- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Protocol for Laser-Assisted Microdissection and tRF & tiRNA Sequencing in Lung Adenocarcinoma
(*Contributed equally to this work, §Technical contact: wangzi@stu.njmu.edu.cn, **Lead contact) Published: Vol 15, Iss 7, Apr 5, 2025 DOI: 10.21769/BioProtoc.5253 Views: 2992
Reviewed by: Ritu GuptaSalah BoudjadiNavnita DuttaHeng Sun

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
snPATHO-seq: A Detailed Protocol for Single Nucleus RNA Sequencing From FFPE
Wani Arjumand [...] Luciano G. Martelotto
May 5, 2025 1936 Views

A Guide to Basic RNA Sequencing Data Processing and Transcriptomic Analysis
Rowayna Shouib [...] Rineke Steenbergen
May 5, 2025 7757 Views

RACE-Nano-Seq: Profiling Transcriptome Diversity of a Genomic Locus
Lu Tang [...] Philipp Kapranov
Jul 5, 2025 2522 Views
Abstract
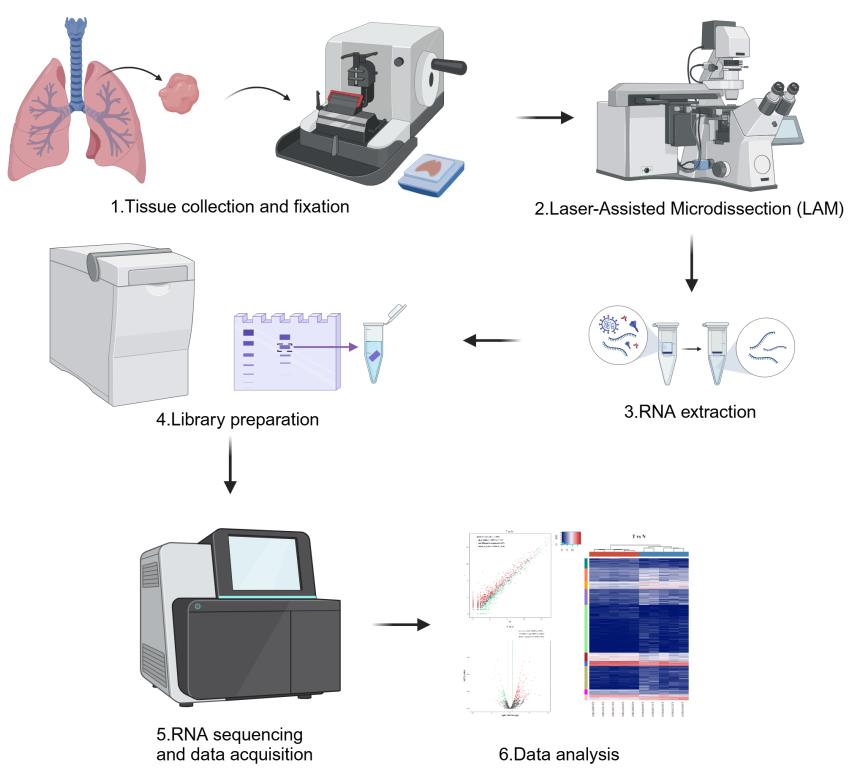
Laser-assisted microdissection (LAM) coupled with next-generation sequencing technologies offers a powerful approach to dissecting the complex cellular heterogeneity within lung adenocarcinoma (LUAD) tumors. This protocol outlines the method for isolating specific high-risk LUAD tissues containing micropapillary/solid (MIP/SOL) patterns, which is linked to poor prognosis. We detail the process of LAM, which involves tissue fixation, microtome sectioning, and the precise dissection and collection of cells of interest under microscopic guidance. The isolated cells are then subjected to RNA extraction, library preparation, and sequencing to profile transfer RNA–derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs), which are emerging as key regulators in cancer. This protocol enables researchers to obtain high-quality transcriptomic data from specific LUAD cell populations, aiming to uncover tRF-Val-CAC-024 and tiRNA-Gly-CCC as potential biomarkers for early diagnosis and therapeutic targets for LUAD treatment.
Key features
• Laser-assisted microdissection (LAM): Utilizes laser precision to isolate high-risk MIP/SOL patterns of LUAD tissues.
• tRF & tiRNA sequencing: Profiles transfer RNA–derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs), reported to play a key role in cancer development.
Keywords: Laser microdissectionGraphical overview
Background
Lung adenocarcinoma (LUAD), a subtype of non-small cell lung cancer, is characterized by its molecular heterogeneity and complex metastatic behavior. Understanding the molecular underpinnings of LUAD is crucial for advancing our knowledge of cancer biology and for the development of targeted therapies. Traditional bulk sequencing approaches provide a comprehensive view of the transcriptome but often mask the cellular heterogeneity within tumors [1]. There is a need for techniques that can isolate and characterize the transcriptomic profiles of specific cell populations within LUAD tumors. Laser-assisted microdissection (LAM) is a microscopy-based technique that allows for the precise isolation of specific cell populations directly from their tissue context. This method has been increasingly used in oncology research to study the molecular signatures of distinct cell types within tumors. LAM offers several advantages over other methods, such as fluorescent activated cell sorting (FACS), as it does not require the generation of marker lines or prior knowledge of a large number of marker genes. Instead, LAM relies solely on the morphological characteristics of the tissues to be collected, making it a versatile tool for studying various cancer types, including LUAD [2]. The combination of LAM with next-generation sequencing has emerged as a powerful approach to dissecting the complex cellular heterogeneity within LUAD tumors. In particular, the study of transfer RNA–derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs) has been gaining momentum, as these small RNA species are implicated in various biological processes, including gene regulation, protein translation, and cellular metabolism, which are often dysregulated in cancer [3].
In this protocol, we describe the method for LAM of LUAD tissues to isolate high-risk components containing micropapillary/solid (MIP/SOL) patterns, which are known to be associated with poor prognosis. We further detail the process of tRF and tiRNA sequencing to characterize the molecular signatures of these high-risk LUAD cell populations. This approach not only enhances our understanding of LUAD biology but also paves the way for the identification of novel therapeutic targets and biomarkers [4].
By following this protocol, researchers can effectively isolate and analyze the transcriptomic profiles of specific LUAD cell populations, contributing to the advancement of personalized medicine. The protocol is designed to be adaptable for various research applications, including the study of other cancer types and complex tissue samples.
Materials and reagents
Biological materials
1. Lung adenocarcinoma (LUAD) tissues and adjacent normal tissues collected during thoracoscopic surgery
Reagents
1. 10% neutral buffered formalin (NBF) (Sigma-Aldrich, catalog number: HT501128) for tissue fixation
2. Xylene (Thermo Fisher Scientific, catalog number: X3S-4)
3. Absolute ethyl alcohol (ethanol) (Sigma-Aldrich, catalog number: E7023)
4. HistoClear II (National Diagnostics, catalog number: HS-202) for tissue clearing and processing
5. Paraplast Plus® (paraffin) (Leica Biosystems, catalog number: 39601006) for tissue embedding
6. Trypsin-EDTA (Thermo Fisher Scientific, catalog number: 25200056) for cell disassociation (if single-cell sequencing is required)
7. PureLink RNA Micro kit (Thermo Fisher Scientific, catalog number: 12183016) for total RNA extraction
8. NextSeq 500/550 v2 kit (Illumina, catalog number: FC-404-2005) for tRF and tiRNA sequencing
9. Small RNA adapters (Illumina, catalog number: 15004197)
Laboratory supplies
1. Sterile Falcon tubes (for sample storage) (Corning, catalog number: 352096)
2. Beakers (for solution mixing) (Pyrex, catalog number: 1000-100)
3. Dissecting forceps [for manual tissue dissection (if not using LAM)] (Fine Science Tools, catalog number: 11050-10)
4. Steel inclusion molds (for tissue embedding) (Leica Biosystems, catalog number: 14046435645)
5. Embedding cassettes without the lid (for tissue embedding) (Thermo Fisher Scientific, catalog number: 15182731)
6. Microscope slides (for tissue sectioning) (Thermo Fisher Scientific, catalog number: 12-550-15)
7. Low-profile disposable blades (for tissue sectioning) (Feather Safety Razor Co., catalog number: 0915)
8. Slide rack and glass staining dish with lid (for tissue staining and deparaffinization) (Thermo Fisher Scientific, catalog number: S90010)
9. TubeCollector 2×200 CM II (for collecting laser micro-dissected tissues) (Zeiss, catalog number: 415190-9081-000)
10. AdhesiveCap 200 clear (for sealing microcentrifuge tubes containing micro-dissected tissues) (Zeiss, catalog number: 415190-9191-000)
11. MembraneSlide 1.0 OPEN (for tissue section adhesion and slide preparation) (Zeiss, catalog number: 415190-9051-000)
12. RNase-free CapSure Macro caps (Zeiss, catalog number: 415190-9191-000)
13. RNase-free wipes (Thermo Fisher Scientific, catalog number: 21-404-303)
Equipment
1. Vacuum pump (Thermo Fisher Scientific, model: R4100649 or Edwards, model: E2M28)
2. Vacuum desiccator (Bel-Art, catalog number: 42022000 or Cole-Parmer, catalog number: EW-06100-20)
3. Mini rotator (Benchmark Scientific, catalog number: H5500 or Thermo Fisher Scientific, model: 88881001)
4. Convection incubator (Thermo Scientific, model: Heratherm IMC18 or Memmert, model: UF110)
5. Paraffin dispenser (Leica Biosystems, model: HistoCore Arcadia H)
6. Microtome (Leica Biosystems, model: RM2235 or Thermo Fisher Scientific, model: HM325)
7. Slide warmer (Leica Biosystems, model: HI1220 or Thermo Fisher Scientific, catalog number: 22-050-263)
8. PALM MicroBeam IV microscope with motorized RoboMover and CapMover (Zeiss, model: PALM MicroBeam IV)
9. NanoDrop (optional) (Thermo Fisher Scientific, model: NanoDrop 2000, NanoDrop One)
10. Covaris S2 ultrasonicator (optional) (Covaris, model: S2)
11. Qubit 4 fluorometer (optional) (Thermo Fisher Scientific, catalog number: Q33238)
12. Agilent Bioanalyzer (Agilent Technologies, model: 2100 Bioanalyzer)
13. Automated gel cutter (Blue-Ray Biotech, catalog number: BLU0001)
Software and datasets
1. PALM Robo software (Zeiss, v4) for operating the LCM system
2. Optional: R studio for RNA-seq data analysis
3. Bioinformatics tools for RNA-seq data processing and analysis, including FastQC, cutadapt, bowtie, and edgeR
4. tRNAscan-SE (v2.0.11, http://lowelab.ucsc.edu/tRNAscan-SE/)
Procedure
A. Tissue collection
1. Patient recruitment and consent: Ensure all procedures are approved by the local ethics committee. The proposal should include:
a. A detailed description of the study objectives and methodology.
b. Justification for the research and its potential scientific and clinical impact.
c. Information on patient recruitment, inclusion/exclusion criteria, and consent procedures.
d. Measures to ensure patient confidentiality, data protection, and secure storage of biological samples.
e. A risk-benefit analysis, outlining potential risks to participants and how they will be mitigated.
f. Plans for handling adverse events or protocol deviations.
Recruit patients who are undergoing thoracoscopic surgery for LUAD. Obtain written consent from each participant after explaining the purpose and procedures of the study.
2. Sample collection
a. Collect tumor tissues and adjacent normal tissues (at least 2 cm away from the tumor) during surgery.
b. Ensure the sampling procedure is controlled within 20 min and strictly adhere to aseptic techniques to prevent contamination.
3. Tissue processing
a. Fix the collected tissues in 10% neutral buffered formalin for 24–48 h at room temperature.
b. Process the tissues through a series of ethanol and xylene washes for dehydration and clearing.
4. Tissue embedding and storage
a. Embed the formalin-fixed, paraffin-embedded (FFPE) tissues in paraffin blocks using steel inclusion molds.
b. Store the paraffin blocks at room temperature in a dry, dust-free environment until sectioning.
5. Sectioning, H&E staining, and pathological review
a. Section preparation for H&E staining:
i. Prepare 4–5 μm thick sections from the FFPE blocks using a microtome.
ii. Mount sections on Superfrost Plus microscope slides.
b. H&E staining: Stain the slides with hematoxylin and eosin following standard protocols.
c. Pathological review: Submit the H&E-stained slides to a qualified pathologist for histological evaluation. Request annotations to demarcate tumor regions and adjacent non-tumoral areas for subsequent microdissection.
d. Preparation of unstained slides for LCM: Based on the pathologist’s annotations, cut additional unstained sections (5–10 μm thick) from the same FFPE blocks. Store unstained slides in an RNase-free, dust-free container at room temperature until laser capture microdissection (LCM).
6. Documentation:
Record detailed clinical characteristics of the enrolled subjects, including demographic, medical history, lifestyle, and other characteristic information.
7. Quality control: Perform quality control checks on the samples, including RNA integrity analysis using an Agilent Bioanalyzer (RIN > 7.0) and concentration measurement using a Nanodrop spectrophotometer (A260/A280 ratio: 1.8–2.1).
8. Data recording: Maintain a record of all samples collected, including patient identifiers, collection dates, and any relevant clinical data.
Notes:
1. All diagnosis and treatment procedures should follow international guidelines.
2. Only patients with stage I and stage II LUAD should be included according to the 8th edition standards set by the International Association for the Study of Lung Cancer (IASLC) regarding lung cancer tumor, node, and metastasis (TNM) staging.
3. Ensure there are no respiratory infections, no history of smoking, and no use of antibiotics, steroids, or probiotic treatment within the past 3 months for all subjects.
B. Laser capture microdissection
Day 1: Slide preparation
1. Deparaffinization
a. Place slides in a slide rack in a glass staining dish filled with 100% HistoClear II at room temperature for 15 min to remove paraffin.
b. Allow slides to air dry at room temperature for approximately 20–30 min.
2. Rehydration
a. Pass slides through a series of ethanol washes: 100%, 95%, 70% (2 min each).
b. Rinse slides in deionized water for 2 min.
Note: Ensure all work surfaces, equipment, and reagents are RNase-free to prevent RNA degradation.
Day 2: Laser capture microdissection
1. Microscope and software setup:
a. Turn on the LCM microscope and launch the LCM software.
b. Ensure the microscope stage is clean and free of debris.
2. Slide mounting
a. Place the deparaffinized and rehydrated slide on the LCM stage.
b. Use the software to move the stage to the loading position and secure the slide.
3. Tissue localization
a. Select the appropriate objective lens (e.g., 10× or 20×) to locate the tissue of interest.
b. Use the autofocus feature to obtain a clear image of the target area.
4. Tissue selection
a. Use the software's drawing tool to outline the region of interest (ROI) around the specific cell population to be captured.
b. Adjust the laser settings (power, focus, and duration) to optimize cutting.
c. Example laser settings:
Laser power: 50–70 mW (adjust based on tissue thickness and density).
Laser focus: 10–20 μm (ensure the focal plane is aligned with the tissue section).
Laser duration: 1–3 ms (adjust for precise cutting without excessive tissue damage).
d. Perform a test run on a small area to confirm optimal settings before capturing the entire ROI.
5. Capture and collection:
a. Position an RNase-free CapSure Macro cap over the ROI.
b. Activate the laser to cut the selected tissue, which will be catapulted into the cap.
c. Repeat this process for all desired ROIs on the slide.
The selective isolation of tumor and adjacent non-tumoral regions via LAM is visually demonstrated in Figure 1.
6. Post-collection handling
a. Carefully remove the cap from the slide and place it into an RNase-free microcentrifuge tube.
b. Immediately immerse the tube in liquid nitrogen to flash-freeze the captured tissue.
c. Store the tube at -80 °C until ready for molecular analysis (e.g., RNA extraction).
7. Cleanup and shutdown
a. Close the LCM software and turn off the microscope.
b. Clean the stage and objective lens with RNase-free wipes.
Notes:
1. The time required for LCM will vary depending on the number of samples and the complexity of the tissue sections.
2. Always wear appropriate personal protective equipment (PPE) when handling hazardous chemicals and when operating the LCM system.
3. Regularly calibrate the LCM system to ensure accurate and consistent cutting.
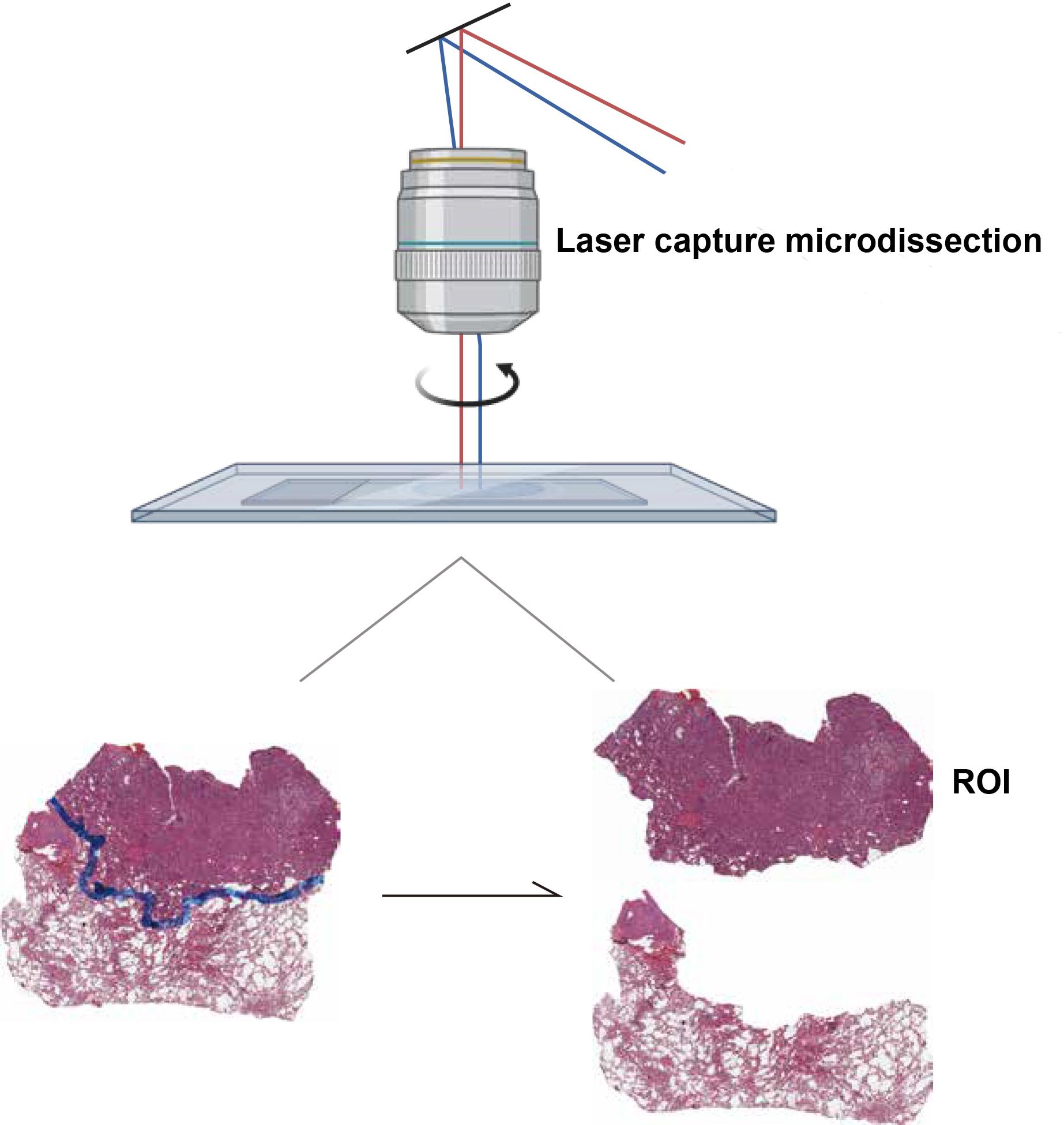
Figure 1. Laser-assisted microdissection (LAM) of lung adenocarcinoma (LUAD) tissue sections. This figure demonstrates the use of LAM to isolate specific regions of interest (ROIs) from LUAD tissue. The image shows the precise cutting of two tissue regions, highlighting the selective capture of tumor and adjacent non-tumoral areas.
C. RNA extraction
After sufficient LCM of the tissue(s) of interest, perform RNA extraction. For this protocol, we used the
PureLinkTM RNA Micro kit.
D. Small RNA library optimization and sequencing
1. Quality control (QC): Check the integrity and quantity of each RNA sample using agarose gel electrophoresis and a Nanodrop spectrophotometer
2. Pretreatment of tRFs and tiRNAs: Perform the following treatments on total RNA samples to remove RNA modifications interfering with library construction:
Deacylate 3′-aminoacyl groups to generate 3′-OH termini for 3′ adapter ligation.
Remove 2′,3′-cyclic phosphate (cP) to generate 3′-OH termini for 3′ adapter ligation.
Phosphorylate 5′-OH groups to 5′-P for 5′ adapter ligation.
Demethylate m1A and m3C modifications to enhance reverse transcription efficiency.
3. Library optimization: Ligate the total RNA of each sample sequentially to 3′ and 5′ small RNA adapters. Synthesize cDNA using Illumina’s proprietary RT primers. Amplify the cDNA with Illumina amplification primers.
Size-select PCR-amplified fragments (~134–160 bp) using an automated gel cutter and purify from PAGE gels. Qualify and absolutely quantify the libraries using an Agilent BioAnalyzer 2100.
4. tRF and tiRNA sequencing: Perform standard small RNA sequencing on an Illumina NextSeq 500 system with 50 bp single-read mode. Denature and dilute libraries to a loading volume of 1.3 mL and concentration of 1.8 pM. Load diluted libraries onto NextSeq 500/550 V2 reagent cartridges and initiate sequencing according to the manufacturer’s instructions.
5. Mature and precursor tRNA sequence annotation: Download cytoplasmic tRNA sequences from GtRNAdb (https://gtrnadb.ucsc.edu/). Predict mitochondrial tRNA sequences using tRNAscan-SE software (v2.0.11, available at http://lowelab.ucsc.edu/tRNAscan-SE/). Generate mature tRNA libraries by removing intronic sequences (if present) and adding a 3′-terminal "CCA" sequence. Construct precursor tRNA libraries by including 40 nucleotides of flanking genomic sequence on both ends of the tRNA.
Data analysis
Image analysis and base calling are performed using Solexa pipeline v1.8 (Off-Line Base Caller software, v1.8). Sequencing quality is examined by FastQC [5] and trimmed reads (pass Illumina quality filter, trimmed 5′, 3′-adaptor bases by cutadapt [6]) are aligned allowing for one mismatch only to the mature tRNA sequences. Then, reads that do not map are aligned allowing for one mismatch only to precursor tRNA sequences with the bowtie software [7]. The remaining reads are aligned allowing for one mismatch only to miRNA reference sequences with miRDeep2 [8]. The integrated workflow for tRF & tiRNA-seq data analysis is summarized in Figure 2. The expression profiling of tRF & tiRNA and miRNA can be calculated based on counts of reads mapped. The differentially expressed tRFs & tiRNAs and miRNAs are screened based on the count value with the R package edgeR [9]. Principal component analysis (PCA), correlation analysis, pie plots, Venn plots, hierarchical clustering, scatterplots, and volcano plots are performed in R or Perl environment for statistical computing and obtaining graphics of the expressed tRF & tiRNA.
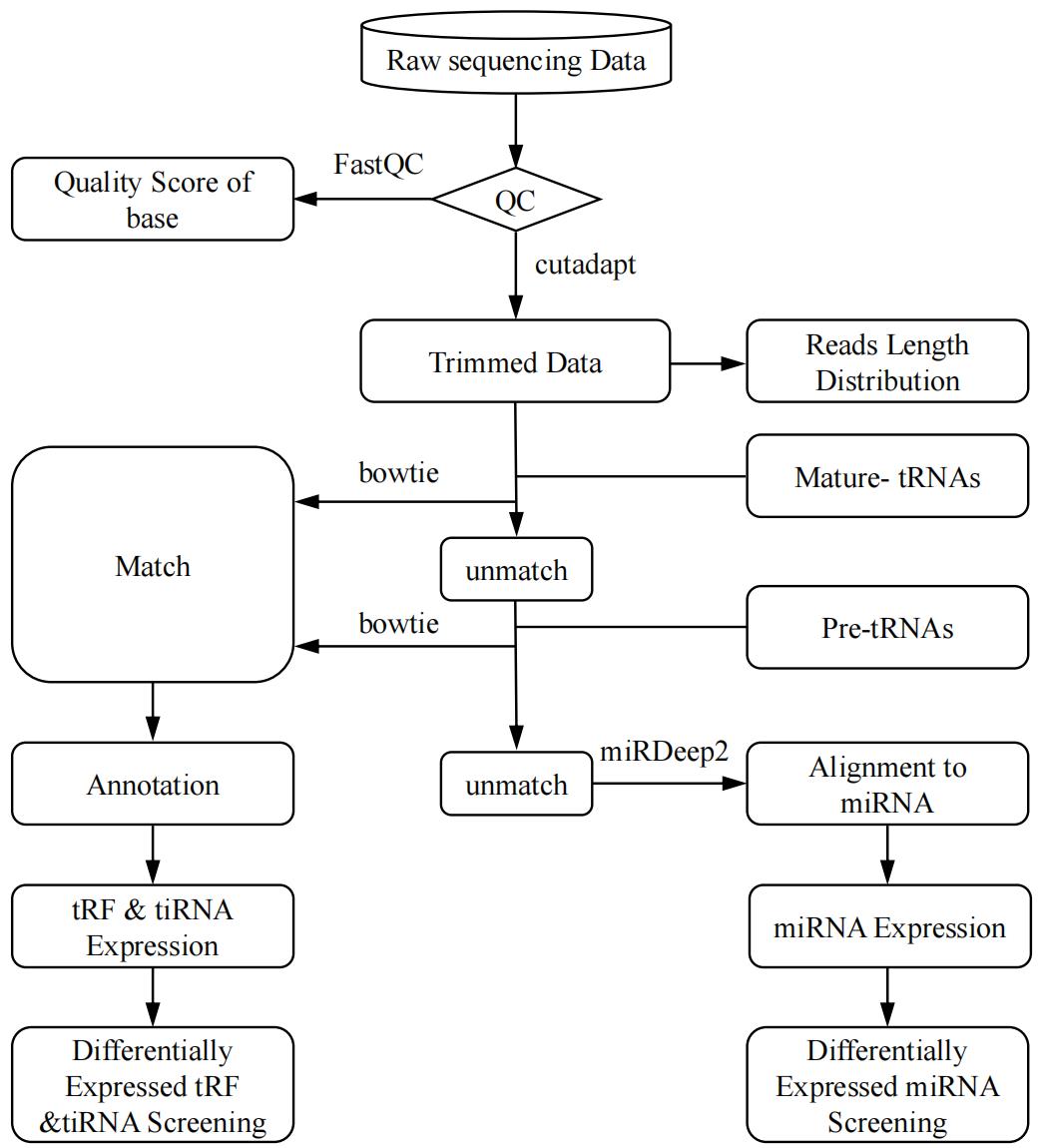
Figure 2. Transfer RNA–derived fragment & tRNA-derived stress-induced RNA (tRF & tiRNA)-seq data analysis workflow. Raw sequencing data generated from Illumina NextSeq 500 that pass the Illumina chastity filter are used for the following analysis. Trimmed reads (trimmed 5′, 3′-adaptor bases) are aligned allowing for one mismatch only to the mature tRNA sequences, then reads that do not map are aligned allowing for one mismatch only to precursor tRNA sequences with the bowtie software. The remaining reads are aligned allowing for one mismatch only to miRNA reference sequences with miRDeep2. Based on alignment statistical analysis (mapping ratio, read length, fragment sequence bias), we determine whether the results can be used for subsequent data analysis. If so, the expression profiling and differentially expressed tRFs & tiRNAs and miRNAs are calculated. Principal component analysis (PCA), correlation analysis, pie plots, Venn plots, hierarchical clustering, scatterplots, and volcano plots are obtained for the expressed tRF & tiRNA in R or Perl environment for statistical computing and graphics construction.
Validation of protocol
This protocol has been used and validated in the following research article:
Wang et al. [10]. A pro-metastatic tRNA fragment drives aldolase A oligomerization to enhance aerobic glycolysis in lung adenocarcinoma. Cell Rep.
The validation of our protocol was substantiated by its application in a study investigating the molecular mechanisms underlying LUAD, particularly focusing on high-risk micropapillary/solid (MIP/SOL) patterns. The protocol’s effectiveness was demonstrated through the successful identification and isolation of specific LUAD cell populations, which were then subjected to tRF and tiRNA sequencing. This approach allowed for the precise characterization of differentially expressed tRFs and tiRNAs associated with high-grade LUAD tissues, as evidenced by the distinct molecular signatures obtained from high-risk and low-risk tissue cohorts.
The clinical relevance of our findings was further validated by the correlation between tRF-Val-CAC-024 expression levels and patient outcomes. High expression levels of tRF-Val-CAC-024 were significantly associated with shorter overall survival, as indicated by Kaplan–Meier survival analysis. Additionally, the expression of tRF-Val-CAC-024 was significantly higher in the later stages of LUAD, reinforcing its potential as a prognostic biomarker and therapeutic target.
Finally, the protocol’s utility in identifying LUAD biomarkers was underscored by the observation that tRF-Val-CAC-024 expression was significantly higher in the plasma of patients with high-risk LUAD components. Receiver operating characteristic (ROC) analysis confirmed the potential of tRF-Val-CAC-024 as a biomarker, with an area under the curve of 0.8596, sensitivity of 90.00%, and specificity of 75.56%. These results highlight the protocol's effectiveness in uncovering molecular signatures that could guide future diagnostic and therapeutic strategies for LUAD.
General notes and troubleshooting
General notes
1. Maintain an RNase-free environment throughout all wet lab experiments. Use gloves, clean work surfaces, pipettes, and centrifuges with ethanol, and utilize sterilized pipette tips and lab coats to minimize contamination.
2. Gain prior experience with the microtome, histological steps, and LAM microscope to ensure precision and efficiency.
3. Be aware that the LAM protocol is time-consuming and requires patience and attention to detail.
4. Refer to the PALM Robo Software 4.5 Quick Guide for detailed instructions on software operation.
5. While this protocol is designed for human LUAD tissues, it may be applicable to other species. Verify if tissue fixation time should be adjusted, as this can be species-specific.
6. Thicker tissues may require increased fixation time to ensure adequate fixation.
Potential limitations of the protocol
1. Variability in LCM efficiency: LCM efficiency can vary depending on tissue type, fixation quality, and operator skills.
Solution: Standardize tissue processing and LCM protocols and provide training for operators.
2. Small sample sizes: LCM typically yields small amounts of RNA, which may limit downstream analyses.
Solution: Pool samples from multiple slides or optimize RNA amplification protocols.
3. RNA degradation: Prolonged fixation or improper handling can lead to RNA degradation.
Solution: Use RNA stabilization reagents and minimize fixation time.
Translation of biomarkers into clinical practice
1. Validate identified biomarkers (e.g., tRF-Val-CAC-024) in larger patient cohorts. Perform functional studies to confirm their role in LUAD pathogenesis.
2. Develop diagnostic assays (e.g., qPCR, RNA-seq) for detecting biomarkers in clinical samples. Optimize assays for sensitivity, specificity, and reproducibility.
3. Explore the potential of biomarkers as therapeutic targets (e.g., small molecule inhibitors, RNA-based therapies). Conduct preclinical studies to evaluate efficacy and safety.
4. Collaborate with regulatory agencies to obtain approval for clinical use. Conduct clinical trials to demonstrate clinical utility.
Troubleshooting
Problem 1: Tissue loss during sectioning.
Possible cause: Tissue is not well fixed.
Solution: Ensure proper fixation by using fresh 10% neutral buffered formalin and fixing tissues for 24–48 h.
If tissue loss persists, collect new tissue and prepare new paraffin blocks.
Problem 2: Low RNA yield or no RNA.
Possible cause(s): RNA degradation during slide preparation or issues during RNA extraction.
Solution(s): Work in RNase-free conditions throughout the protocol. Use DEPC-treated water and RNase-free reagents. Perform RNA extraction immediately after LCM to minimize degradation.
Problem 3: Laser cutting or catapulting issues.
Possible cause(s): Incorrect settings in the PALM software, such as focus, energy, and speed.
Solution: Adjust the settings in the PALM software to optimize cutting and catapulting. Perform test runs on control tissues to determine optimal parameters.
Problem 4: Lack of data reproducibility.
Possible cause(s): Variations in tissue stages used or lack of tissue-specificity during LAM.
Solution(s): Ensure consistency in tissue selection and LAM procedures. If necessary, repeat the LAM and RNA sequencing experiments.
Problem 5: Insufficient tissue recovery.
Possible cause: Laser settings may be too low, or the tissue may not be properly adhered to the slide.
Solution: Adjust laser settings and ensure that the tissue is properly adhered to the slide before proceeding with LAM.
Problem 6: RNA integrity issues.
Possible cause: Inadequate sample preservation or handling.
Solution: Keep samples on ice during RNA extraction, ensure rapid freezing and storage at -80 °C, and use RNA stabilization reagents (e.g., RNAlater) if immediate freezing is not possible.
Problem 7: Sequencing library preparation failures.
Possible cause: Inconsistent or failed adapter ligation, or PCR amplification issues.
Solution: Verify the quality of adapters and enzymes, and optimize the PCR conditions (e.g., annealing temperature, cycle number).
Problem 8: Poor sequencing results.
Possible cause: Low library quality or contamination.
Solution: Re-prepare the sequencing library with additional purification steps and quality checks.
Problem 9: Bioinformatics analysis challenges.
Possible cause: Incorrect parameters or software bugs.
Solution: Review the analysis pipeline, adjust parameters, and consult with bioinformatics experts if necessary.
Problem 10: Equipment failures.
Possible cause: Malfunction or lack of maintenance.
Solution: Regularly maintain and calibrate equipment and follow the manufacturer's guidelines for troubleshooting.
Acknowledgments
This method was described by Wang et al. [10].
Competing interests
The authors declare no competing interests.
References
- Siegel, R. L., Miller, K. D., Wagle, N. S. and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J Clin. 73(1): 17–48. https://doi.org/10.3322/caac.21763
- Langmead, B., Trapnell, C., Pop, M. and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3): R25. https://doi.org/10.1186/gb-2009-10-3-r25
- Herbst, R. S., Morgensztern, D. and Boshoff, C. (2018). The biology and management of non-small cell lung cancer. Nature. 553(7689): 446–454. https://doi.org/10.1038/nature25183
- Travis, W. D., Brambilla, E., Noguchi, M., Nicholson, A. G., Geisinger, K. R., Yatabe, Y., Beer, D. G., Powell, C. A., Riely, G. J., Van Schil, P. E., et al. (2011). International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol. 6(2): 244–285. https://doi.org/10.1097/jto.0b013e318206a221
- Simone, N. L., Bonner, R. F., Gillespie, J. W., Emmert-Buck, M. R. and Liotta, L. A. (1998). Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 14(7): 272–276. https://doi.org/10.1016/s0168-9525(98)01489-9
- Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W. and Rajewsky, N. (2011). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40(1): 37–52. https://doi.org/10.1093/nar/gkr688
- Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1): 10. https://doi.org/10.14806/ej.17.1.200
- Robinson, M. D., McCarthy, D. J. and Smyth, G. K. (2009). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1): 139–140. https://doi.org/10.1093/bioinformatics/btp616
- Wang, Q., Song, X., Zhang, Y., Liang, S., Zhang, M., Wang, H., Feng, Y., Li, R., Ding, H., Chen, Y., et al. (2024). A pro-metastatic tRNA fragment drives aldolase A oligomerization to enhance aerobic glycolysis in lung adenocarcinoma. Cell Rep. 43(8): 114550. https://doi.org/10.1016/j.celrep.2024.114550
Article Information
Publication history
Received: Nov 23, 2024
Accepted: Feb 17, 2025
Available online: Mar 10, 2025
Published: Apr 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Wang, Z., Wang, Q., Xu, L., Mao, Q. and Jiang, F. (2025). A Protocol for Laser-Assisted Microdissection and tRF & tiRNA Sequencing in Lung Adenocarcinoma. Bio-protocol 15(7): e5253. DOI: 10.21769/BioProtoc.5253.
Category
Cancer Biology > Cancer biochemistry > Cancer metabolism
Systems Biology > Transcriptomics > RNA-seq
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









