- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Optimized Protocol for DNA Extraction in Three Theobroma Species
Published: Vol 15, Iss 9, May 5, 2025 DOI: 10.21769/BioProtoc.5297 Views: 2043
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Optimized CTAB Method for Genomic DNA Extraction from Freshly-picked Pinnae of Fern, Adiantum capillus-veneris L.
Yi Shu [...] Fang Yu-Han
Jul 5, 2018 18032 Views

Safe DNA-extraction Protocol Suitable for Studying Tree-fungus Interactions
Susanna Keriö [...] Jared M. LeBoldus
Jun 5, 2020 8039 Views
Quantification of Botrytis cinerea Growth in Arabidopsis thaliana
Patricia Scholz [...] Athanas Guzha
Aug 20, 2023 2822 Views
Abstract
DNA extraction is a crucial step in molecular biology research, particularly for genetic and genomic analyses. These studies require a high concentration of high-quality DNA, which is often a challenge for underexplored species or when the available plant material consists of aged tissue. To address these challenges, the cetyltrimethylammonium bromide (CTAB)-based DNA extraction method has been optimized to improve efficiency and yield. The process begins with an overnight incubation of plant tissue macerated with liquid nitrogen in a solution containing a high concentration of CTAB (4%). Subsequently, the mixture undergoes two washes with chloroform: isoamyl alcohol. The nucleic acids are then precipitated using isopropanol, followed by a wash with 70% ethanol to ensure purity. Finally, the purified DNA is resuspended in ultrapure water. This optimized procedure produces high-quality DNA suitable for various downstream applications, including PCR and sequencing, even from older leaves of the three Theobroma species: T. cacao, T. bicolor, and T. grandiflorum. Additionally, this protocol significantly enhances throughput and allows for the parallel processing of a substantially larger number of samples compared to conventional techniques.
Key features
• An efficient CTAB-based DNA extraction protocol provides high-quality nucleic acids from older leaves by increasing CTAB concentration and incubation time in lysis buffer.
• Provides reliable yields in the three Theobroma species: T. cacao, T. bicolor, and T. grandiflorum.
• A high-throughput workflow reduces processing time and increases daily sample capacity, supporting large-scale genomic investigations.
Keywords: TheobromaGraphical overview
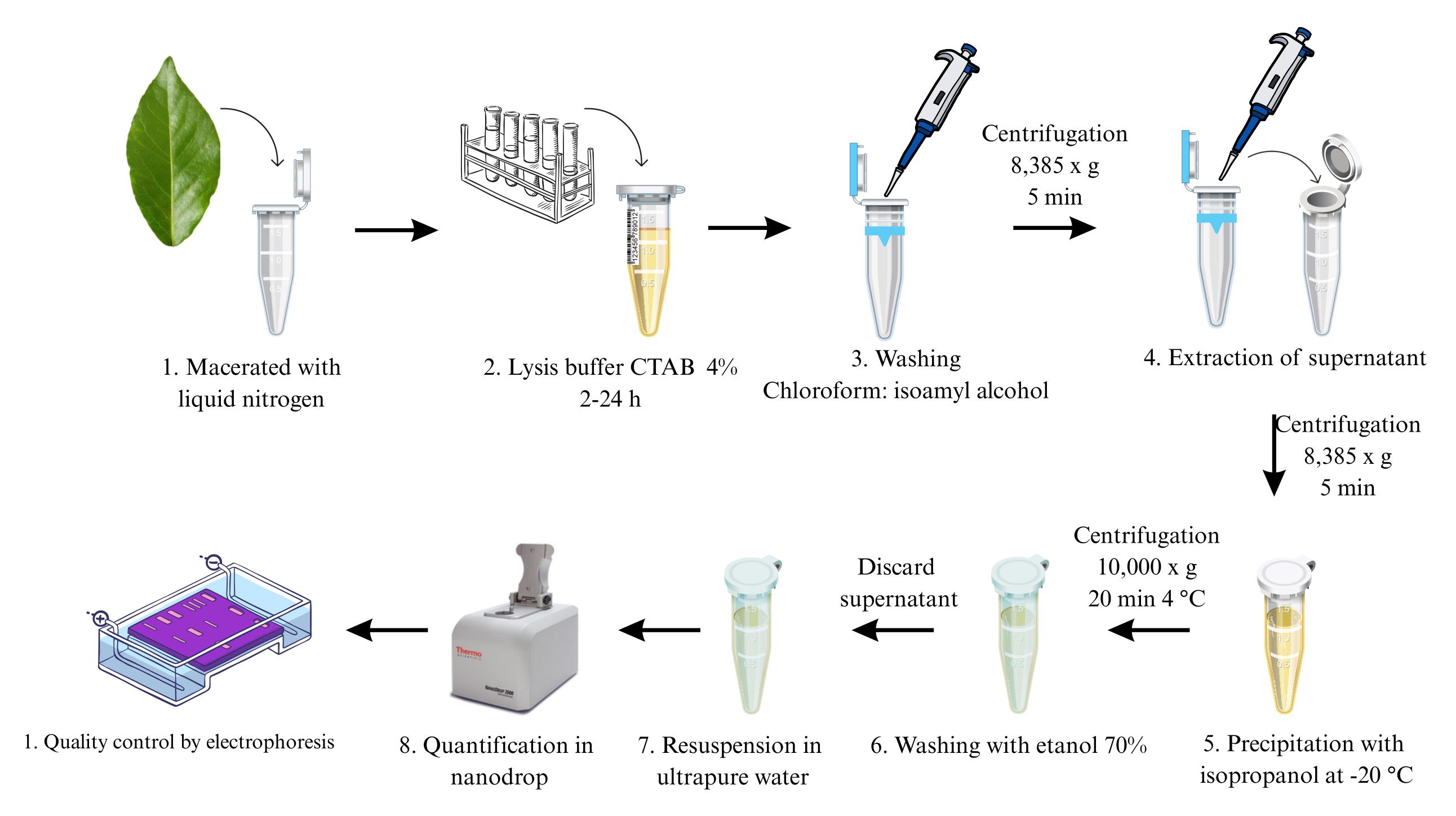
Extraction, quantification, and assessment of DNA integrity
Background
The extraction of high-quality DNA is a fundamental step in molecular biology and genomics. DNA isolation often requires high levels of concentration, purity, and integrity, particularly for library preparation and sequencing applications [1]. Despite significant advancements in DNA extraction protocols, obtaining high‐molecular‐weight DNA from challenging plant materials, such as older leaves, remains problematic [2], especially when dealing with underexplored species like T. grandiflorum and T. bicolor. Therefore, there is a need to evaluate protocols capable of extracting high-quality DNA from Theobroma species, which have promising applications. Furthermore, obtaining young tissue for DNA extraction is often impractical, particularly in native Theobroma species, which are frequently decades old and grow to considerable heights [3]. Traditional methodologies, such as the cetyltrimethylammonium bromide (CTAB)-based DNA extraction protocol, are widely used due to their adaptability and efficiency in isolating plant DNA. However, conventional CTAB protocols often yield suboptimal results when applied to samples with high levels of contaminants or structural complexity [4]. These limitations underscore the need for protocol modifications to improve DNA yield and purity, especially in underexplored species such as T. bicolor and T. grandiflorum, as well as in economically significant species like T. cacao. The protocol described here addresses these challenges by increasing the CTAB concentration in the lysis buffer and extending the incubation period. These adjustments enhance cell wall disruption and facilitate the removal of secondary metabolites, resulting in higher DNA yield and quality [5]. Additionally, the optimized protocol significantly enhances throughput, enabling the processing of a greater number of samples compared to conventional techniques. This makes it particularly suitable for large-scale genomic studies, such as comparative genomics and population genetics in cacao-related species. Beyond its use in Theobroma species, this protocol may also be adapted for other plant species with similar extraction challenges. By providing a more efficient and reliable method for isolating high-quality DNA, this protocol facilitates the exploration of genetic diversity and supports a range of molecular and genomic investigations.
Materials and reagents
Biological materials
1. Leaves of T. grandiflorum
2. Leaves of T. cacao
3. Leaves of T. bicolor
Reagents
1. Liquid nitrogen
2. β-mercapto-ethanol (BME) (Thermo Scientific, CAS: 60-24-2)
3. Chloroform (Merck, CAS: 67-66-3)
4. Isoamyl alcohol (CARLO ERBA, CAS: 123-51-3)
5. Isopropanol, AR purity (Honeywell, CAS: 67-63-0)
6. 70% ethanol, AR purity (Sigma-Aldrich, CAS: 64-17-5)
7. Ultrapure water
8. CTAB (Sigma-Aldrich, CAS: 57-09-0)
9. Sodium chloride (CARLO ERBA, CAS: 7647-14-5)
10. Ethylenediaminetetraacetic acid (EDTA) (PanReac-ITW Reagents, CAS: 6381-92-6)
11. Tris(hydroxymethyl)aminomethane (TRIS) (Thermo Fisher Scientific, CAS: 76-86-1)
12. CTAB 2%, 3%, 4% (Sigma-Aldrich, CAS: 57-09-0)
13. Proteinase K (Canvax, CAS: 39450-01-6)
14. Boric acid (Scharlau, CAS: 10043-35-3)
15. Ethidium bromide (Thermo Scientific, CAS: 1239-45-8)
16. Agarose (AMRESCO, LLC, CAS: 90-12-36-6)
17. HyperLadder 1 kb (Bioline, catalog number: BIO-33025)
18. Ethanol (Merck, catalog number: 100983)
Solutions
1. Lysis buffer CTAB 4% m/v (see Recipes)
2. CIA (see Recipes)
Recipes
1. Lysis buffer CTAB 4% m/v
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CTAB | 4% m/v | 5 g |
| EDTA | 0.02 M | 1.46 g |
| Sodium chloride | 1.4 M | 20.45 g |
| TRIS | 0.1 M | 3.028 g |
| Total | n/a | 250 mL |
2. CIA
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Chloroform | 96% (v/v) | 48 mL |
| Isoamyl alcohol | 4% (v/v) | 2 mL |
| Total | n/a | 50 mL |
Laboratory supplies
1. 2.0 mL Eppendorf tubes (Citotest, catalog number: 4610-1913)
2. 1.5 mL Eppendorf tubes (Citotest, catalog number:4610-1842)
3. 50 mL Falcon tubes (GenFollower, catalog number: CTB50YE)
4. Pipette tips (Thermo Scientific, model: Finnpipette F1)
Equipment
1. Pipettes (Thermo Scientific, model: Finnpipette F1, catalog numbers: 4641010N, 4641070N, 4641100N, and 4641120)
2. Stainless steel spatula (8") (VWR, catalog number: 82027-532)
3. Mortar (diameter: 100mm) and pestle (size: 117 mm) (DURAN, catalog number: 2120074)
4. High-speed centrifuge (Thermo Scientific, model: MicroCL 21R, catalog number: 75002470)
5. Refrigerator (4°C) (Panasonic Biomedical, model: MPR-721, catalog number: MPR-721-PA)
6. Oven (65°C) (Memmert, model: UN30, catalog number: UN30)
7. Magnetic stirrer (IKA, model: C-MAG HS 7, catalog number: 0020002690)
8. Electrophoresis system (Labnet International, model: ENDUROTM Horizontal Gel Box, catalog number: E1010-E)
9. Photodocumentation system (Labnet International, model: ENDUROTM GDS Touch, catalog number: GDS-1300)
Software and datasets
1. Canvas LMS (Versión 2025.4.10). The chart is available at the following link: https://www.canva.com/design/DAGkQWGQxUM/_eq90lI0a2AZSLoaquijpw/edit?utm_content=DAGkQWGQxUM&utm_campaign=designshare&utm_medium=link2&utm_source=sharebutton
2. RStudio (Posit Software, PBC, © 2009–2024)
3. ImageJ (Fiji, Version 1.52)
Procedure
A. Sample preparation
1. Collect approximately 100 mg of fresh plant tissue.
2. Carefully remove the main vein from the leaf tissue. After vein removal, weigh exactly 40 mg of the remaining leaf tissue and transfer it into a 2 mL Eppendorf tube.
3. Immediately freeze the tissue in liquid nitrogen to preserve DNA integrity. Thoroughly grind the frozen sample using a pre-chilled mortar and pestle until a fine powder is obtained.
Note: Ensure the tissue is thoroughly ground into a fine powder to maximize cell lysis and DNA yield.
B. DNA extraction
1. Transfer the ground tissue to a 2 mL microcentrifuge tube.
2. Add 650 μL of CTAB lysis buffer (4% prewarmed to 65 °C) as per experimental treatment (see Graphical overview).
Notes:
1. For old leaves, the CTAB concentration can be increased to 4% to enhance efficiency, as indicated in the protocol (Figure 1A).
2. Always prepare fresh CTAB buffer and add β-mercaptoethanol immediately before use to maintain reducing conditions.
3. Gently mix by inversion to ensure complete coverage of the tissue.
4. Incubate the tube at 65 °C for 120 min with occasional mixing (overnight for old leaves).
Note: When working with old or tough leaves, the initial lysis step may be shortened to 40 min. After this, remove the used buffer and replace it with 650 µL of fresh prewarmed CTAB buffer, followed by an additional 120 min incubation at 65 °C.
5. Add an equal volume (approximately 650 μL) of chloroform: isoamyl alcohol (24:1) to the tube.
6. Vortex briefly to form an emulsion and then centrifuge at 10,000× g for 10 min at room temperature.
7. Carefully transfer the clear, aqueous phase (upper layer) to a new 1.5 mL microcentrifuge tube, avoiding disturbance of the interphase.
Note: If the sample contains visible contaminants or pigments, repeating the CIA extraction step may significantly improve DNA purity, as mentioned for cacao leaves.
8. Add 1 volume of ice-cold isopropanol to the aqueous phase.
9. Incubate the mixture at -20 °C for at least 30 min (or overnight for maximum yield).
10. Centrifuge at 8,385× g for 5 min at 4 °C.
11. Carefully discard the supernatant and retain the pellet.
12. Rinse the DNA pellet with 500 μL of 70% ethanol.
Note: Repeat this step if necessary to ensure the removal of residual salts or impurities.
13. Centrifuge at 8,385× g for 5 min at 4 °C.
14. Discard the supernatant and briefly air-dry the pellet, ensuring it does not over-dry (which can make DNA difficult to dissolve).
Note: Air-drying is considered complete when no visible droplets of isopropanol remain inside the Eppendorf tube and the pellet is not completely dry or tightly stuck to the tube walls.
15. Dissolve the pellet in 50–100 μL of TE buffer or nuclease-free water.
16. Store the DNA at -20 °C for long-term preservation.
Note: Evaluation of DNA integrity on agarose gel (Figure 2C).

Figure 1. Collected samples. (A) Theobroma grandiflorum. (B) Theobroma bicolor. (C) Theobroma cacao 1. (D) Theobroma cacao 2.
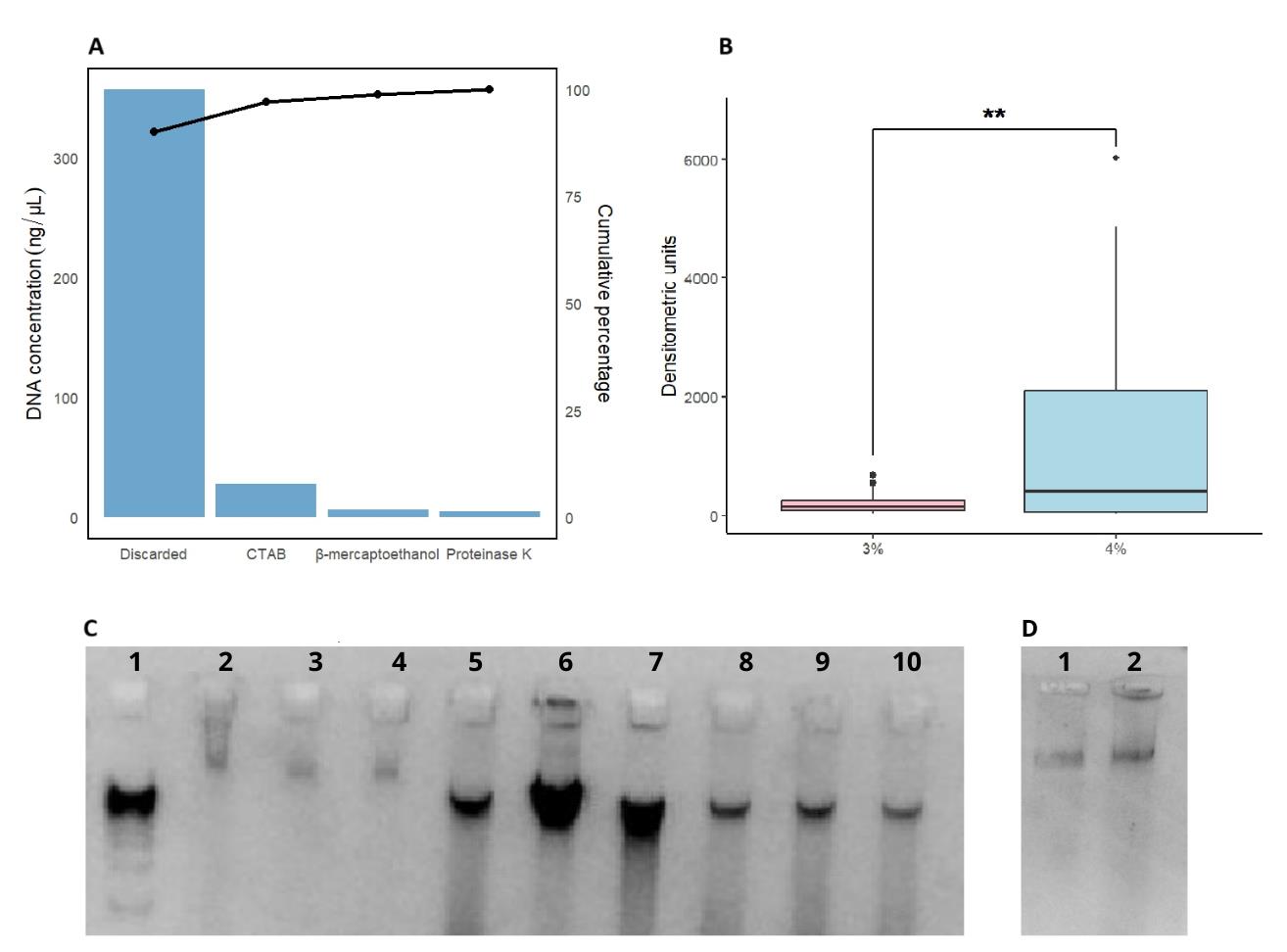
Figure 2. Comparative analysis of efficiency, yield, and quality of DNA extracted using different CTAB protocols in Theobroma species. (A) Pareto chart showing the DNA concentration (ng/µL) obtained using different extraction protocols (discarded, CTAB, β-mercaptoethanol, and proteinase K). The bars represent the individual contribution of each protocol, while the line indicates the cumulative percentage of extraction efficiency. (B) Comparison of DNA yield between CTAB concentrations (3% and 4%), evaluated by densitometric units on a 1% agarose gel. Asterisks (**) denote statistically significant differences between the two groups. (C) Agarose gel electrophoresis showing the integrity and quality of extracted DNA; (1) 1 kb DNA ladder, (2, 3, 4) DNA from Theobroma cacao, (5, 6, 7) DNA from Theobroma grandiflorum, (8, 9, 10) DNA from Theobroma bicolor. (D) Agarose gel electrophoresis showing DNA from Theobroma cacao subjected to a double CIA extraction step, (1,2) DNA from Theobroma cacao.
Data analysis
DNA extraction was quantified using a Nanodrop spectrophotometer and image analysis of electrophoresis results, employing the ImageJ software to quantify densitometric units. The latter values were more consistent with the experimental results. Subsequently, a study of variance (ANOVA) was conducted to determine which of the 48 proposed treatments (see Table 1 for full treatment list and results) exhibited significant differences (p < 0.05). The analysis identified notable variations in the concentration of CTAB. Tukey’s post hoc test indicated that the highest DNA yield was obtained using the highest CTAB concentration (4%).
Table 1. Experimental treatments and densitometric unit measurements across different species and conditions
| Treat. | Rep. | Species | SDS | CTAB | Proteinase K | Densitometric units |
|---|---|---|---|---|---|---|
| 1 | 1 | Genotype 1 | 10% | 4% | 2 mg/mL | 1389.18 |
| 2 | 1 | Genotype 1 | 10% | 3% | - | 105.61 |
| 3 | 1 | Genotype 1 | - | 4% | 2 mg/mL | 27.61 |
| 4 | 1 | Genotype 1 | - | 3% | - | 233.09 |
| 5 | 1 | Genotype 2 | 10% | 4% | 2 mg/mL | 25.95 |
| 6 | 1 | Genotype 2 | 10% | 3% | - | 97.02 |
| 7 | 1 | Genotype 2 | - | 4% | 2 mg/mL | 30.66 |
| 8 | 1 | Genotype 2 | - | 3% | - | 185.85 |
| 9 | 1 | Genotype 3 | 10% | 4% | 2 mg/mL | 50.49 |
| 10 | 1 | Genotype 3 | 10% | 3% | - | 679.06 |
| 11 | 1 | Genotype 3 | - | 4% | 2 mg/mL | 6022.61 |
| 12 | 1 | Genotype 3 | - | 3% | - | 534.1 |
| 13 | 1 | Genotype 4 | 10% | 4% | 2 mg/mL | 452.08 |
| 14 | 1 | Genotype 4 | 10% | 3% | - | 203.97 |
| 15 | 1 | Genotype 4 | - | 4% | 2 mg/mL | 344.18 |
| 16 | 1 | Genotype 4 | - | 3% | - | 36.95 |
| 17 | 2 | Genotype 1 | 10% | 4% | 2 mg/mL | 121.26 |
| 18 | 2 | Genotype 1 | 10% | 3% | - | 58.36 |
| 19 | 2 | Genotype 1 | - | 4% | 2 mg/mL | 3888.75 |
| 20 | 2 | Genotype 1 | - | 3% | - | 66.61 |
| 21 | 2 | Genotype 2 | 10% | 4% | 2 mg/mL | 2669.32 |
| 22 | 2 | Genotype 2 | 10% | 3% | - | 229.09 |
| 23 | 2 | Genotype 2 | - | 4% | 2 mg/mL | 37.95 |
| 24 | 2 | Genotype 2 | - | 3% | - | 179.44 |
| 25 | 2 | Genotype 3 | 10% | 4% | 2 mg/mL | 49.66 |
| 26 | 2 | Genotype 3 | 10% | 3% | - | 42.66 |
| 27 | 2 | Genotype 3 | - | 4% | 2 mg/mL | 1124.43 |
| 28 | 2 | Genotype 3 | - | 3% | - | 542.46 |
| 29 | 2 | Genotype 4 | 10% | 4% | 2 mg/mL | 3146.08 |
| 30 | 2 | Genotype 4 | 10% | 3% | - | 276.68 |
| 31 | 2 | Genotype 4 | - | 4% | 2 mg/mL | 4863.78 |
| 32 | 2 | Genotype 4 | - | 3% | - | 490.51 |
| 33 | 3 | Genotype 1 | 10% | 4% | 2 mg/mL | 23.54 |
| 34 | 3 | Genotype 1 | 10% | 3% | - | 78.61 |
| 35 | 3 | Genotype 1 | - | 4% | 2 mg/mL | 282.92 |
| 36 | 3 | Genotype 1 | - | 3% | - | 38.78 |
| 37 | 3 | Genotype 2 | 10% | 4% | 2 mg/mL | 56.61 |
| 38 | 3 | Genotype 2 | 10% | 3% | - | 110.44 |
| 39 | 3 | Genotype 2 | - | 4% | 2 mg/mL | 108.85 |
| 40 | 3 | Genotype 2 | - | 3% | - | 76.73 |
| 41 | 3 | Genotype 3 | 10% | 4% | 2 mg/mL | 657.7 |
| 42 | 3 | Genotype 3 | 10% | 3% | - | 220.85 |
| 43 | 3 | Genotype 3 | - | 4% | 2 mg/mL | 1867.45 |
| 44 | 3 | Genotype 3 | - | 3% | - | 553.51 |
| 45 | 3 | Genotype 4 | 10% | 4% | 2 mg/mL | 1904.88 |
| 46 | 3 | Genotype 4 | 10% | 3% | - | 93.61 |
| 47 | 3 | Genotype 4 | - | 4% | 2 mg/mL | 3914.41 |
| 48 | 3 | Genotype 4 | - | 3% | - | 31.07 |
Genotype 1, T. cacao 1; genotype 2, T. cacao 2; genotype 3, T. grandiflorum; genotype 4, T. bicolor.
Validation of protocol
This protocol is currently being used for the phenotypic and molecular characterization of Theobroma grandiflorum diversity in Caquetá, Colombia (Velasquez-Vasconez et al. [6]; https://doi.org/10.1016/j.ijagro.2025.100034), as part of the project funded by the Ministry of Science, Technology and Innovation – MINCIENCIAS, grant number 106866 and program number 106735.
Acknowledgments
Conceptualization, A.F.R.E.; Investigation, A.F.R.E., B.A.T.C., and M.I.C.Z.; Writing—Original Draft, A.F.R.E. and B.A.T.C.; Writing—Review & Editing, A.F.R.E. and P.A.V.-V.; Funding acquisition, P.A.V.-V. and J.C.Z.A.; Supervision, P.A.V.-V. and J.C.Z.A.
This research was funded by the Ministerio de Ciencia, Tecnología e Innovación (MINCIENCIAS), grant number 106866 and program number 106735. This protocol was used in [6].
Competing interests
The authors declare no conflicts of interest.
References
- Alves, R. M., de Abreu, V. A. C., Oliveira, R. P., Almeida, J. V. d. A., de Oliveira, M. d. M., Silva, S. R., Paschoal, A. R., de Almeida, S. S., de Souza, P. A. F., Ferro, J. A., et al. (2024). Genomic decoding of Theobroma grandiflorum (cupuassu) at chromosomal scale: evolutionary insights for horticultural innovation. GigaScience. 13: giae027. https://doi.org/10.1093/gigascience/giae027
- Kang, M., Chanderbali, A., Lee, S., Soltis, D. E., Soltis, P. S. and Kim, S. (2023). High‐molecular‐weight DNA extraction for long‐read sequencing of plant genomes: An optimization of standard methods. Appl Plant Sci. 11(3): e11528. https://doi.org/10.1002/aps3.11528
- Lagneaux, E., Andreotti, F. and Neher, C. M. (2021). Cacao, copoazu and macambo: Exploring Theobroma diversity in smallholder agroforestry systems of the Peruvian Amazon. Agrofor Syst. 95(7): 1359–1368. https://doi.org/10.1007/s10457-021-00610-0
- Rivas-García, T., Córdova-Pérez, D., Arredondo-Espinoza, R. C., Murillo-Amador, B., González-Estrada, R. R., Reyes-Pérez, J. J., Torres-Rodriguez, J. A., Martínez-Camacho, R. A. and Alejandre-Rosas, J. A. (2024). Optimization of DNA Extraction for ITS/U4U3 analysis of Rhipsalis baccifera. J Prof Assoc Cactus Dev. 26: 107–118. https://doi.org/10.56890/jpacd.v26i.563
- Martínez-López, A. A., Lesher, J. M., Jiménez-García, M. E., et al. (2013). Comparación de tres métodos para la extracción de ARN total a partir de hojas de cacao (Comparison of three total RNA extraction methods from cacao leaves). Biotecnología Vegetal. 13(2): 93–98.
- Velasquez-Vasconez, P. A., Castro-Zambrano, M. I., Rodríguez-Cabal, H. A., Castro, D., Arbelaez, L. and Zambrano, J. C. (2025). Exploring Theobroma grandiflorum diversity to improve sustainability in smallholdings across Caquetá, Colombia. Ital J Agron. 20(2): 100034. https://doi.org/10.1016/j.ijagro.2025.100034
Article Information
Publication history
Received: Jan 22, 2025
Accepted: Mar 30, 2025
Available online: Apr 20, 2025
Published: May 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Riascos-España, A. F., Cuastumal, B. T. A., Zambrano, M. C. I., Arteaga, J. Z. C. and Velasquez-Vasconez, P. A. (2025). Optimized Protocol for DNA Extraction in Three Theobroma Species. Bio-protocol 15(9): e5297. DOI: 10.21769/BioProtoc.5297.
Category
Plant Science > Plant molecular biology > DNA > DNA extraction
Molecular Biology > DNA > DNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










