- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Baculovirus and Plasmid Vector-Mediated Gene Transfer in Daphnia magna Cells in Primary Embryonic In Vitro Cultures in an Optimized Microenvironment: Methods and Protocols
Published: Vol 15, Iss 7, Apr 5, 2025 DOI: 10.21769/BioProtoc.5266 Views: 1592
Reviewed by: Alberto RissoneSrestha DasguptaKeisuke Tabata

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Endothelin-1-Induced Persistent Ischemia in a Chicken Embryo Model
Neha Kumari [...] Syed S. Raza
Sep 5, 2024 1441 Views
Using the Sleeping Beauty Transposon System for Doxycycline-inducible Gene Expression in RAW264.7 Macrophage Cells to Study Phagocytosis
Parsa Kamali and Gregory D. Fairn
Feb 5, 2025 1839 Views
In Vivo Retroviral Transduction of Cardiac Myofibroblasts Using Intramyocardial Injection Immediately Post-myocardial Infarction
Satsuki Ono [...] Michio Nakaya
Nov 20, 2025 1368 Views
Abstract
Daphnia magna is a well-established model organism in ecotoxicology, environmental monitoring, and genetics due to its sensitivity to pollutants, its pivotal role in freshwater ecosystems, and its well-characterized genome. Despite its extensive use in these fields, there is a notable lack of established protocols for developing primary cell culture systems and conducting transgenic studies in Daphnia spp. This study addresses these gaps by optimizing a medium and standardizing protocols for primary cell culture and transgenic experiments in D. magna. Primary cell cultures were established from both D. magna embryos and whole organisms, with medium optimization verified using XTT assay. Cell viability was sustained for over two months using a modified Schneider’s insect medium enriched with FBS, glucose, MEM vitamin mix, and selenium. DNA replication and cell proliferation were confirmed through BrdU labeling. Both mechanical and enzymatic passaging methods were compared, resulting in 20% and 10% cell attachment, respectively. For transgenic applications, this study successfully standardized plasmid-mediated lipofection and baculovirus-mediated transduction, achieving success rates of 52% and 45%. These findings represent a pioneering effort in D. magna embryonic cell culture, offering a reliable in vitro platform for future biological research, including ecotoxicological and epigenetic investigations. The established protocols and optimized cell culture medium have significant implications for advancing crustacean cell line research and transgenic model development, enhancing our understanding of biological processes in controlled laboratory environments.
Key features
• Optimized primary embryonic cell culture system for Daphnia magna ensures extended viability and minimized contamination compared to adult-derived cultures.
• Modified Schneider’s insect medium (MSIM) with precise supplement concentrations enhances Daphnia cell viability, supporting applications in toxicity testing and genetic modifications.
• Efficient transfection and baculovirus-based transduction protocols enable robust transgene expression, validated through fluorescence microscopy and promoter-specific activity.
• Comparative subculture techniques demonstrate improved reattachment rates with gentle pipetting, providing insights into non-enzymatic handling for fragile aquatic cells.
Keywords: Daphnia magnaGraphical overview
Primary cell culture development from Daphnia magna embryos
Background
Crustaceans, encompassing ~67,000 species, include Daphnia, a keystone species in freshwater ecosystems known for their adaptability, parthenogenetic reproduction, and utility in evolutionary and ecotoxicological studies. Despite Daphnia’s potential as a model organism, the lack of established cell lines limits in vitro research and pathogen studies [1]. Challenges in crustacean cell culture primarily stem from the absence of suitable media [2,3], which has prompted modifications of commercially available media to optimize cell viability and microenvironments.
Previous attempts to develop crustacean cell lines have faced poor cell survival [1–3]. Notable successes include hybrid lines like PmLyO-Sf9 [4], but efforts for Daphnia magna have yielded limited results [5,6], emphasizing the need for optimized protocols. Advances in Daphnia genomic tools and transgenic methods, including RNAi [7], targeted mutagenesis [8], and genome editing [9], have paved the way for genetic manipulation. Cellfectin has shown higher efficiency than lipofectamine in Arthropoda [10]. The EF1α-DsRed plasmid, modified as pRCS21-EF1α1(UTR), achieved 0.67% transgenic D. magna via microinjection [11]. Baculovirus expression systems, mainly Autographa californica MNPV, enhance protein expression using polyhedrin or p10 promoters. However, challenges persist in transfection and transduction due to low cell numbers and attachment efficiency in novel cultures.
This study establishes a standardized protocol for developing primary D. magna embryonic cell cultures using a modified medium to enhance cell survival. Transfection and transduction protocols are optimized to create an immortalized D. magna cell line, offering broad applications in crustacean research and biotechnology.
Materials and reagents
Biological materials
1. Daphnia magna: Daphnia Research Facility (DRF), National Centre for Aquatic Animal Health (NCAAH; https://www.ncaah.ac.in), Cochin University of Science and Technology (CUSAT; https://cusat.ac.in/), Kerala, India
2. pCS-EF1α1-DSRed2 vector, obtained from Dr. Hajime Watanabe, Osaka University, Japan [10]
3. Recombinant baculovirus expression system (BacIe1-GFP and BacP2-GFP), developed in NCAAH, CUSAT [2,3]
4. SF9 cells, maintained at NCAAH, CUSAT
Reagents
1. MEM vitamins (MEM) (Gibco, catalog number: 11120052)
2. 1× PBS (Gibco, catalog number: 10010023)
3. Schneider’s insect medium (Gibco, catalog number: 21720024)
4. Fetal bovine serum (FBS) (Gibco, catalog number: A5670701)
5. 1× Leibovitz’s L-15 medium (L-15) (Sigma-Aldrich, catalog number: L1518)
6. TNM-FH insect medium (TNMFH) (Sigma-Aldrich, catalog number: T1032)
7. Grace’s insect medium (GIM) (Sigma-Aldrich, catalog number: G9771)
8. Minimal Essential Medium (MEM) (Sigma-Aldrich, catalog number: 56416C)
9. Dulbecco’s modified Eagle medium (DMEM) (Sigma-Aldrich, catalog number: D6546)
10. Roswell Park Memorial Institute Medium (RPMI) (Sigma-Aldrich, catalog number: R8758)
11. XTT (Invitrogen, catalog number: X6493)
12. Glucose, ≥99.5% (Sigma-Aldrich, catalog number: G7021)
13. Trehalose, ≥99% (Sigma-Aldrich, catalog number: T0167)
14. Sucrose, ≥99% (Sigma-Aldrich, catalog number: S5390)
15. Amino acids mix (MEM) (Gibco, catalog number: 11130036)
16. Cholesterol (Sigma-Aldrich, catalog number: C4951)
17. Sodium selenite, ≥98% (Sigma-Aldrich, catalog number: S5261)
18. Trypsin-EDTA solution (Sigma-Aldrich, catalog number: T4049)
19. Acridine orange (AO) (Invitrogen, catalog number: A1301)
20. Ethidium bromide (EB) (Invitrogen, catalog number: 15585011)
21. BrdU (5-Bromo-2’-deoxyuridine) (TCI, catalog number: B1575)
22. Mouse monoclonal anti-BrdU antibody (Sigma-Aldrich, catalog number: SAB4700630)
23. Alexa Fluor 488 (Jackson ImmunoResearch Labs, catalog number: 115-545-003)
24. DAPI (Thermo Fisher Scientific, catalog number: D3571)
25. Cellfectin (CellfectinTM II) (Invitrogen, catalog number: 10362100)
26. Selenium dioxide (SeO2) (Sigma-Aldrich, catalog number: 204315)
27. Penicillin (100 IU/mL) and streptomycin (100 μg/mL) solution (Roche, Merk, catalog number: 11074440001)
Solutions
1. ADaM media (see Recipes)
2. 0.05% sodium hypochlorite (see Recipes)
3. 70% ethanol (see Recipes)
4. 2 M HCl (see Recipes)
5. 0.1 M sodium borate (see Recipes)
6. 0.2% Triton X-100 (see Recipes)
7. 3% BSA (see Recipes)
8. 0.05% trypsin-EDTA (see Recipes)
Recipes
1. ADaM media
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CaCl2·2H2O | 0.2705 g/L | 23 mL of Stock A (117.6 g/L CaCl2·2H2O) |
| NaHCO3 | 0.0554 g/L | 22 mL of Stock B (25.2 g/L NaHCO3) |
| SeO2 | 7 μg/L | 1 mL of Stock C (0.07 g/L SeO2) |
| Distilled water | Up to 10 L |
2. 0.05% sodium hypochlorite
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sodium hypochlorite powder | 0.05% (w/v) | 0.05 g (for 10 mL) |
| Distilled water | n/a | Up to 10 mL |
| Total | n/a | 10 mL |
3. 70% ethanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanol (100%) | 70% (v/v) | 7 mL |
| Distilled water | n/a | 3 mL |
| Total | n/a | 10 mL |
4. 2 M HCl
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HCl (12 M) | 2 M | 1.67 mL |
| Distilled water | n/a | 8.33 mL |
| Total | n/a | 10 mL |
5. Sodium borate (0.1 M)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sodium borate | 0.1 M | 0.382 g |
| Distilled water | n/a | 9.618 mL |
| Total | n/a | 10 mL |
6. 0.2% Triton X-100
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Triton X-100 | 0.2% | 20 mg |
| Distilled water | n/a | 10 mL |
| Total | n/a | 10 mL |
7. 3% BSA
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 3% BSA | 3% | 0.3 g |
| Distilled water | n/a | 10 mL |
| Total | n/a | 10 mL |
8. 0.05% trypsin-EDTA
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Trypsin-EDTA stock | 0.25% | 2 mL |
| Distilled water | n/a | 8 mL |
| Total | n/a | 10 mL |
Laboratory supplies
1. 1.5 mL microcentrifuge tubes (MCTs) (Tarson, catalog number: 500010)
2. 2 mL microcentrifuge tubes (MCTs) (Tarson, catalog number: 500020)
3. Needles (Sigma-Aldrich, catalog number: Z117013)
4. Glass homogenizer (Sigma-Aldrich, catalog number: D8938)
5. 96-well plate (TPP, catalog number: Z707902)
6. 24-well plate (TPP, catalog number: Z707791)
7. Coverslips (Sigma-Aldrich, catalog number: Z355445)
8. Pipette tips: 1–10, 10–100, 100–1,000 μL (Sigma-Aldrich, catalog numbers: Z678295, Z678325, AXYT1000CR)
Equipment
1. Fluorescent microscope (Invitrogen, model: AMF7000, EVOSTM M7000 Imaging System)
2. Centrifuge (Eppendorf, model: 5424R, catalog number: 5404000138)
3. Dark incubator, 28 °C (Thermo Fisher Scientific, catalog number: 3120, model: Forma Series II)
4. Laminar airflow (Thermo Scientific, catalog number: 51025413, MSC-AdvantageTM Class II Biological Safety Cabinets)
Software and datasets
1. SPSS® software (version 21; SPSS Inc., Chicago, IL, USA)
Procedure
A. Establishing primary cell cultures from Daphnia individuals and embryos
1. Collection and selection of specimens
a. Collect approximately 80 gravid female Daphnia carrying embryos raised from a single clone into a 1.5 mL MCT (Figure 1A).
Note: Select individuals with embryos before clear morphological features (cell differentiation) become visible (Figure 1B).
c. Individual Daphniacan be observed under a microscope on a microscopic slide to assess the morphological features of the embryos. Transfer selected gravid females to 1.5 mL MCTs containing fresh ADaM media using a plastic dropper pipette.
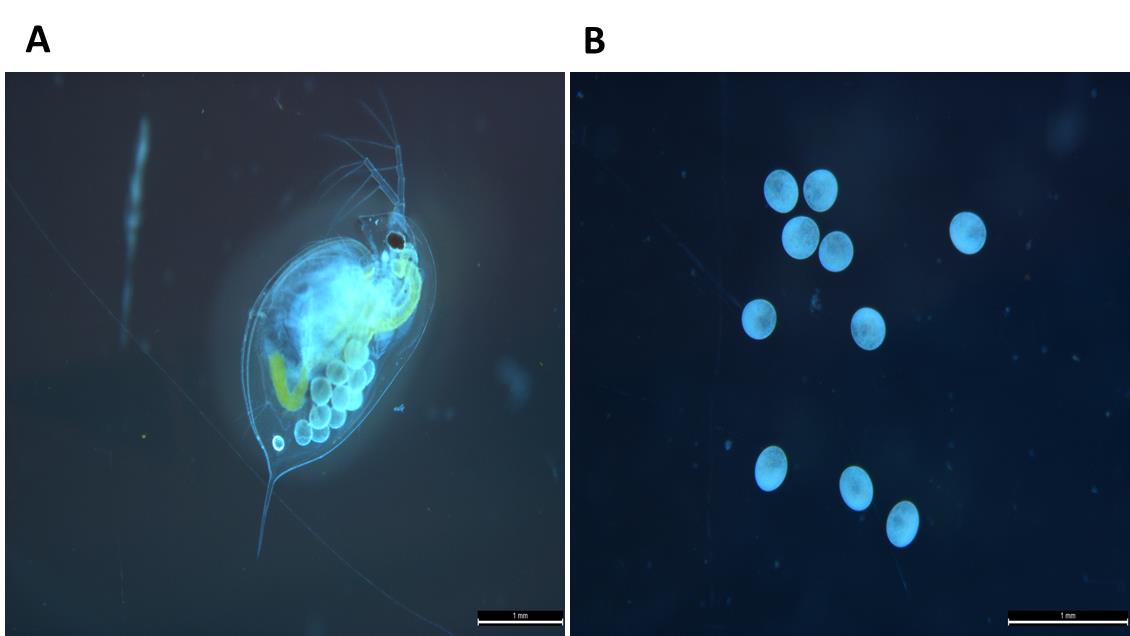
Figure 1. Experimental animal Daphnia magna. (A) Adult female Daphnia magna exhibiting parthenogenetic embryos within the brood chamber. (B) D. magna embryos used for developing primary embryonic cell culture. Scale bar: 1 mm.
2. Surface sterilization of Daphnia
a. Immerse gravid females sequentially in the following solutions to sterilize their surface: 0.05% sodium hypochlorite for 30 s; 1× PBS for 50 s; 70% ethanol for 30 s; 1× PBS for 50 s; serum-free Schneider's insect medium for 1 min; and 1× PBS for 10 s.
Critical: Ensure precise timing for each step to avoid overexposure, which may harm the samples.
3. Isolation of embryos
a. Aseptically remove embryos from the brood chamber of sterilized gravid females using two sterile syringe needles:
i. Position one needle anteriorly below the heart to stabilize the Daphnia.
ii. Gently press the other needle over the brood chamber to release intact embryos.
Critical: Avoid excessive pressure to prevent damage to the embryos.
b. Transfer 800–1,000 embryos into a 1.5 mL MCT containing freshly prepared and filtered 0.05% sodium hypochlorite solution.
4. Sterilization and washing of embryos
a. Sterilize embryos by repeating step A2.
b. Perform a final thorough wash with Schneider's insect medium (pH 6.8) without FBS.
5. Preparation of embryonic cell suspension
a. Transfer sterilized embryos to a fresh MCT containing Schneider’s insect medium.
b. Gently macerate embryos using a sterile homogenizer. Move the homogenizer up and down without twisting or touching the bottom of the MCT.
Critical: Avoid excessive fragmentation to preserve the integrity of cell clusters.
c. Centrifuge the homogenized suspension at 1,000× g for 5 min.
d. Resuspend the cell pellet in 500 μL of fresh Schneider's insect medium containing 10% FBS andantibioticsby adding 0.5–1 mL of penicillin-streptomycin solution to 100 mL of Schneider's insect medium [penicillin (100 IU/mL) and streptomycin (100 μg/mL) solution].
6. Seeding and maintenance of embryonic primary cell cultures
a. Seed 100 μL of cell suspension (~12,000 cells/well) into each well of a 96-well plate.
b. Incubate the plate at 28 °C in a dark incubator.
c. Change the cell culture medium weekly as follows:
i. After three weeks, transfer half of the existing medium into a new 96-well plate.
ii. Add an equal volume of fresh medium to both wells.
Note: Minimize frequent medium changes to maintain culture stability and reduce disruptions to cell growth and adherence.
7. Establishing primary cell cultures from whole Daphnia
a. Collect 30–60 Daphnia individuals without embryos and transfer them to fresh ADaM media.
b. Starve the animals for 24 h.
c. Sterilize the animals using the same washing steps as described in step A2.
d. Homogenize the sterilized animals with minimal pressure to avoid excessive fragmentation.
Critical: Preserve the integrity of the cells during homogenization.
e. Centrifuge the resulting suspension at 1,000× g for 5 min.
f. Resuspend the cell pellet in Schneider's insect medium containing FBS (10%) and antibiotics [penicillin (100 IU/mL) and streptomycin (100 μg/mL)].
8. Seeding and maintenance of whole-body primary cell cultures
a. Seed 50–100 μL of cell suspension into a 96-well plate.
b. Incubate the plate at 28 °C in a dark incubator.
c. Change the cell culture medium weekly:
i. Transfer half of the existing medium into a new 96-well plate.
ii. Add an equal volume of fresh medium to both wells.
Critical: Ensure precise timing during sterilization steps. Minimize mechanical stress during embryo and animal homogenization.Avoid frequent medium changes during the initial culture period to maintain cell health.
Troubleshooting:
a. If cell adherence or growth is poor, confirm the pH of the medium and reduce the frequency of medium changes.
b. D. magna embryos (Figure 2A) were found to be the ideal source for cell culture development compared to adult animals (Figure 2B). Even though using whole animal cells was relatively easier for cell culture, we faced regular contamination and high debris (parts of the Daphnia body) in the culture. In contrast, embryonic cell cultures maintained a healthy state for almost two months (Figure 2C).
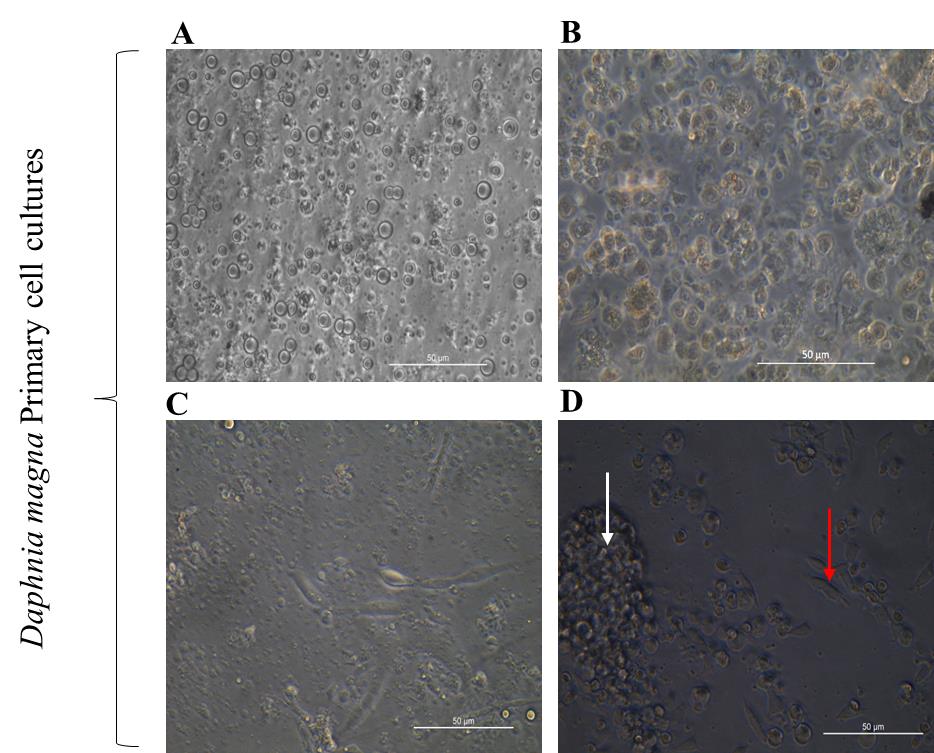
Figure 2. Primary cell cultures developed from Daphniamagna. (A) 24-h-old primary cell culture in modified Schneider’s insect medium (MSIM) derived from D. magna embryos. (B) 24-h-old primary culture developed from the whole body of D. magna in MSIM. (C) Two-month-old embryonic cells in MSIM. (D) One-week-old embryonic cells adhered in MSIM, with a white arrow denoting cell aggregates and a red arrow indicating attached cells. Scale bars: 50 μm.
B. Screening of suitable media for the primary cell culture developed from D. magna
Overview of the optimization process
The optimization of the medium involves a systematic, stepwise selection process conducted over seven phases of metabolic activity–based scoring using XTT assays. The basic principle of this assay relies on the cleavage of a yellow tetrazolium salt into an orange-colored formazan by the mitochondrial enzyme, which functions in only metabolically active cells. Each phase utilizes 96-well plates, with each well seeded with ~12,000 cells; triplicates of the samples are analyzed.
Phase 1: Screening basal media
1. Perform the XTT assay to evaluate basal media for supporting maximum cell viability.
2. Use seven commercially available tissue culture media (with 20% FBS):
• 1× Leibovitz’s L-15 Medium (1× L-15)
• Schneider’s Insect Medium (SIM)
• TNM-FH Insect Medium (TNMFH)
• Grace’s Insect Medium (GIM)
• Minimal Essential Medium (MEM)
• Dulbecco’s Modified Eagle Medium (DMEM)
• Roswell Park Memorial Institute Medium (RPMI)
In our study, SIM was identified as the most effective medium after this screening, so it was selected for further fine-tuning (Figure 3A).
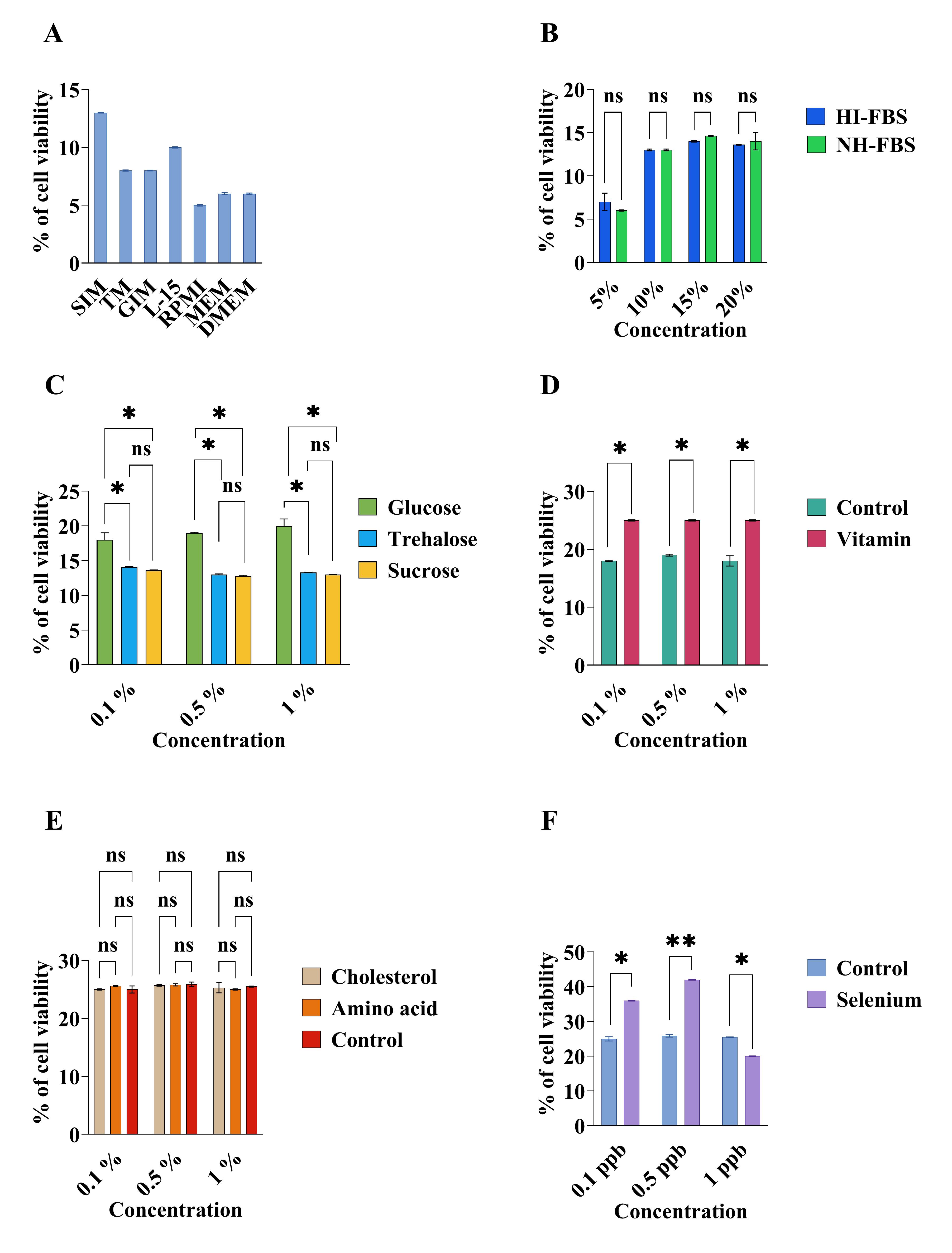
Figure 3. Comparison and optimization of media compositions for Daphnia magna primary embryonic cell culture by XTT assay. (A) Comparison of basal medium. (B) Effects of heat-inactivated (HI-FBS) and non-heat-inactivated FBS (NH-FBS). (C) Effects of carbohydrates. (D) Effects of vitamins. (E) Effects of amino acids and cholesterol. (F) Effects of selenium. The data were explored using a two-way analysis of variance followed by Tukey’s post hoc test. The experiment was conducted in triplicates, and the error bars represent the standard deviation of the samples. The asterisk (*) indicates a significant difference among the sample.
Phase 2: Optimizing FBS concentration
1. Use SIM without FBS as the basal medium.
2. Test varying concentrations of FBS (5%, 10%, 15%, and 20%).
3. Assess the impact of heat-inactivated FBS (H-FBS) versus non-heat-inactivated FBS (NH-FBS).
In our case, no differences between H-FBS and NH-FBS were found for Daphnia cells, and 10% FBS was enough for cells. As such, SIM with 10% FBS was selected for further fine-tuning (Figure 3B).
Phase 3: Screening carbohydrate concentrations
1. Evaluate glucose, trehalose, and sucrose at 0.1%, 0.5%, and 1% concentrations.
In our case, 0.1% glucosewas selected, and SIM with 10% FBS and 0.1% glucose was selected for further fine-tuning (Figure 3C).
Phase 4: Evaluating vitamin mix concentrations
1. Test varying concentrations of MEM vitamins (0.1%, 0.5%, and 1%).
In our case, 0.1% vitamins were selected;the selected mediumwas SIM with 10% FBS, 0.1% glucose, and 0.1% vitamins (Figure 3D).
Phase 5: Optimizing amino acids mix concentrations
1. Assess the performance of varying concentrations of an amino acids mix.
In our case, no significant effects of amino acids on cells were found, so amino acidswere not used for further selection.The final medium after this screening was SIM with 10% FBS, 0.1% glucose, and 0.1% vitamins (Figure 3E).
Phase 6: Testing cholesterol concentrations
1. Evaluate cholesterol at 0.1%, 0.5%, and 1% concentrations.
In our case, no significant effects were found. So, cholesterol was not added (Figure 3E).
Phase 7: Evaluating selenium concentrations
1. Test sodium selenite at 0.1, 0.5, and 1 ppb concentrations.
In our case, selenium at 0.5 ppb had the highest significance. Thus, SIM with 10% FBS, 0.1% glucose, 0.1% vitamins, and 0.5 ppb selenium was selected for cell culture development and further experiment (Figure 3F).
Final optimized medium
The final optimized medium, designated as modified Schneider’s insect medium (MSIM), contains 10% FBS, 0.1% glucose, 0.1% vitamins, and 0.5 ppb selenium, and incorporates the best-performing concentrations of all tested components.
C. Subculture techniques
1. Mechanical method
a. Use 1-month-old cell culture triplicates (Figure 4A).
b. Wash cells with PBS and gently triturate using a pipette (6–8 cycles).
c. Resuspend detached cells in fresh MSIM and maintain at 28 °C.
d. Monitor viability periodically.
2. Enzymatic method
a. Use a 1-month-old cell culture (Figure 4B).
b. Wash cells with PBS and treat with 0.05% trypsin-EDTA.
c. Incubate at RT for 1 min, observe detachment, and neutralize trypsin with MSIM containing 10% FBS.
d. Subculture cells and maintain at 28 °C.
In our case, a maximum of 15% of cellsinitiated reattachment to the surface after one week. The first attachment was observed on the third day of passaging by pipetting (Figure 4C), whereas a maximum of 10% of cells were reattached following trypsinization after one week (Figure 4D). After post hoc analysis, significant differenceswere found between trypsinization and pipetting (p = 0.00005) (Figure 4E). Over time, the attached cells progressively detached, and the number of viable cells steadily declined, culminating in complete cell death within one month after passaging.
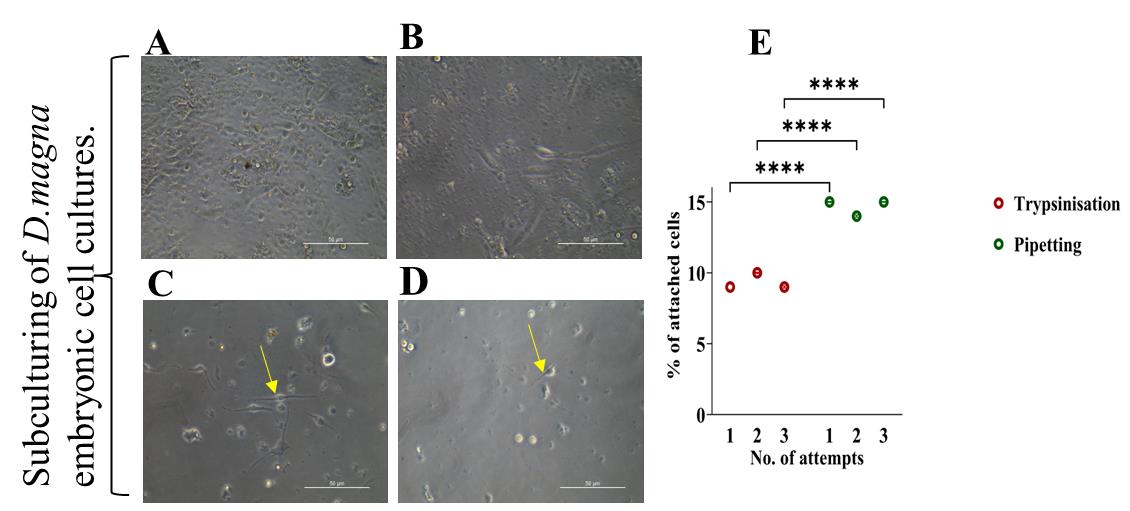
Figure 4. Comparison of passaging in primary cell cultures (in triplicate) developed from Daphnia magna. (A) One-month-old primary embryonic cell culture just before passaging by pipetting. (B) One-month-old primary embryonic cells just before passaging by trypsinization. (C) Cells after one week of passaging by pipetting. The yellow arrow indicates reattached cells. (D) Cells after one week of passaging by trypsinization. The yellow arrow indicates reattached cells. (E) Comparison between subculturing methods on cell attachment.Scale bars: 50 μm. The data were explored using a one-way analysis of variance followed by Tukey’s post hoc test. The experiment was conducted in triplicates, and the error bars represent the standard deviation of the samples. The asterisk (*) indicates a significant difference among the sample.
D. Transgenic experiments in primary embryonic cell culture
1. Standardization of transfection
a. Optimize Cellfectin reagent and plasmid concentrations: Test 3–10 μL of Cellfectin II and 1–5 μg of plasmid DNA and use the pCS-EF1α1-DSRed2 plasmid for expression studies [9].
b. Protocol:
i. Seed cells in 96-well plates with MSIM.
ii. Prepare serum-free MSIM with Cellfectin reagent and plasmid.
iii. Incubate cells with the transfection mixture for 3–9 h at 28 °C.
iv. Replace the medium with FBS-containing MSIM and observe DSRed2 expression under fluorescence microscopy.
As shown in Figure 5, maximal transfection efficiency (54%) was obtained using 3 μg of plasmid with 7 μL of Cellfectin and an incubation time of 7 h.
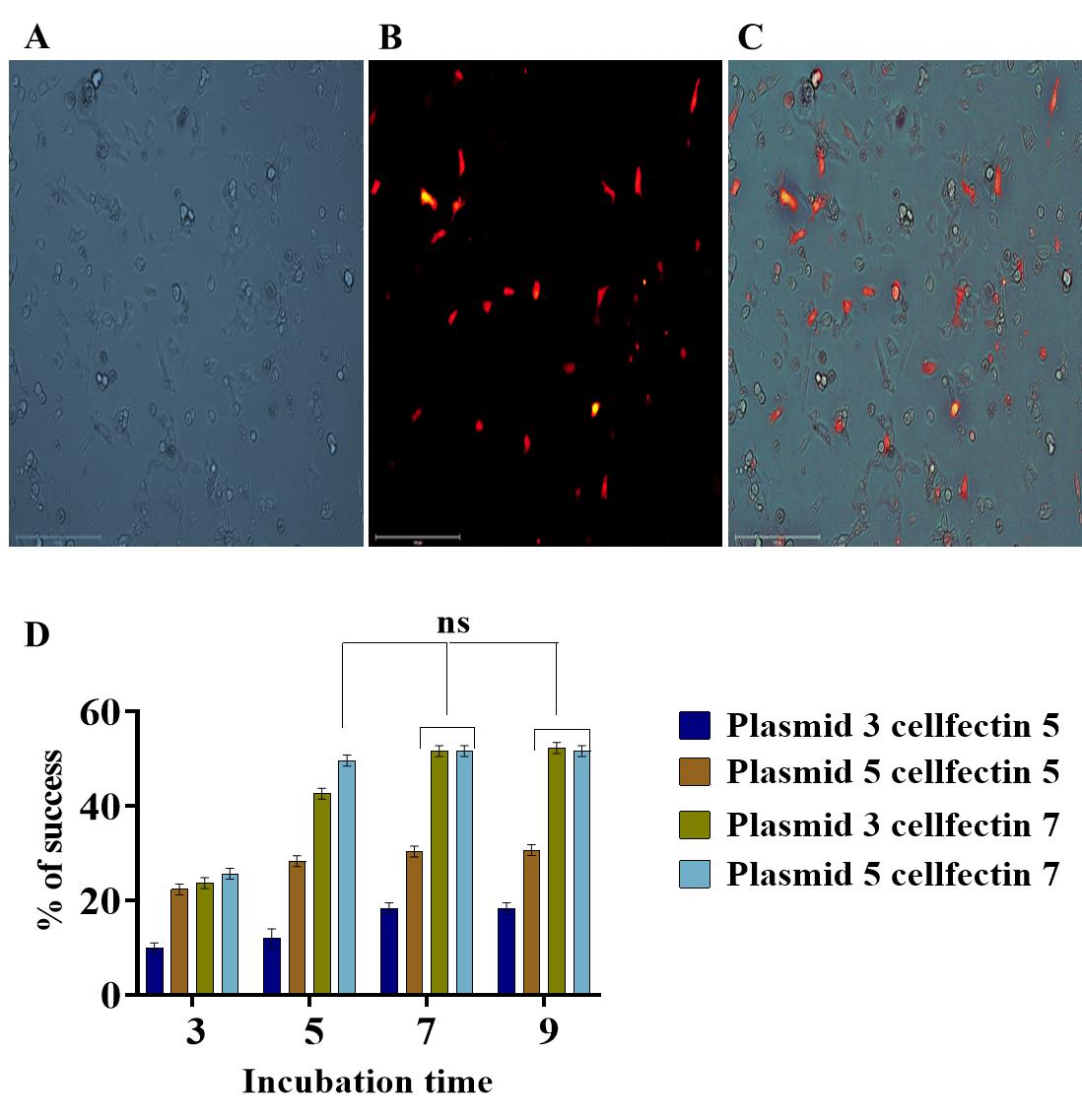
Figure 5. Transfection experiment in Daphnia magna primary embryonic cell cultures using a Daphnia-specific expression vector (pCS-EF1α1-DSRed2). (A) Daphnia embryonic primary culture used in lipofection. (B) Red fluorescent protein expression from the reporter gene DSRed2 under the control of Daphnia-specific promoter EF1α1 in D. magna primary embryonic cells. (C) Merged image. Scale bar: 150 μm. (D) Graphical representation depicting the influence of factors such as plasmid concentrations (PC), Cellfectin volume (CV), and incubation time on transfection efficiency (TE) in D. magna embryonic primary cell culture. The data were explored using a three-way analysis of variance followed by Tukey’s post hoc test. The experiment was conducted in triplicates, and the error bars represent the standard deviation of the samples. ns represent the non-significant difference among the sample means.
2. Standardization of transduction
a. Use recombinant baculovirus vectors (BacIe1-GFP and BacP2-GFP) with shrimp-specific promoters [2,3].
i. Seed 50–100 μL of Daphnia cells in 96-well plates.
ii. Replace MSIM with a mixture of fresh MSIM and recombinant baculovirus.
iii. Observe GFP signals every 6 h for 24 h and daily for a week under fluorescence microscopy.
iv. Use non-infected Daphnia cells as negative controls and SF9 cells as positive controls.
Green fluorescence in Daphnia magna primary embryonic cells confirmed GFP protein expression (Figure 6A) under the PH-Ie1 hybrid promoter in the recombinant baculovirus vector. In contrast, the baculovirus with the P2 promoter (IHHNV) did not induce GFP expression (Figure 6B). GFP expression appeared on Day 3 in Sf9 cells (positive control) and Day 4 in Daphnia cells, with ~30% of cells expressing GFP, increasing to 45% after one week. The absence of expression in the negative control validated the efficacy of the BacIe1-GFP system in Daphnia cells.
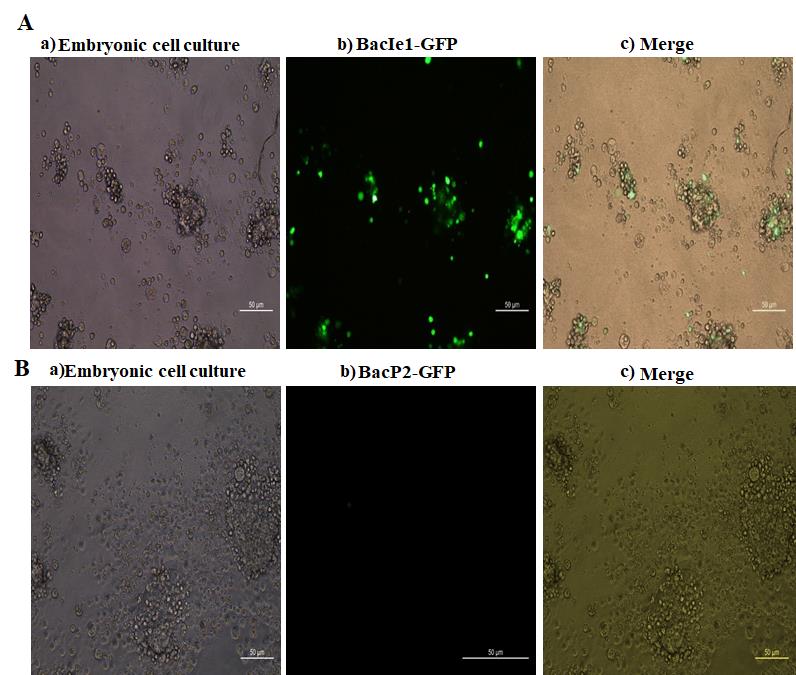
Figure 6. Transduction experiment in Daphnia magna primary embryonic cell cultures using a recombinant baculovirus expression vector system with PH-Ie1 (Ie1 promoter from WSSV) and PH-P2 (P2 promoter from IHHNV) hybrid promoters. (A): a) Primary embryonic cells. b) Green fluorescent protein (GFP) expression in Daphnia magna primary embryonic cells after transduction with PH-Ie1 hybrid promoter. c) Merged image. (B): a) Primary embryonic cells. (b) No expression of green fluorescent protein in D. magna primary embryonic cells after transduction with PH-P2 hybrid promoter. (c) Merged image. Scale bar: 50 μm.
Data analysis
To analyze the developed primary culture and to ensure the robustness of cells, we performed two assays, namely cell viability assessments, and DNA synthesis.
A. Assessment of cell viability
Note: Acridine orange (AO) and ethidium bromide (EB) dual staining is a method to differentiate live and dead cells. AO stains live cells green, and EB stains dead cells orange-red.
1. Seed 24-h-old primary embryonic cells in 50 μL of MSIM in a 96-well plate.
2. Centrifuge and resuspend cells in 10 μL of MSIM.
3. Add 10 μL of 1× staining solution (100 μg/mL AO and EB prepared in PBS).
4. Incubate for 2 min at room temperature in the dark.
5. Centrifuge at 1,000× g for 10 min, discard the supernatant, and resuspend the pellet in 20 μL of PBS.
6. Load 10 μL on a slide, cover it with a coverslip, and observe under a fluorescent microscope.
In our work, after AO/EB dual staining, approximately 90% of cells in MSIM displayed green fluorescence, signifying a predominance of viable cells at 24 h of growth (Figure 7).
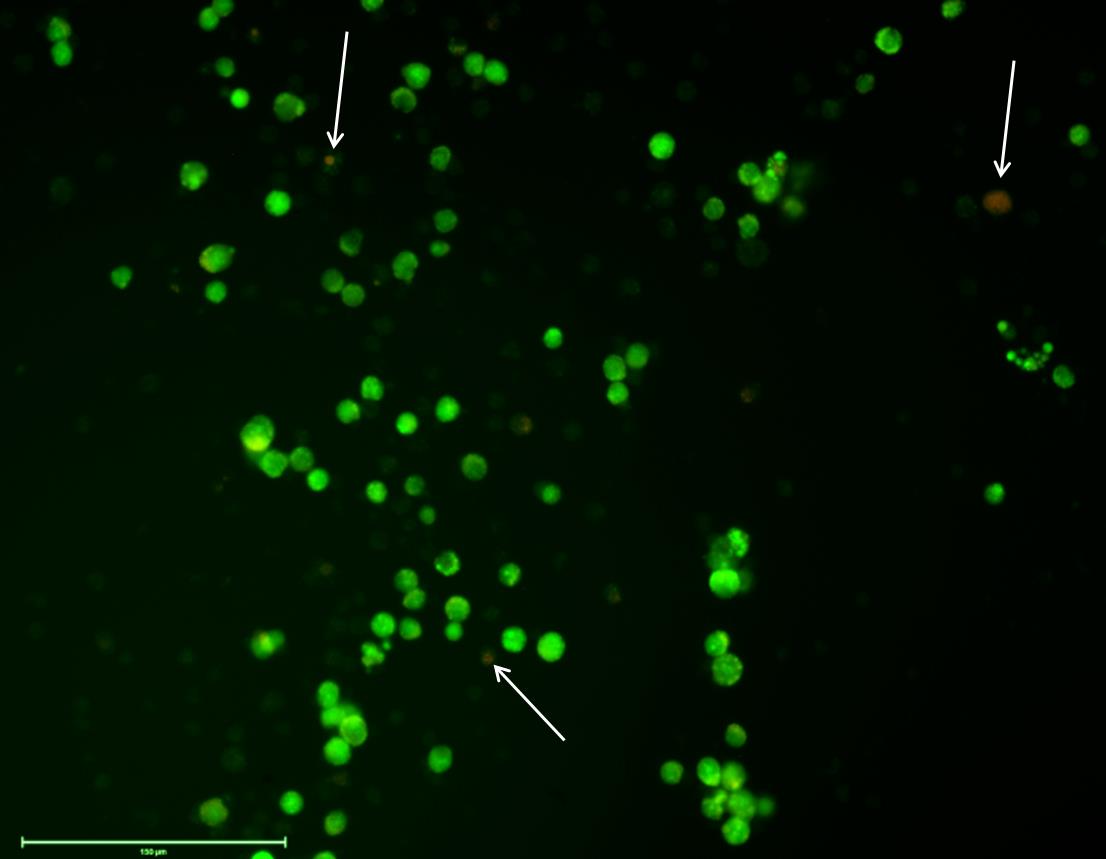
Figure 7. Viability test to determine the number of live/dead cells of Daphnia magna primary embryonic cell cultures in the modified medium (MSIM). Acridine orange (AO) and ethidium bromide (EB) dual staining shows live cells (green) and dead cells (yellow/orange/red). White arrows indicate dead cells. Scale bar: 150 μm.
B. Evaluation of DNA synthesis
Note: The BrdU incorporation assay detects cells in the S-phase of the cell cycle.
1. Treat coverslips for cell adhesion and seed cells in 24-well plates.
2. Add 180 μL of filtered 10 μM BrdU solution to test wells; use untreated wells as controls.
3. Incubate overnight at 28 °C.
4. Fix, permeabilize, and block cells using the detailed supplementary protocol (File S1).
a. Fix cells with 4% paraformaldehyde in PBS (pH 7.4) for 15 min at RT and then wash three times with PBS for 5 min each.
b. Subsequently, add 2 M HCl to each well and incubate for 20 min at RT.
c. Neutralize the acid with 0.1 M sodium borate (pH 8.5) for 2 min, followed by three washes with PBS (each for 2 min).
d. Permeabilize cells with PBS containing 0.2% Triton X-100 and 3% BSA for 5 min and then block with 3% BSA in PBS for 1 h at RT.
5. Incubate with anti-BrdU antibody (1:1,000) and Alexa Fluor 488 secondary antibody (1:400).
6. Stain with DAPI and observe under a fluorescence microscope.
In our work, D. magna primary embryonic cells incubated in 10 μM BrdUsolution for 24 h exhibited enlarged nuclei, emitting Alexa Fluor 488 fluorescence signals. 23% of cellsexhibiteda positive response to BrdU incorporationwithin24 h. Blue fluorescence of DAPI was observed in the cell nuclei (Figure 8).
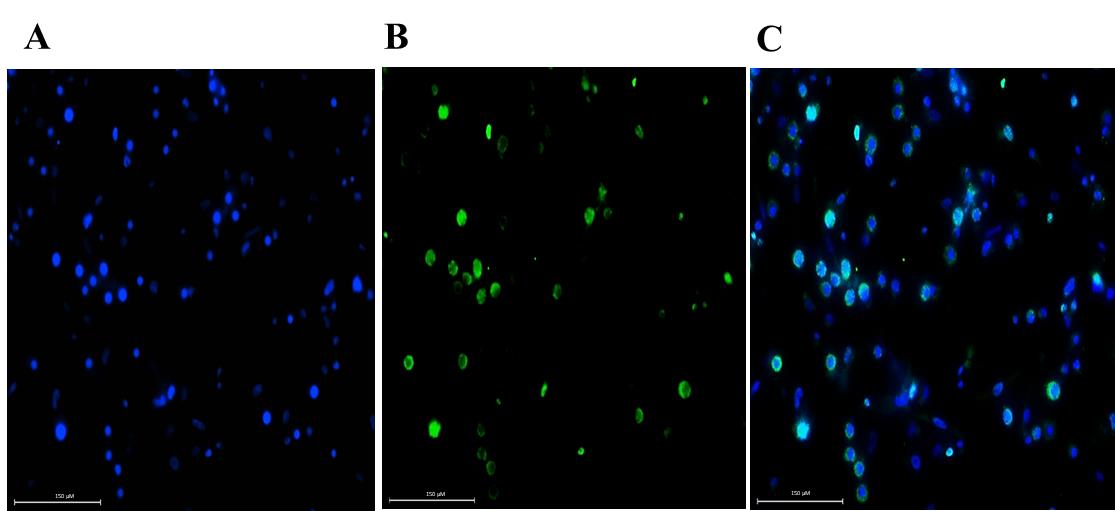
Figure 8. Immunofluorescent detection of BrdU incorporation in the DNA of Daphnia magna embryonic cells (in vitro). BrdU incorporation confirms that cells are in the S-phase of the cell cycle. (A) DAPI-stained nucleus. (B) BrdU-incorporated DNA stained with a mouse monoclonal anti-BrdU primary antibody, linked to an Alexa Fluor 488 secondary antibody. (C) Merged image. Scale bar: 150 μm.
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
• Sreevidya et al. [5]. Establishment of a cell culture fromDaphnia magnaas anin vitromodel for (eco)toxicology assays: Case study using Bisphenol A as a representative cytotoxic and endocrine disrupting chemical, Aquatic Toxicology, Volume 278, January 2025, 107173.
• Sreevidya et al. [6]. BIF-induced ROS-mediated cytotoxicity and genotoxicity in embryonic cell culture of Daphnia magna. Aquatic Toxicology. 107285.
General notes and troubleshooting
General notes
1. Variability in cell viability and growth rates may arise from differences in embryo quality, isolation methods, and initial cell density. Use embryos at a similar developmental stage to ensure consistency.
2. The protocol is optimized for Daphnia magna embryonic cells and may not be directly applicable to other Daphnia species or aquatic organisms without modifications. Optimization of medium composition and culture conditions is necessary for other species.
3. Maintaining sterility throughout the process is critical to preventing microbial contamination, which can drastically affect cell viability and assay outcomes. Use autoclaved instruments and perform procedures in a sterile environment.
4. The modified Schneider’s insect medium (MSIM) should be freshly prepared and stored at 4 °C for no longer than two weeks to avoid degradation of components such as FBS, vitamins, and selenium.
5. Transfection and transduction efficiencies are highly dependent on reagent quality, incubation time, and plasmid purity. It is recommended to use endotoxin-free plasmid preparations to improve results.
6. Reduced reattachment and viability after subculturing are expected due to the delicate nature of embryonic cells. Minimize handling stress by avoiding excessive pipetting or prolonged enzymatic treatments. The difference in reattachment propensity between trypsinization and trituration in our result could be attributed to the extent of cellular damage incurred during the process. Trypsinization, being an enzymatic method, may affect adhesion proteins critical for cell attachment, whereas trituration, a mechanical dissociation method, might preserve these proteins better, leading to improved reattachment.
7. This protocol can serve as a reference for developing primary cell cultures in other small aquatic invertebrates, though adjustments in enzymatic treatments, media, and supplements may be necessary.
Troubleshooting
Problem 1: Contamination in cultures
Cause: Non-sterile technique during embryo collection or culture setup.
Solution: Ensure all instruments and reagents are autoclaved, use sterile gloves, and work in a laminar flow hood. Perform media changes carefully to minimize contamination.
Problem 2: Poor cell viability
Cause: Inappropriate media composition or prolonged storage of medium or more pressure during egg homogenization.
Solution: Use freshly prepared MSIM and verify the concentration of supplements (10% FBS, 0.1% glucose, 0.1 ppb selenium). Avoid storing the medium for longer than two weeks and standardize the pressure used for homogenization.
Supplementary information
The following supporting information can be downloaded here:
1. File S1. Protocol for BrdU staining in D. magna embryonic primary cell culture
Acknowledgments
S.C.P. carried out the formal analysis and investigation. S.C.P. and S.B. conducted the writing original draft. J.P. conceptualized, designed the methodology, and supervised S.C.P. for all the experiments.
The authors thank the Department of Biotechnology (DBT), Govt. of India, for the Innovative Young Biotechnologist Award (DBT-IYBA; BT/09/IYBA/2015/11) to Dr. Jayesh Puthumana and the funding for this research work. The first author thanks DBT for the fellowship.
This protocol was first described and validated in: Robinsonet al. [12]; Sreevidya et al. [5]; Sreevidya et al. [6].
Competing interests
The authors declare that there are no conflicts of interest.
Ethical considerations
All investigations were conducted as per guidelines provided by the Institutional Biosafety Committee (IBSC) and Ethical Committee of NCAAH, CUSAT in Kerala, India.
References
- Claydon, K. (2009). Advances in crustacean cell culture. Dissertation, James Cook University.
- Jayesh, P. (2013). Development of lymphoid cell culture system from Penaeus monodon and molecular approaches for its transformation. Ph.D. Thesis, Cochin University of Science and Technology, India.
- Jayesh, P., Jose, S., Philip, R. and Bright Singh, I. S. (2013). A novel medium for the development of in vitro cell culture system from Penaeus monodon. Cytotechnology. 65(3): 307–322. https://doi.org/10.1007/s10616-012-9491-9
- Bhaskaran Sathyabhama, A., Puthumana, J., Kombiyil, S., Philip, R. and Bright Singh, I. S. (2021). ‘PmLyO-Sf9 - WSSV complex’ could be a platform for elucidating the mechanism of viral entry, cellular apoptosis and replication impediments. Virology. 553: 102–110. https://doi.org/10.1016/j.virol.2020.10.014
- Sreevidya, C. P., Kumar, M., Balakrishnan, S., Kunjiraman, S., Sarasan, M., Magnuson, J. T. and Puthumana, J. (2025). Establishment of a cell culture from Daphnia magna as an in vitro model for (eco)toxicology assays: Case study using Bisphenol A as a representative cytotoxic and endocrine disrupting chemical. Aquat Toxicol. 278: 107173. https://doi.org/10.1016/j.aquatox.2024.107173
- Sreevidya, C. P., Ajitha, V., Kumar, M., Manomi, S., Bhavya, K., Singh, I. B. and Puthumana, J. (2025). BIF-induced ROS-mediated cytotoxicity and genotoxicity in embryonic cell culture of Daphnia magna. Aquat Toxicol. 280: 107285. https://doi.org/10.1016/j.aquatox.2025.107285
- Kato, Y., Shiga, Y., Kobayashi, K., Tokishita, S. I., Yamagata, H., Iguchi, T. and Watanabe, H. (2011). Development of an RNA interference method in the cladoceran crustacean Daphnia magna. Dev Genes Evol. 220: 337–345. https://doi.org/10.1007/s00427-011-0353-9
- Nakanishi, T., Kato, Y., Matsuura, T. and Watanabe, H. (2014). CRISPR/Cas-Mediated Targeted Mutagenesis in Daphnia magna. PLoS One. 9(5): e98363. https://doi.org/10.1371/journal.pone.0098363
- Kumagai, H., Nakanishi, T., Matsuura, T., Kato, Y. and Watanabe, H. (2017). CRISPR/Cas-mediated knock-in via non-homologous end-joining in the crustacean Daphnia magna. PLoS One. 12(10): e0186112. https://doi.org/10.1371/journal.pone.0186112
- Smith, G. E., Summers, M. D. and Fraser, M. J. (1983). Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 3(12): 2156–2165. https://doi.org/10.1128/mcb.3.12.2156
- Kato, Y., Matsuura, T. and Watanabe, H. (2012). Genomic Integration and Germline Transmission of Plasmid Injected into Crustacean Daphnia magna Eggs. PLoS One. 7(9): e45318. https://doi.org/10.1371/journal.pone.0045318
- Robinson, C. D., Lourido, S., Whelan, S. P., Dudycha, J. L., Lynch, M. and Isern, S. (2005). Viral transgenesis of embryonic cell cultures from the freshwater microcrustacean Daphnia. J Exp Zool A Comp Exp Biol. 62–67. https://doi.org/10.1002/jez.a.250
Article Information
Publication history
Received: Dec 25, 2024
Accepted: Mar 2, 2025
Available online: Mar 24, 2025
Published: Apr 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Sreevidya, C. P., Balakrishnan, S. and Puthumana, J. (2025). Baculovirus and Plasmid Vector-Mediated Gene Transfer in Daphnia magna Cells in Primary Embryonic In Vitro Cultures in an Optimized Microenvironment: Methods and Protocols. Bio-protocol 15(7): e5266. DOI: 10.21769/BioProtoc.5266.
Category
Environmental science > Parasites
Cell Biology > Model organism culture > Embryo selection
Cell Biology > Cell-based analysis > Gene expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








