image quality.
B. Optimize RNA tailing conditions
B1. Preparation of reagents
1. Prepare a 0.5 M EDTA solution (see Recipe 6) and dilute it to 50 mM (10× dilution) for the experiments.
2. Prepare a control linear RNA for optimizing RNA tailing.
Notes:
a. It is recommended to design or synthesize a linear RNA of a length comparable to the target circRNA to serve as a control. RNA could be synthesized using any commercial kit, e.g., RiboMAXTM Large Scale RNA Production System.
b. Purify RNA for tailing reactions beforehand (any method that removes RNase R can be used).
B2. Reaction of RNA tailing
1. Add 1.25 μg of purified RNA to each tube for the tailing reaction (see Recipe 7).
2. Incubate at 37 °C for different durations (30, 60, and 120 min here, for example).
Notes:
1. Replacing the enzyme with DEPC-treated water could be used here for a technical negative control.
2. This step optimizes reaction conditions, and RNA tailing duration can be adjusted as needed.
3. Centrifuge at 14,000 rpm (~18,000× g) for 1 min to spin down.
4. Add 10 mM EDTA stop solution to each reaction and mix gently (see Recipe 8).
Note: The reaction stops at this step, and samples can be stored at -80 °C until further processing.
5. Prepare 100–200 ng of RNA samples and 200 ng of markers with 2× RNA loading dye in equal volumes.
6. Heat RNA samples to 70 °C for 5 min, then load the samples into a 1% denaturing formaldehyde agarose gel or a 1% non-denaturing TBE agarose gel (Figure 1).
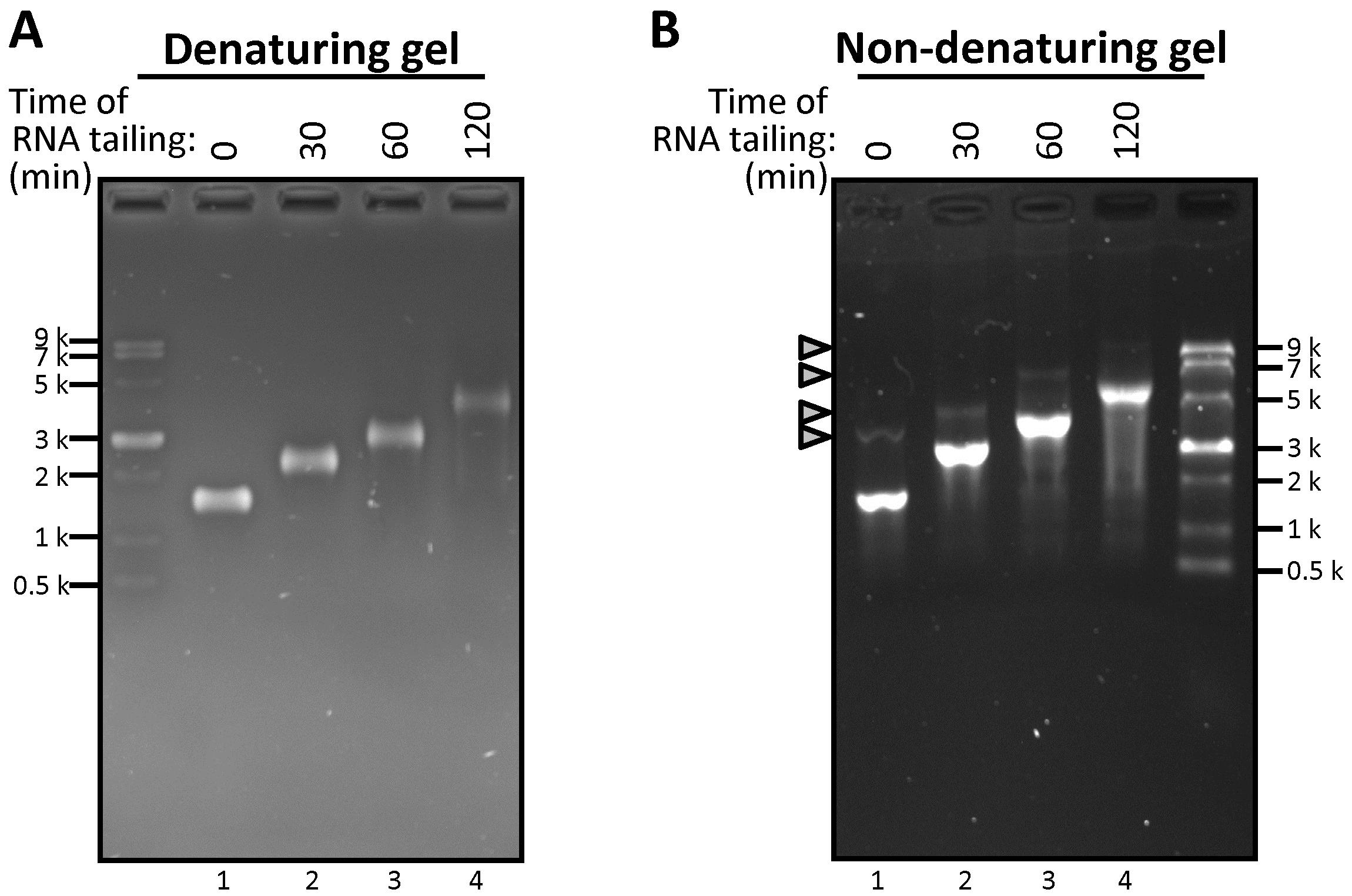
Figure 1. Comparison of linear RNA mobility changes post-RNA tailing using denaturing or non-denaturing agarose gel electrophoresis. (A,B) Equal amounts (1.25 μg) of control linear RNA (1,584 nt) transcript served as the template for the 3' end polyA tailing assay. The reactions were incubated at 37 °C for the indicated times and analyzed by electrophoresis using 1% denaturing formaldehyde gel (A) or 1% non-denaturing TBE gel (B).
C. Qualify circRNA in RNA pools
C1. Linear RNA removal by RNase R digestion
1. Treat RNA products with RNase R at 37 °C for 1 h to remove uncircularized RNA (see Recipe 9).
Notes:
1. RNase R selectively degrades linear RNA while sparing circRNA.
2. To assess the quality and purity of circRNA, prepare RNA samples by dividing one sample into two equal portions: one subjected to RNase R digestion (+) and the other untreated (-).
2. Stop digestion by removing RNase R with the MEGAclear Transcription Clean-Up kit.
C2. RNA sample preparation (using MEGAclear Transcription Clean-Up kit)
1. Add 350 μL of binding solution to the reaction mixture and then add 250 μL of 100% ethanol to promote RNA binding to the filter cartridge.
2. Transfer the RNA mixture to the cartridge and centrifuge at 14,000 rpm (~18,000× g) for 1 min.
3. Add 500 μL of wash solution to the column and centrifuge at 14,000 rpm for 1 min. Repeat this step twice.
4. Elute RNA using 50 μL of hot nuclease-free water.
Note: RNA can be stored at -80 °C or used immediately for the RNA tailing assay.
C3. CircRNA assessment by RNA tailing assay
1. Aliquot 1.25 μg of purified RNA (with or without RNase R digestion) for the RNA tailing assay (see Recipe 7).Note: For the non-tailing group, replace RNA polymerase with DEPC-treated water. Process the control group under the same conditions.
2. Incubate at 37 °C for 60 min.
3. Stop the reaction by adding 10 mM EDTA solution (see Recipe 8).
4. Mix RNA samples (100–200 ng) and markers (200 ng) with an equal volume of 2× loading dye.
5. Heat RNA samples at 70 °C for 5 min and then load the samples into a 1% denaturing formaldehyde agarose gel (Figure 2).
D. Analyze RNA band intensity by ImageJ
This approach provides an image analysis option for general reference. Users should determine their RNA quality according to their specific experimental design and requirements.
1. Convert the RNA gel image to a negative film and save it as a TIFF file.
2. Open the image in ImageJ and select the analysis area based on the gel electrophoresis results [9].
Note: Ensure the image is horizontally aligned before analysis.
3. Utilize the Straight function to identify and remove the background.
4. Select the zone for analysis and compute the area ratio.
Notes:
a. The band shift, more than its intensity, is relatively valuable for the circRNA assessment (see Troubleshooting).
b. This method qualitatively distinguishes linear and circRNA. For more quantification approaches of the linear-to-circular RNA ratio, an alternative method is recommended [10].
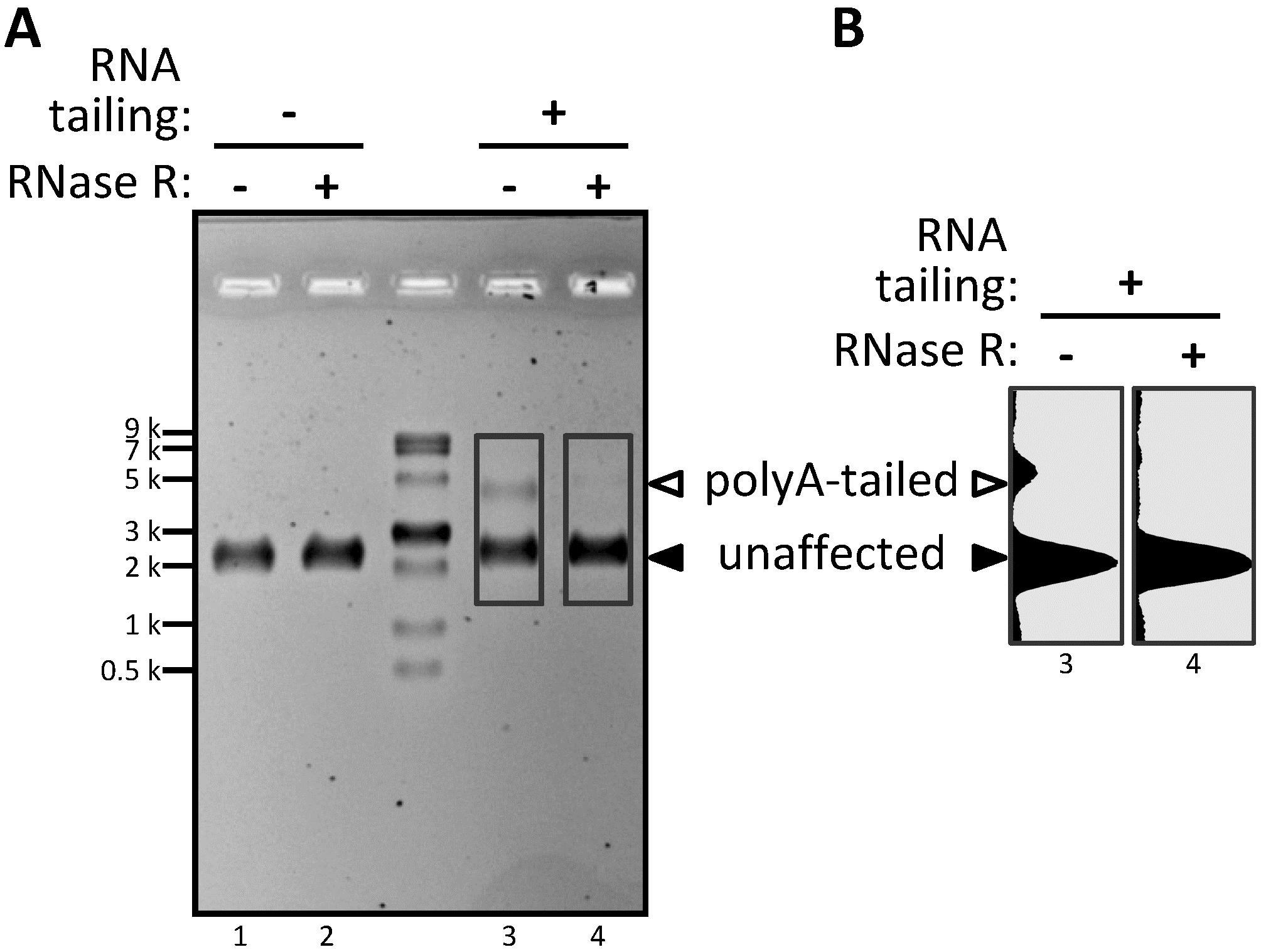
Figure 2. Visualization of non-circular RNA in a circRNA pool using the tailing assay. (A) Pooled mixture of circular (major) and linear (minor) RNA species, both with a length of 2,527 nt, treated with RNase R (+) or not (−), and sampled for RNA tailing assay or not at 37 °C for 1 h. The end-tailed RNA products were analyzed by denaturing gel electrophoresis. (B) RNA band intensity was analyzed by ImageJ. White triangle: polyA-tailed non-circRNA; black triangle: unaffected circRNA.
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Su et al. [10] A cis-acting ligase ribozyme generates circular RNA in vitro for ectopic protein functioning. Nat Commun. (Figure 1h, i).
General notes and troubleshooting
General notes
1. Based on the availability of a free 3' end for end labeling, the RNA polyA tailing assay could be repurposed to identify a wide range of non-circRNA in an RNA pool, regardless of the circularization strategy used.
2. Optimizing electrophoresis conditions for high-resolution RNA separation in a gel is a prerequisite for accurately reading the tailing assay results.
3. Denaturing gel electrophoresis is preferable for achieving better resolution. It prevents RNA degradation under nuclease-free conditions and provides superior resolution compared to non-denaturing TBE gels by eliminating misleading band shifts (Figure 1B, gray triangle).
4. Synthesizing linear RNA, with a length corresponding to the target circRNA, could primarily aid in optimizing the experimental conditions to meet the requirements of each RNA tailing assay tailored for a specific circRNA target.
5. RNA tailing raises the molecular weight to assess linear RNA content, with its effectiveness gradually decreasing as RNA length increases.
6. Increased reaction time and reduced RNA molecules with a free 3' OH group in the tailing assay could result in a longer poly A tail at the RNA 3' end, which may significantly affect experimental outcomes.
7. Conduct experiments in a nuclease-free environment to reduce RNase contamination. CircRNA resists exonucleases but not all nucleases. Any RNA degradation may incorrectly produce a free 3' OH group available for tailing, which can lead to misleading conclusions.
8. Evaluate RNase R activity using synthetic linear RNA as a control, as enzyme activity can vary between batches from different manufacturers and may decline over time.
Troubleshooting
Problem 1: A faint or undetectable 3' end tailing is observed in gel electrophoresis.
Possible cause: An excessively long RNA may be hard to distinguish after RNA tailing because the amplitude of length changes is marginal. An abundance of RNA templates could cause rapid ATP depletion, which decreases tailing efficiency.
Solution: Reduce the RNA amount or extend the reaction time in the tailing assay.
Problem 2: Degraded RNA bands are apparent in the denaturing gel.
Possible cause: i) Repeated freeze-thaw cycles may cause RNA degradation.
Solution: i) Aliquot samples before freezing to minimize freeze and thaw cycles and maintain RNA integrity.
Possible cause: ii) Environmental or sample-derived ribonucleases can degrade RNA.
Solution: ii) Utilize sterile, RNase-free materials and adhere to strict aseptic techniques. Disinfect all equipment (e.g., pipettes, tube racks, electrophoresis tanks) and rinse with DEPC-treated water prior to electrophoresis to reduce nuclease contamination.
Acknowledgments
This work was supported by grants to C.Y.Y. from the National Health Research Institutes (NHRI), Taiwan (IV-114-PP-14 and IV-114-SP-08) to support C.I.S. and H.I.W., and from the National Science and Technology Council (NSTC), Taiwan to support Z.S.C. (113-2320-B-400-020). We also acknowledge our previous work published in Nat Commun (2024), DOI: 10.1038/s41467-024-51044-y, which serves as the basis for the current protocol [10].
Competing interests
The authors declare no conflicts of interest.
References
- Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 21(8): 475–490. https://doi.org/10.1038/s41580-020-0243-y.
- Liu, J., Zhang, X., Yan, M. and Li, H. (2020). Emerging Role of Circular RNAs in Cancer. Front Oncol. 10: 663. https://doi.org/10.3389/fonc.2020.00663.
- Jeck, W. R. and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat Biotechnol. 32(5): 453–461. https://doi.org/10.1038/nbt.2890.
- Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., Marzluff, W. F. and Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19(2): 141–157. https://doi.org/10.1261/rna.035667.112.
- Panda, A. C. and Gorospe, M. (2018). Detection and Analysis of Circular RNAs by RT-PCR. Bio Protoc. 8(6). https://doi.org/10.21769/BioProtoc.2775.
- Yehudai-Resheff, S. and Schuster, G. (2000). Characterization of the E.coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 28(5): 1139–1144. https://doi.org/10.1093/nar/28.5.1139.
- Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A. and Mayeda, A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34(8): e63. https://doi.org/10.1093/nar/gkl151.
- Passmore, L. A. and Coller, J. (2022). Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol. 23(2): 93–106. https://doi.org/10.1038/s41580-021-00417-y.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9(7): 671–675. https://doi.org/10.1038/nmeth.2089.
- Su, C. I., Chuang, Z. S., Shie, C. T., Wang, H. I., Kao, Y. T. and Yu, C. Y. (2024). A cis-acting ligase ribozyme generates circular RNA in vitro for ectopic protein functioning. Nat Commun. 15(1): 6607. https://doi.org/10.1038/s41467-024-51044-y.
Article Information
Publication history
Received: Mar 10, 2025
Accepted: Apr 16, 2025
Available online: May 5, 2025
Published: May 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
How to cite
Chuang, Z., Su, C., Wang, H. and Yu, C. (2025). RNA PolyA Tailing Assay to Qualitatively Analyze Circular RNA Manufacturing. Bio-protocol 15(10): e5310. DOI: 10.21769/BioProtoc.5310.
Category
Molecular Biology > RNA > RNA Modification
Biochemistry > RNA > RNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









