- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Human iPSC-Derived Neuron and Oligodendrocyte Co-culture as a Small-Molecule Screening Assay for Myelination
Published: Vol 15, Iss 9, May 5, 2025 DOI: 10.21769/BioProtoc.5227 Views: 3398
Reviewed by: Xiaokang WuShanmugaPriyaa MadhukaranKrishna Murthy NakuluriMithun Santra

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
An Automated Imaging Method for Quantification of Changes to the Endomembrane System in Mammalian Spheroid Models
Margaritha M. Mysior and Jeremy C. Simpson
Jun 5, 2025 1646 Views

Quantifying Intracellular Distributions of HaloTag-Labeled Proteins With SDS-PAGE and Epifluorescence Microscopy
Julia Shangguan and Ronald S. Rock
Jul 20, 2025 2497 Views
Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1319 Views
Abstract
Neurons and oligodendrocytes are the building blocks of the brain. Neurons form synaptic connections and transmit signals, while oligodendrocytes, including oligodendrocyte precursor cells (OPCs) and their derivatives, are vital for central nervous system maintenance and myelination. The demand for human-specific neuron-oligodendrocyte model systems to study these interactions has grown, yet co-culture protocols remain limited. Recent advancements in the field provide methods for deriving co-cultures of neurons and OPCs from human induced pluripotent stem cells (hiPSC), each with distinct benefits and challenges. This study presents a time-efficient, reproducible method to derive neurons and O4-expressing oligodendrocytes, followed by a straightforward co-culture system that minimizes astrocyte differentiation and ensures robust neuron and oligodendrocyte populations.
Key features
• Reliable, stable generation of neurons and O4-expressing oligodendrocytes within a practical timeframe.
• Co-culture system utilizing hIPSC-derived neurons and O4-expressing oligodendrocytes.
• Maturation of neurons and oligodendrocytes achieved within 10 days of co-culturing.
Keywords: hiPSCBackground
Neurons and oligodendrocytes are pivotal cell types in the brain, supporting the central nervous system and participating in white matter formation [1–4]. This is critical for higher brain functions such as social and cognitive learning [5–8]. Neurons establish synaptic connections and transmit signals across the brain [9,10], while oligodendrocyte lineage cells—including oligodendrocyte precursor cells (OPCs), myelinating oligodendrocytes, and transitional cell types—perform essential functions for central nervous system (CNS) maintenance and myelination of axons [3,11,12]. Beyond myelination, oligodendrocytes support neuronal energy metabolism, help buffer potassium [13], and promote synaptic plasticity through the secretion of neurotrophic factors such as BDNF [14,15]. Early in life, the disruption of the oligodendrocyte-neuron unit results in axonal dysfunction and impairs neurodevelopment [16,17]; later, it can be observed in traumatic injuries, Alzheimer’s disease, demyelinating diseases such as multiple sclerosis [18–20], and psychiatric disorders such as bipolar disorders and schizophrenia [21].
Consequently, there is an increasing demand for the development of human-based neuron-oligodendrocyte model systems for studying these interactions in both healthy and diseased brains. Compared to equivalent models derived from mice, primary cultures, or animal-derived co-cultures [22,23], the induced pluripotent stem cell (iPSC) technology allows the generation of several cell types that can respond to the full spectrum of human nutritional and hormonal stimuli.
Although numerous methods exist for deriving neurons or OPCs individually from iPSCs [24,25], protocols for their co-culture are limited. Over the past five years, there has been a rise in neuron-oligodendrocyte co-culture studies. Notably, Dooves et al. [26], Assetta et al. [27], and von der Bey et al. [28] have developed comprehensive protocols for deriving neurons and OPCs from human iPSCs. Each protocol offers distinct advantages and drawbacks. Dooves et al. [26] presented a straightforward but time-consuming method involving small molecule-directed differentiation, achieving mature neurons by day 37 and mature oligodendrocytes by day 67, followed by a 28-day co-culture period. In contrast, Assetta et al. and von der Bey et al. achieved neuron and oligodendrocyte maturation and subsequent co-culture in a shorter timeframe, utilizing pro-myelinating compounds (small molecules that promote myelination by the direct interaction with the oligodendrocytes) or complex media changes [27,28].
In this study, we generate well-characterized populations of neurons and O4-expressing oligodendrocytes separately from the same iPSC line and from neural progenitor cells (NPCs) in a time-efficient, robust, and reproducible manner. Additionally, we aimed to establish a straightforward co-culture system using these defined cell types. Neurons were derived using a standardized small molecule-driven differentiation protocol, incorporating the CultureOneTM Supplement (Gibco) to minimize astrocyte production. This method yielded stable neuronal cultures after 29 days of differentiation. O4-expressing oligodendrocytes, cells that are considered pre-myelinating oligodendrocytes, were generated by integrating methodologies from Ehrlich et al. [25] and Dooves et al. [26]. A tetracycline-inducible lentivirus transduction method [25,29] was combined with simplified media changes [26], achieving stable O4-expressing oligodendrocytes after 28 days of differentiation. Compared to other protocols, this approach ensures that most of the cell population differentiates into a high percentage of neurons or oligodendrocyte lineage cells. In addition, the experimenter can control the percentage of astrocytes in co-culture. While the presence of astrocytes maintains neurons in culture for longer [30], they also decrease the percentage of neurons after differentiation and might represent a confounding factor in studying the exclusive interaction between neurons and oligodendrocytes [31].
This co-culture demonstrates the potential for future studies investigating neuron-oligodendrocyte crosstalk in various human myelin disorders [32]. The co-culture also offers high translational potential to humans and broad scientific impact. Since neurons and oligodendrocytes retain the genome of the donor [33], they could be used to develop drug screening platforms for targeted drug discovery and personalized medicine in a reasonable time.
Materials and reagents
Biological materials
1. Lentiviral vector (provided by Prof. Dr. Kuhlman [25,29])
Reagents
1. 4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, catalog number: D9542)
2. Accutase (Sigma-Aldrich, catalog number: A6964-500ML)
3. Antibiotic antimycotic (Merck, Sigma-Aldrich, catalog number: A5955-20ML)
4. Anti-forkhead box protein G1 (FoxG1) (dilution 1:400; working concentration: 2.25 μg/mL) (Abcam, catalog number: Ab18259)
5. Anti-galactosylceramidase (GalC) (dilution 1:250; working concentration: 4 μg/mL) (Millipore, catalog number: MAB342)
6. Anti-glial fibrillary acidic protein (GFAP) (dilution 1:3,500; working concentration: 0.5 μg/mL) (Novusbio, catalog number: NBP1-05198)
7. Anti-myelin-associated protein (MAG) (dilution 1:400; working concentration: 5 μg/mL) (Merck, catalog number: MAB1567)
8. Anti-microtubule-associated protein 2 (MAP2) (dilution 1:900; working concentration: 0.84 μg/mL) (Abcam, catalog number: Ab183830)
9. Anti-myelin basic protein (MBP) (dilution 1:1,500; initial concentration not disclosed from the manufacturer) (Invitrogen, catalog number: PA1-10008)
10. Anti-nestin (dilution 1:250; working concentration: 4 μg/mL) (Merck, Sigma-Aldrich, catalog number: MAB5326)
11. Anti-neuronalNuclei (NeuN) (dilution 1:500; working concentration: 2 μg/mL) (Merck, catalog number: ABN78)
12. Anti-neurofilament heavy (NFH) (dilution 1:400; working concentration: 20 μg/mL) (Merck, catalog number: N4142)
13. Anti-neural/glial antigen 2 (NG2) (dilution 1:100; working concentration: 10 μg/mL) (Invitrogen, catalog number: 14-6504-82)
14. Anti-oligodendrocyte marker O4 (O4) (dilution 1:400; working concentration: 1.25 μg/mL) (Merck, catalog number: MAB1326)
15. Anti-oligodendrocyte transcription factor 2 (Olig2) (dilution 1:250; initial concentration not disclosed from the manufacturer) (Merck, catalog number: AB9610)
16. Anti-paired box protein (Pax6) (dilution 1:100; working concentration: 20 μg/mL) (BioLegend, catalog number: 901301)
17. Anti-Sox10 (dilution 1:250; working concentration: 4 μg/mL) (Millipore, catalog number: AB5727)
18. Anti-class III-beta tubulin (Tuj1) (dilution 1:750; working concentration: 1.32 μg/mL) (BioLegend, catalog number: MMS-435P)
19. Ascorbic acid (AA) (Sigma-Aldrich, catalog number: A4403-100MG)
20. B27 with vitamin A (Thermo Fisher Scientific, catalog number: 17504044)
21. B27 without vitamin A (Thermo Fisher Scientific, catalog number: 12587010)
22. Basic fibroblast growth factor (bFGF or FGF) (Sigma-Aldrich, catalog number: SRP4037-50UG)
23. Biotin (Sigma-Aldrich, catalog number: B4501)
24. Brain-derived neurotrophic factor (BDNF) (PeproTech, catalog number: AF-450-02-10UG)
25. CHIR99021 (Cayman Chemicals, catalog number: CAY13122-10mg)
26. Cholest-4-en-3-one (Sigma-Aldrich, catalog number: 188174)
27. Clemastine fumarate salt (Sigma-Aldrich, catalog number: SML0445)
28. CultureOneTM supplement (100×) (Thermo Fisher Scientific, catalog number: A3320201)
29. Cyclic adenosine monophosphate (Merck, catalog number: D0260-25UG)
30. DMEM/F12 – L-glutamine (Thermo Fisher Scientific, Gibco, catalog number: 21331-020)
31. DMEM/F12 +/+ L-glutamine/sodium bicarbonate (Thermo Fisher Scientific, Gibco, catalog number: 11320-033)
32. DMEM/F12 + GlutaMAX (Thermo Fisher Scientific, Gibco, catalog number: 31331-093)
33. Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2438)
34. Donkey-anti-rabbit (Alexa 488) (Jackson Immuno Research, catalog number: 711-545-152)
35. Donkey-anti-chicken (Alexa 647) (Jackson Immuno Research, catalog number: 703-175-155)
36. Donkey-anti-mouse (Cy3) (Jackson Immuno Research, catalog number: 715-165-150)
37. Dorsomorphin (Cayman Chemicals, catalog number: 11967-5mg)
38. Doxycycline (Sigma-Aldrich, catalog number: D3447-500MG)
39. DPBS (-/- Ca2+ and Mg2+) (Gibco, catalog number: 14190-094)
40. DPBS (+/+ Ca2+ and Mg2+) (Gibco, catalog number: 14040-133)
41. Epidermal growth factor (EGF) (Sigma-Aldrich, catalog number: SRP3027-500UG)
42. Fluoromount G (Invitrogen, catalog number: 00-4958-02)
43. Geltrex (Thermo Fisher Scientific, catalog number: A1413202)
44. Glial cell line–derived neurotrophic factor (GDNF) (PeproTech, catalog number: 450-10)
45. GlutaMAX (Thermo Fisher Scientific, catalog number: 35050-038)
46. Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148)
47. Human leukemia inhibitory factor (hLIF) (PeproTech, catalog number: AF-300-05-25UG)
48. Insulin (Sigma-Aldrich, catalog number: I9278-5mL)
49. BioLaminin (LN521) (Biolaminin, catalog number: LN521-02)
50. MEM non-essential amino acids (Thermo Fisher Scientific, catalog number: 11140-035)
51. Miconazole nitrate salt (Sigma-Aldrich, catalog number: M3512)
52. Mouse laminin (L2020) (Sigma-Aldrich, catalog number: L2020-1MG)
53. N1 supplement (Sigma-Aldrich, catalog number: N6530-5ml)
54. N2 supplement (Thermo Fisher Scientific, catalog number: 17052048)
55. Neurobasal media (Thermo Fisher Scientific, Gibco, catalog number: 21103-049)
56. Noggin (PeproTech, catalog number: 120-10C-50UG)
57. Normal donkey serum (Merck, catalog number: S30-100ml)
58. NSC freezing media (Merck, catalog number: SCM007)
59. NT3 (Merck, catalog number: CS204470)
60. Papain (Worthington Biochemistry, catalog number: LK003178)
61. Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: 441244)
62. PBS (Gibco, catalog number: 10010-015)
63.Poly-L-ornithine solution (PLO) (Sigma-Aldrich, catalog number: P4957-50mL)
64. Potassium phosphate (Sigma-Aldrich, catalog number: P5655-500G)
65. Protamine sulfate (Sigma-Aldrich, catalog number: 110123)
66. Purmorphamine (Calbiochem, catalog number: 540220-5mg)
67. Puromycin (Thermo Fisher Scientific, catalog number: J67236)
68. Rock inhibitor (also known as Y-27632) (Lubioscience, catalog number: HB2297-5MG)
69. SB431542 (Sigma-Aldrich, catalog number: S4317)
70. Smoothened agonist (SAG) (Calbiochem, catalog number: 566660)
71. Sodium hydroxide solution (NaOH) (Fuka, catalog number: 35255)
72. Sodium phosphate (ACROS, catalog number: 204855000)
73. Triiodothyronine (T3) (Merck, catalog number: T6397-100MG)
74. Triton X-100 (Sigma-Aldrich, catalog number: 93443-100ml)
75. β-Mercaptoethanol (Sigma-Aldrich, catalog number: M3148-25ml)
Solutions
1. Neuronal differentiation media with CultureOneTM supplement for neurons (NDMC) (see Recipes)
2. Neural maintenance media for oligodendrocyte progenitor cells (OPC) and oligodendrocytes (NMM) (see Recipes)
3. Neural stem cell maintenance media for NPCs maintenance (NSMM) (see Recipes)
4. Neuroglia co-culture media (see Recipes)
5. NPC transduction media for transduction of NPCs to obtain (OPC) (NTM) (see Recipes)
6. Clemastine or miconazole treatment
7. Sodium phosphate buffer (see Recipes)
8. 4% PFA (see Recipes)
9. Blocking buffer (PBS+) (see Recipes)
10. Permeabilization buffer (see Recipes)
11. Poly-L-ornithine/BioLaminin (PLO/LN521) coating (see Recipes)
12. Poly-L-ornithine/mouse Laminin (PLO/L2020) coating (see Recipes)
13. Geltrex coating (see Recipes)
Recipes
Media recipes
1. Neuronal differentiation media with CultureOneTM supplement for neurons (NDMC)
| Reagent | Final concentration | Amount |
|---|---|---|
| Neurobasal media | N/A | 474 mL |
| GlutaMAX (100×) | 1× | 5 mL |
| B27 supplemented with vitamin A (50×) | 1× | 10 mL |
| Antibiotic antimycotic (100×) | 1× | 5 mL |
| Ascorbic acid (100 mM) | 200 μM | 1 mL |
| CultureOneTM supplement (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
To prepare a 100 mM stock solution of ascorbic acid, dissolve 100 mg of the powder in 5.67 mL of sterile ddH2O.
Mix reagents and filter (pore size 0.4 μm or smaller) media. Media can be stored for 2 weeks at 4 °C.
Note: The CultureOneTM supplement minimizes astrocyte differentiation. It can be introduced during a later phase of differentiation to obtain the desired number of astrocytes.
2. Neural maintenance media for oligodendrocyte progenitor cells and oligodendrocytes (NMM)
| Reagent | Final concentration | Amount |
|---|---|---|
| Neurobasal media | N/A | 241 mL |
| DMEM/F12 + GlutaMAX | N/A | 241 mL |
| GlutaMAX (100×) | 0.5× | 2.5 mL |
| MEM non-essential amino acids (100×) | 0.5× | 2.5 mL |
| B27 supplement without vitamin A (50×) | 0.5× | 5 mL |
| N2 supplement (100×) | 1× | 2.5 mL |
| Antibiotic antimycotic (100×) | 1× | 5 mL |
| Insulin (10.5 mg/mL) | 5 μg/mL | 238 μL |
| β-Mercaptoethanol (143 mM) | 10 μM | 35 μL |
| Total | N/A | 500 mL |
Mix reagents and filter (pore size 0.4 μm or smaller) media. Media can be stored for 2 weeks at 4 °C.
3. Neural stem cell maintenance media for NPC maintenance (NSMM)
| Reagent | Final concentration | Amount |
|---|---|---|
| Neurobasal media | N/A | 237.5 mL |
| DMEM/F12 +/+ L-glutamine/sodium bicarbonate | N/A | 237.5 mL |
| GlutaMAX (100×) | 1× | 5 mL |
| B27 supplemented with vitamin A (50×) | 1× | 10 mL |
| N2 supplement (100×) | 1× | 5 mL |
| Antibiotic antimycotic (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Mix reagents and filter (pore size 0.4 μm or smaller) media. Media can be stored for 2 weeks at 4 °C.
4. Neuroglia co-culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| Neurobasal media | N/A | 237.5 mL |
| DMEM/F12 – L-glutamine | N/A | 237.5 mL |
| MEM non -essential amino acids (100×) | 1× | 5 mL |
| B27 supplemented with vitamin A (50×) | 1× | 10 mL |
| N1 supplement (100×) | 1× | 5 mL |
| Antibiotic antimycotic (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Mix reagents and filter (pore size 0.4 μm or smaller) media. Media can be stored for 2 weeks at 4 °C.
5. NPC transduction media for transduction of NPCs to obtain oligodendrocyte progenitor cells (NTM)
| Reagent | Final concentration | Amount |
|---|---|---|
| Neurobasal media | N/A | 241.25 mL |
| DMEM/F12 +/+ L-glutamine/sodium bicarbonate | N/A | 241.25 mL |
| GlutaMAX (100×) | 1× | 5 mL |
| B27 supplement without vitamin A (50×) | 0.5× | 5 mL |
| N2 supplement (100×) | 0.5× | 2.5 mL |
| Antibiotic antimycotic (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Mix reagents and filter (pore size 0.4 μm or smaller) media. Media can be stored for 2 weeks at 4 °C.
6. Clemastine or miconazole treatment
Clemastine: Prepare a 100 mM stock solution dissolving 45.9 mg of clemastine in 1 mL of DMSO. Store at -20 °C.
Miconazole: Prepare a 100 mM stock solution by dissolving 47.914 mg of miconazole in 1 mL of DMSO. Store at -20 °C.
Immunostaining recipes
7. Sodium phosphate buffer, 0.4 M stock, pH 7.4–7.6
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium phosphate | N/A | 0.53 g |
| Potassium phosphate | N/A | 0.28 g |
| Deionized water | N/A | ~50 mL |
| HCl | 1 M | As needed |
| NaOH | 1 M | As needed |
| Total | N/A | 50 mL |
Dissolve in ~90% of deionized water, adjust pH to 7.4–7.6 with 1 M HCl or 1 M NaOH, and then bring to the final volume.
8. 4% Paraformaldehyde fixative (PFA), pH 7–7.4
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde | N/A | 2 g |
| Deionized water | N/A | ~37.5 mL |
| 0.4 M sodium phosphate buffer | 0.4 M | 12.5 mL |
| HCl | 1 M | As needed |
| Total | N/A | 50 mL |
a. Add 2/3 deionized water to a flask and heat to 60 °C, keeping below 70 °C.
b. Add the paraformaldehyde with constant stirring for 10 min.
c. Slowly add a few drops of 10 M NaOH to the cloudy mixture, stirring until the solution clears.
d. Stir a few minutes longer, adding more NaOH if necessary to clear.
e. Remove from heat and cool to room temperature.
f. Filter with #3 Whatman® filter paper.
g. Add the 0.4 M sodium phosphate buffer.
h. Adjust pH to 7–7.4 with 1 M HCL and check with a pH meter.
i. Bring to the final volume with deionized water.
Notes:
1. Paraformaldehyde will never appear to be dissolving until the NaOH is added.
2. Be patient, allow time for the NaOH to cause the paraformaldehyde to dissolve before adding more.
3. There is always a little colloidal “fluff’ in an unfiltered state.
4. Do not weigh paraformaldehyde in plastic weigh boats; instead, use a glass beaker to minimize static dispersion of paraformaldehyde dust.
5. Wash out the filter paper with tap water before discarding, rinse everything that was in contact with paraformaldehyde, and make sure you collect it in a special container.
9. Blocking buffer (PBS+)
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | N/A | 45 mL |
| Normal donkey serum (100%) | 10% | 5 mL |
| Total | N/A | 50 mL |
Mix reagents and filter (pore size 0.2 μm). Blocking buffer can be stored at 4 °C for 48 h.
10. Permeabilization buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Blocking buffer (PBS+) | N/A | 49 mL |
| Triton X-100 (10%) | 0.2 % | 1 mL |
| Total | N/A | 50 mL |
Use the filtered PBS+. Permeabilization buffer can be stored at 4 °C for 48 h.
Coating of plates
11. Poly-L-ornithine/BioLaminin (PLO/LN521) coating
a. Place coverslips into a 24-well plate.
b. Dilute PLO in PBS (500 μL/well = 75 μL PLO + 425 μL PBS).
c. Incubate at room temperature for 2 h.
d. Remove PLO and wash three times with PBS.
e. Allow coverslips to dry slightly before the next step.
f. Add 10 μg/mL of LN521 + cold DPBS ++ [50 μL BioLaminin + 450 μL DPBS+ Ca2+ and Mg2+(DPBS++)].
g. Cover with parafilm.
h. Incubate at 37 °C overnight or at room temperature for 48 h.
i. Remove BioLaminin and immediately add medium with the cells.
Notes:
1. PLO should be stored at -20 °C in smaller aliquots upon arrival.
2. Thaw LN521 on ice, aliquot, and store at -20 °C.
3. PLO/LN521 coverslips can be prepared in advance. To store, add cold DPBS to the wells, wrap the plate with parafilm, and keep it at 4 °C for up to two weeks. Before use, place the plate in the incubator for 30 minutes. Ensure that there is sufficient liquid to cover the wells to prevent them from drying out.
12. Poly-L-ornithine/mouse Laminin (PLO/L2020) coating
a. Place coverslips into a 24-well plate.
b. Dilute PLO in PBS (500 μL/well = 75 μL PLO + 425 μL PBS).
c. Incubate at room temperature for 2 h.
d. Remove PLO and wash three times with PBS.
e. Allow coverslips to dry slightly before the next step.
f. Add L2020 + cold DPBS ++ (50 μL L2020 + 4.95 mL DPBS++).
g. Cover with parafilm.
h. Incubate at 37 °C overnight or at room temperature for 48 h.
i. Remove L2020 and immediately add medium with the cells.
Notes:
1. Thaw L2020 on ice, aliquot, and store at -20 °C.
2. PLO/L2020 coverslips can be prepared in advance. To store, add cold DPBS to the wells, wrap the plate with parafilm, and keep it at 4 °C for up to two weeks. Before use, place the plate in the incubator for 30 min. Ensure that there is sufficient liquid to cover the wells to prevent them from drying out.
13. Geltrex coating
a. Dilute Geltrex 1:1 with cold DMEM/F12 – L-glutamine and aliquot in appropriate sizes.
b. Store aliquots at -20 °C.
c. Dilute 1:100 with cold DMEM/F12 – L-glutamine.
d. Add the appropriate amount to flask or well (5 mL per T75 flask; 1 mL per 6-well plate).
e. Incubate at 37 °C for 1 h.
f. Remove Geltrex and immediately add medium with the cells.
CRITICAL: Geltrex must be handled at low temperatures to prevent polymerization. It is recommended to pre-cool laboratory consumables, such as tubes for aliquots, pipette tips, and Falcon tubes, prior to diluting and aliquoting. Ideally, perform this work on a cooling block or on ice.
Laboratory supplies
1. 0.2 μm filters (Sarstedt, catalog number: 83.1826.001)
2. 1.5 mL Eppendorf tubes (Eppendorf, catalog number: 0030120.086)
3. 10 μL pipette tips (Tip One, catalog number: S1111-3700)
4. 1,000 μL pipette tips (Tip One, catalog number: S1111-6001)
5. L graduated pipette (Greiner Bio-One, catalog number: 607180)
6. 15 mL Falcon (Cellstar Tubes, catalog number: 188271N)
7. 200 μL pipette tips (Tip One, catalog number: S1111-0006)
8. 24-well plate (Falcon, catalog number: 353047)
9. 50 mL Falcon (Cellstar Tubes, catalog number: 227261)
10. 5 mL graduated pipette (Greiner Bio-One, catalog number: 606180)
11. 6-well plates (Falcon, catalog number: 353046)
12. Whatman® qualitative filterpapier (Merck, catalog number: WHA1001090)
13. Coverslips (VWR, catalog number: 631-1578)
14. Microscope slides (25 mm × 75 mm × 1.0 mm, super frost) (Epredia, catalog number: J1800AMNZ)
15. One-way 20 mL syringes (Injekt, catalog number: 4606205V
16. 500 mL vacuum filter, 0.45 μm (Sarstedt, catalog number: 83.3941)
17. T75 flask (Sarstedt, catalog number: 833911002)
Equipment
1. Pipet boy (Integra, catalog number: 155000)
2. Incubator (Binder, catalog number: 9040-0131)
3. Benchtop centrifuge 420R (Hettich, model: Rotina 420R)
4. Benchtop centrifuge (Eppendorf, model: 5430/5430R)
5. 4 °C refrigerated storage unit (Liebherr, model: K3130Index20C/001)
6. -20 °C ProFiline freezer (Liebherr, model: GG5210Index40A/006)
7. 37 °C water bath (Sun Lab, model: SU1811)
8. Widefield microscope Zeiss, Axio Observer 1 (Center for Microscopy and Image Analysis, University of Zurich)
Software and datasets
1. Fiji ImageJ-win64
2. GraphPad Prism V10
3. ZEN V3.1 (blue edition)
Procedure
Point 1:
The protocol involves several steps of Accutase-based cell passaging. Follow the standard enzymatic dissociation as outlined:
1. Prewarm DMEM/F12 + GlutaMAX in a 37 °C water bath for a maximum of 10 min (for neutralization).
2. Aspirate the used media and wash 1× with DPBS (5 mL per T75 flask).
3. Add cold Accutase (4 mL per T75 flask).
4. Incubate at 37 °C for 3–5 min. Check after 3 min.
5. By gently tapping the plate, the cells will visibly come off.
6. Neutralize the Accutase by adding 4 mL of DMEM/F12 + GlutaMAX and transfer everything into a 15 mL Falcon tube.
7. Centrifuge at 200 rcf for 5 min at room temperature (RT).
8. Aspirate the supernatant and resuspend the cells in the suggested specific media for cell type.
A. Neural progenitor cell (NPC) maintenance
This protocol starts with NPCs since they serve as a critical intermediate cell type, capable of differentiating into neurons and OPCs under specific conditions. It outlines robust and reproducible methods for NPC maintenance, neuronal differentiation, and OPC generation, followed by their co-culture for studying neuron-oligodendrocyte interactions.
NPCs, provided by the Institute of Regenerative Medicine (IREM), Zurich, were derived from peripheral blood nuclear cells (PBMCs) via iPSC reprogramming as described previously [34]. Quality control for iPSCs [permission from the cantonal ethics commission of Zurich, Switzerland (KEK-ZH-2014-0430)] has been performed at RNA and protein level for pluripotent markers, in vitro differentiation, and karyotyping.
1. Thawing and seeding NPCs: thaw NPCs onto Geltrex-coated T75 flasks in 10 mL of NSMM (Recipe 3) + 10 μM Y (Rock inhibitor) + factors [5 ng/mL FGF, 10 ng/mL hLIF, 4 μM CHIR99021 + 3 μM SB431542].
a. Prepare Geltrex-coated flasks (Recipe 13).
b. Prewarm NSMM in a 37 °C water bath for a maximum of 10 min (for neutralization and seeding).
c. Thaw frozen cells in a 37 °C water bath (for no longer than 1 min).
d. Gently transfer cells into a 15 mL Falcon and dropwise add 10 mL of prewarmed NSMM to the side of the Falcon tube.
Note: NPCs are sensitive; add the media on the flask’s corner without touching the cells.
e. Spin down at 200 rcf for 5 min at RT.
f. Gently aspirate away the supernatant.
g. Resuspend cells in 1 mL of prewarmed NSMM + factors + Y-27632.
h. Remove Geltrex and immediately seed cells in a total of 10 mL of NSMM + factors + Y-27632.
i. Place the flask into the incubator and gently move back to front and left to right to homogenously distribute the cells and avoid clumping.
2. Daily media change.
a. Prewarm NSMM in a 37 °C water bath.
b. Add factors (5 ng/mL FGF, 10 ng/mL hLIF, 4 μM CHIR99021, and 3 μM SB431542) to prewarmed NSMM.
c. Aspirate 3/4 of the media from the flask.
d. Gently add fresh prewarmed NSMM + factors to the side of the flask.
3. Splitting NPCs: Typically, NPCs reach 90%–100% confluency within 3–4 days, depending on initial plating density and culture conditions. When they are confluent, enzymatically split them using Accutase.
a. Prepare Geltrex-coated T75 flasks (Recipe 13).
b. Prewarm NSMM and DMEM/F12 + GlutaMAX in a 37 °C water bath for a maximum of 10 min (for seeding and neutralization, respectively).
c. Aspirate the supernatant and resuspend the cells in 1 mL of NSMM + factors + Y-27632.
d. Remove Geltrex and immediately plate 2.5–3 million cells per T75 flask in 10 mL of NSMM + factors + Y-27632.
e. Place the flask into the incubator and gently move back to front and left to right to homogenously distribute the cells and avoid clumping.
Note: NPCs should be split at least once before being used for differentiation into neurons or oligodendrocyte progenitor cells.
4. Assessing proliferation capacity: To ensure NPC quality and purity before further differentiation, the expression of two specific NPC markers, PAX6 and Nestin [35], was assessed. To this aim, plate the cells onto coverslips coated with PLO/LN521 at a density of 10,000–15,000 cells per coverslip. Allow the cells to attach for 24 h and then fix them with 4% PFA (Recipe 8) for 12 min at RT. Wash the cells 3× with PBS, leaving the final wash on the cells. Use the coverslips for immunostaining within a timeframe of 24 h to two weeks (Figure 1).
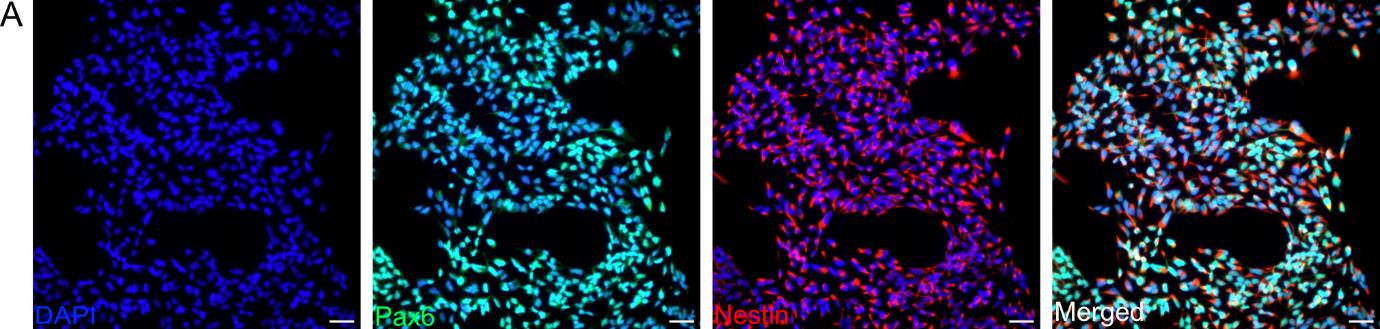
Figure 1. Characterization of neural progenitor cells (NPCs) derived from human induced pluripotent stem cells (hiPSCs). Representative immunofluorescence images showing different proliferative NPC markers at passage 10. Nestin (red) marks intermediate filaments and Pax6 (green) stains a specific transcription factor. Nuclei are counterstained with DAPI (blue). Scale bar, 50 μm. Images are representative of three biological replicates. Images were acquired and processed using identical microscope settings. Abbreviations are as follows: DAPI: 4’,6-diamidin-2-phenylindol; Pax6: paired box protein 6.
B. Neurons derived from NPCs
Differentiate neurons according to the manufacturer’s user guideline (CultureOneTM Supplement, Gibco) as follows:
1. On day 0, passage NPCs at a density of 3 × 106 cells onto PLO/L2020-coated T75 flasks in 10 mL of NDMC (Recipe 1) + 10 μM Y-27632 (Rock inhibitor) and factors (10 ng/mL BDNF and 10 ng/mL GDNF). Rock inhibitor is essential for preventing apoptosis and improving cell survival during and after dissociation [36]. BDNF and GDNF promote neuronal survival and differentiation [37]. By day 10–14, cells begin extending neurites, and early neuronal markers such as Tuj1 can be detected. Mature markers like MAP2 and NeuN are expressed by day 29.
a. Prepare the PLO/L2020-coated T75 flasks (Recipe 12) at least 24 h in advance.
b. Aspirate the supernatant and resuspend the cells in 1 mL of NDMC + factors + Y-27632.
c. Remove L2020 and immediately plate 2.5–3 million cells per T75 flask in 10 mL of NDMC + factors + Y-27632.
d. Place the flask into the incubator and gently move back to front and left to right to homogenously distribute the cells and avoid clumping.
2. Change media every two to three days.
a. Prewarm NDMC in a 37 °C water bath (maximum of 10 min).
b. Add factors (10 ng/mL BDNF and 10 ng/mL GDNF) to prewarmed NDMC.
c. Aspirate 2/3 of the media from the T75 flask.
d. Gently add fresh prewarmed NDMC + factors to the side of the flask.
Note: Avoid touching the cells during media changes to prevent detachment, which could reduce differentiation efficiency.
3. Following the initial plating of NPCs, the cells are maintained under neurodifferentiation conditions with regular media changes to support their maturation into neurons. Under optimal conditions, neurons typically reach 90%–100% confluency within 5–7 days after initial plating. When they are confluent, enzymatically split them using papain (135 u/vial) and Accutase. Papain enhances neuronal survival during dissociation by gently dissociating cells while preserving their viability. Prepare PLO/L2020-coated T75 flasks (Recipe 12) at least 24 h in advance.
a. Prepare 30 min before use. Dissolve 1 vial of papain in 10 mL of cold Accutase according to the manufacturer’s recommendation and store at 4 °C.
b. Prewarm NDMC and DMEM/F12 + GlutaMAX in a 37 °C water bath for a maximum of 10 min (for seeding and neutralization, respectively).
c. Aspirate the used media and wash once with DPBS (5 mL per T75 flask).
d. Add cold papain/Accutase (4 mL per T75 flask).
e. Aspirate the supernatant and resuspend the cells in 1 mL of NDMC + factors + Y-27632.
f. Remove L2020 and immediately plate 2–3 million cells per T75 flask in 10 mL of NDMC + factors + Y-27632.
Note: As differentiation progresses, the number of cells needed for seeding increases due to the lack of proliferating cells. Ideally, neurons should only be split within the first 14 days or when they are used for the assessment of lineage markers or for co-culture.
4. Continue changing media until day 29 or later, following the instructions in step B2.
5. At day 24/25, seed 25,000–30,000 neurons onto coverslips coated with PLO/LN521 to facilitate final maturation and subsequent immunostaining for marker validation.
a. Prepare PLO/LN521-coated coverslips (Recipe 11) at least 24 h in advance.
b. Perform papain/Accutase treatment as outlined in step A3.
c. Remove LN521 and immediately plate 25,000–30,000 cells per coverslip in 500 μL of NDMC + factors + Y-27632.
Note: The number of cells depends on the clumping and attachment of the specific neurons generated.
d. Place the plate into the incubator and gently move back to front and left to right to homogenously distribute the cells and avoid clumping.
6. Fix the coverslips on day 28 or 29.
a. Aspirate the used media and wash once with PBS.
b. Add 500 μL of 4% PFA and fix for 12 min at RT.
c. Wash 3× with PBS, leaving the last wash on the cells.
d. Use coverslips for immunostaining within a timeframe of 24 h to two weeks (Figure 2).
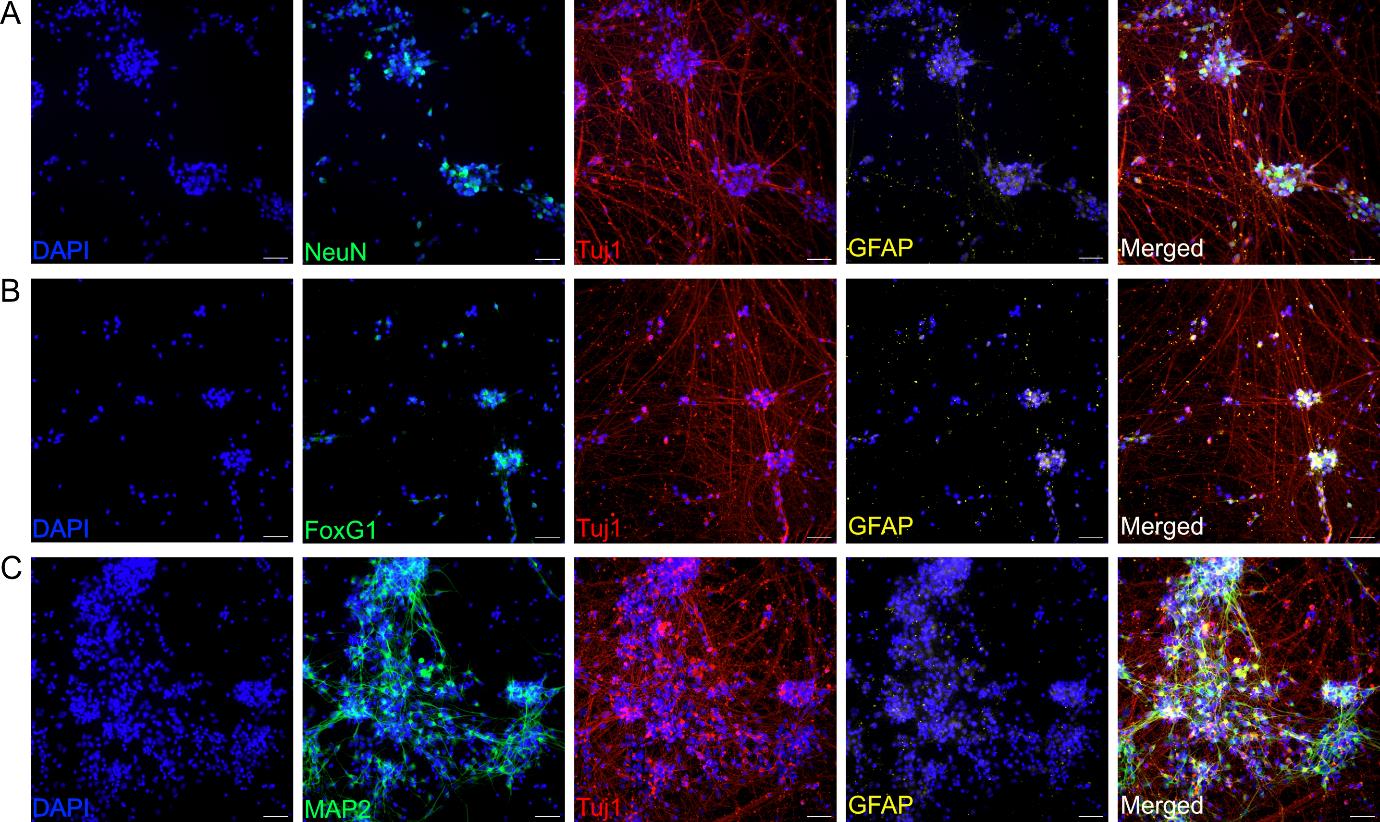
Figure 2. Characterization of day-29 neurons derived from neural progenitor cells (NPCs). Representative immunofluorescence images showing the expression of specific neuronal markers. (A) NeuN (green), Tuj1 (red), astrocyte marker GFAP (yellow), and merged (white). NeuN is a marker of mature neuronal differentiation [38,39]. (B) FoxG1 (green), Tuj1 (red), GFAP (yellow), and merged (white). FoxG1 is a marker for early cortical neurons [40]. (C) MAP2 (green), Tuj1 (red), GFAP (yellow), and merged (white). MAP2 is a marker of neuronal differentiation [41]. Nuclei are counterstained with DAPI (blue). Scale bar, 50 μm. Images are representative of three biological replicates. Images were acquired and processed using identical microscope settings. DAPI: 4’,6-diamidin-2-phenylindol; FoxG1: Forkhead box protein G1; GFAP: glial fibrillary acidic protein; MAP2: microtubule-associated protein 2; NeuN: also known as Fox-3, RNA binding protein; Tuj1: class III-beta tubulin.
C. Oligodendrocyte progenitor cells (OPCs) derived from NPCs
OPCs were generated from NPCs by combining two previously published protocols [25,26]. The differentiation protocol begins on day -6 with the seeding of NPCs into specific conditions to induce oligodendrocyte lineage commitment. The timeline includes lentiviral transduction (Day 0) and subsequent media changes designed to promote OPC maturation and marker expression, which peaks around day 28.
1. On day -6, passage NPCs at a density of 3 × 106 cells onto Geltrex-coated T75 flask in NSMM + factors (3 μM CHIR99021, 10 μM SB431542, 1 μM dorsomorphin, 0.5 μM purmorphamine).
a. Prepare Geltrex-coated T75 flasks (Recipe 13).
b. Enzymatically detach NPCs from the flask as outlined in Procedure, point 1.
c. Prewarm NSMM and DMEM/F12 + GlutaMAX in a 37 °C water bath for a maximum of 10 min (for seeding and neutralization, respectively).
d. As a final step of Accutase dissociation, aspirate the supernatant and resuspend the cells in 1 mL of NSMM media + factors + Y-27632.
e. Remove Geltrex and immediately plate 2.5–3 million cells per T75 flask in 10 mL of NSMM media + factors + Y-27632.
2. On day -4, NPC media is changed to NTM (Recipe 5) + factors (10 μM SB431542, 3 μM CHIR99021, 1 μM dorsomorphin, 0.5 μM purmorphamine) to support early differentiation of NPCs into pre-OPC stages.
a. Prewarm NTM in a 37 °C water bath for a maximum of 10 min.
b. Add factors to prewarmed NTM.
c. Aspirate the used media and gently add fresh NTM + factors to the side of the flask.
3. On day -2, split NPC onto Geltrex-coated 6-well plates at a density of 500,000 cells per well in NTM + factors [3 μM CHIR99021, 150 μg/mL ascorbic acid (AA), and 0.5 μM purmorphamine].
a. Prepare Geltrex-coated 6-well plates (Recipe 13).
b. Prewarm NTM and DMEM/F12 + GlutaMAX in a 37 °C water bath for a maximum of 10 min (for seeding and neutralization, respectively).
c. Aspirate the supernatant and resuspend the cells in 1 mL of NTM + factors + Y-27632.
d. Remove Geltrex and immediately plate 500,000 cells per well in 2.5 mL of NTM + factors + Y-27632.
4. On day 0, transduce cells with the lentivirus vector [multiplicity of infection (MOI) 50] in fresh NTM supplemented with 3 μM CHIR99201 + 0.5 μM smoothened agonist + 150 μg/mL AA + 5 μg/mL protamine sulfate. Lentiviral transduction is performed to enhance OPC-specific gene expression [25,29]. CHIR99021 (GSK3 inhibitor) promotes Wnt signaling [42] to support oligodendrocytes lineage commitment. Noggin inhibits BMP signaling, ensuring cells remain in the oligodendrocyte lineage [43].
a. Prewarm NTM media.
b. Add factors and lentivirus vector to warmed NTM.
c. Remove all used media and add fresh NTM + factors + lentivirus vector to the side of the wells.
CAUTION: (Risk-mitigation strategies) Lentiviral transduction must be performed in a BSL2 laboratory in a biological safety cabinet. Proper disposal of contaminated materials, such as pipettes and media, should follow institutional biosafety protocols. Always wear appropriate PPE, including gloves, a lab coat, and safety glasses.
5. On day 1, 24 h after transduction, change media to NMM (Recipe 2) + factors [5 ng/mL epidermal growth factor (EGF) + 5 ng/mL fibroblast growth factor (FGF) + 1 μg/mL L2020 + 50 μg/mL AA + 2 μg/mL doxycycline + 0.5 μg/mL puromycin].
a. Prewarm NMM in a 37 °C water bath.
b. Add factors to NMM.
c. Remove transduction media and gently add fresh NMM + factors to the side of the wells.
6. Change media every two days.
Note: Media composition is similar to day 1 (step C5).
7. On day 5, media should contain new factors to promote differentiation.
a. Prewarm NMM in a 37 °C water bath.
b. Add 5 ng/mL EGF + 5 ng/mL FGF + 1 μg/mL L2020 + 50 μg/mL AA + 2 μg/mL doxycycline + 0.5 μg/mL puromycin + 50 ng/mL noggin to NMM.
c. Aspirate the used media and replace with the new prepared media from step C7b.
8. On day 7, end puromycin selection by removing puromycin. Do this by replacing the media with fresh NMM + factors (1 μg/mL L2020 + 50 μg/mL AA + 50 ng/mL noggin + 2 μg/mL doxycycline).
a. Prewarm NMM in a 37 °C water bath.
b. Add factors to NMM.
c. Aspirate the used media and replace with the new prepared media from step C8b.
Note: Once cells have reached 80%–90% confluency, they can be pulled into a T75 flask. Accutase passaging is as described in Procedure, point 1; use 1 mL of Accutase per well. Make sure to use the correct media composition.
9. On day 10, remove doxycycline by replacing media with fresh NMM + factors (1 μg/mL L2020 + 50 μg/mL AA + 50 ng/mL noggin).
a. Prewarm NMM in a 37 °C water bath.
b. Add factors to NMM.
c. Aspirate the used media and replace with the new prepared media from step C9b.
10. Change media every other day and replace with fresh NMM + factors (1 μg/mL L2020+ 50 μg/mL AA + 50 ng/mL noggin).
11. Between days 14 and 18, perform a second puromycin selection to promote the full selection of transduced cells.
a. Prewarm NMM in a 37 °C water bath.
b. Add 1 μg/mL L2020 + 50 μg/mL AA + 50 ng/mL noggin + 0.5 μg/mL puromycin to NMM.
c. Aspirate the used media and replace with the new prepared media from step C11b.
12. At day 20, end the puromycin selection by replacing media with fresh NMM +1 μg/mL L2020 + 50 μg/mL AA + 50 μg/mL noggin.
a. Prewarm NMM in a 37 °C water bath.
b. Add factors to NMM.
c. Aspirate the used media and replace with prepared media from step C12b.
13. Continue changing media every second day, keeping oligodendrocytes in cultures in the media of step C9 until needed for quality control or co-culture.
Note: OPCs typically reach 85%–95% confluency within 4–6 days, depending on initial seeding density and culture conditions. Split at a 1:2–1:3 ratio for optimal growth. Cell proliferation decreases as cells mature. Follow the general enzymatic detachment procedure using Accutase as outlined in Point 1. From day 10, use NMM supplemented with 1 μg/mL L2020, 50 μg/mL ascorbic acid (AA), and 50 μg/mL noggin.
14. Check the oligodendrocytes for specific lineage markers at day 28 by plating 15,000–20,000 oligodendrocyte progenitor cells onto coverslips coated with PLO/LN521.
a. Prepare PLO/LN521-coated coverslips (Recipe 11) at least 24 h in advance.
b. Prewarm NMM and DMEM/F12 + GlutaMAX in a 37 °C water bath for a maximum of 10 min (for seeding and neutralization, respectively).
c. Aspirate the media and resuspend the cells in 1 mL of NMM + factors + Y-27632.
d. Remove LN521 and plate 15,000–20,000 cells per coverslip in 500 μL of NMM + factors + Y-27632.
15. Fix the coverslips 24–48 h after seeding.
a. Aspirate the used media and wash once with PBS.
b. Add 500 μL of 4% PFA and fix for 12 min at RT.
c. Wash 3× with PBS, leaving the last wash on the cells.
d. Use coverslips for immunostaining within a timeframe of 24 h to two weeks (Figure 3).
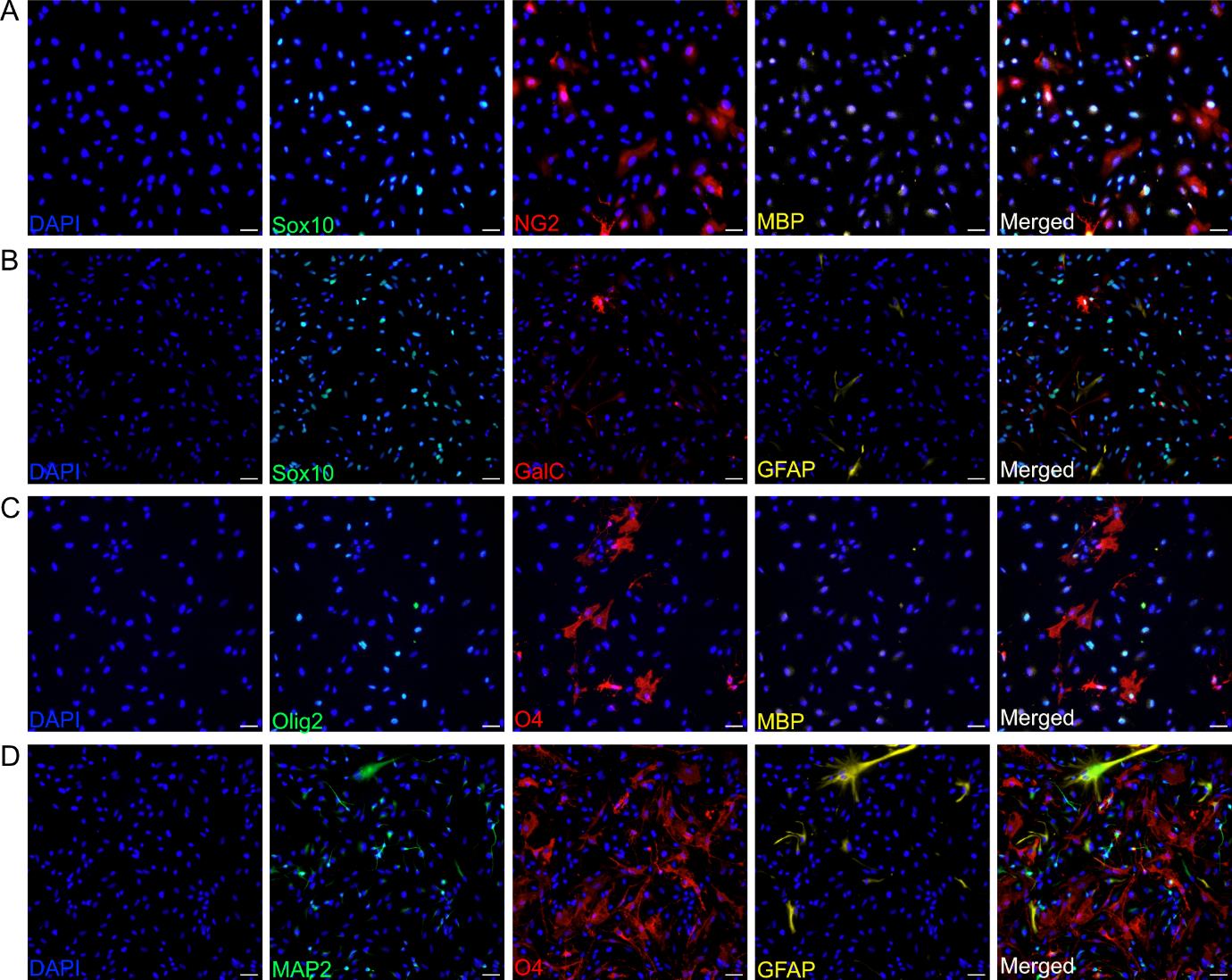
Figure 3. O4-expressing oligodendrocytes derived from neural progenitor cells (NPCs). Representative immunofluorescence images showing the expression of specific O4-expressing oligodendrocytes. (A) Sox10 (green), NG2 (red), MBP (yellow), and merged (white). (B) Sox10 (green), GalC (red), GFAP (yellow), and merged (white). (C) Olig2 (green), O4 (red), MBP (yellow), and merged (white). (D) Sox10 (green), NG2 (red), MBP (yellow), and merged (white). DAPI (blue) counterstains the nucleus. Nuclei are counterstained with DAPI (blue). Scale bar, 50 μm. Images are representative of three biological replicates. Images were acquired and processed using identical microscope settings. DAPI: 4’,6-diamidin-2-phenylindol; GalC: Galactosylceramidase; GFAP: glial fibrillary acidic protein; MAP2: microtubule-associated protein 2; MBP: myelin basic protein; NG2: neural/glial antigen 2; Olig2: oligodendrocyte transcription factor 2.
16. Assess the ability of O4+oligodendrocytes to respond to clemastine or miconazole, two well-known pro-myelinating compounds.
a. Prepare Geltrex-coated T75 flasks (Recipe 13).
b. Plate 5 million cells per flask and add 10 mL of fresh NMM + factors (1 μg/mL L2020 + 50 μg/mL AA + 50 ng/mL noggin) plus 2.5 μM or 5 μM miconazole or clemastine treatment (Recipe 6).
c. Change media every other day and add treatment every day for 10 days.
d. After 10 days of treatment, treat the cells as outlined in steps C14 and C15 (Figure 4).
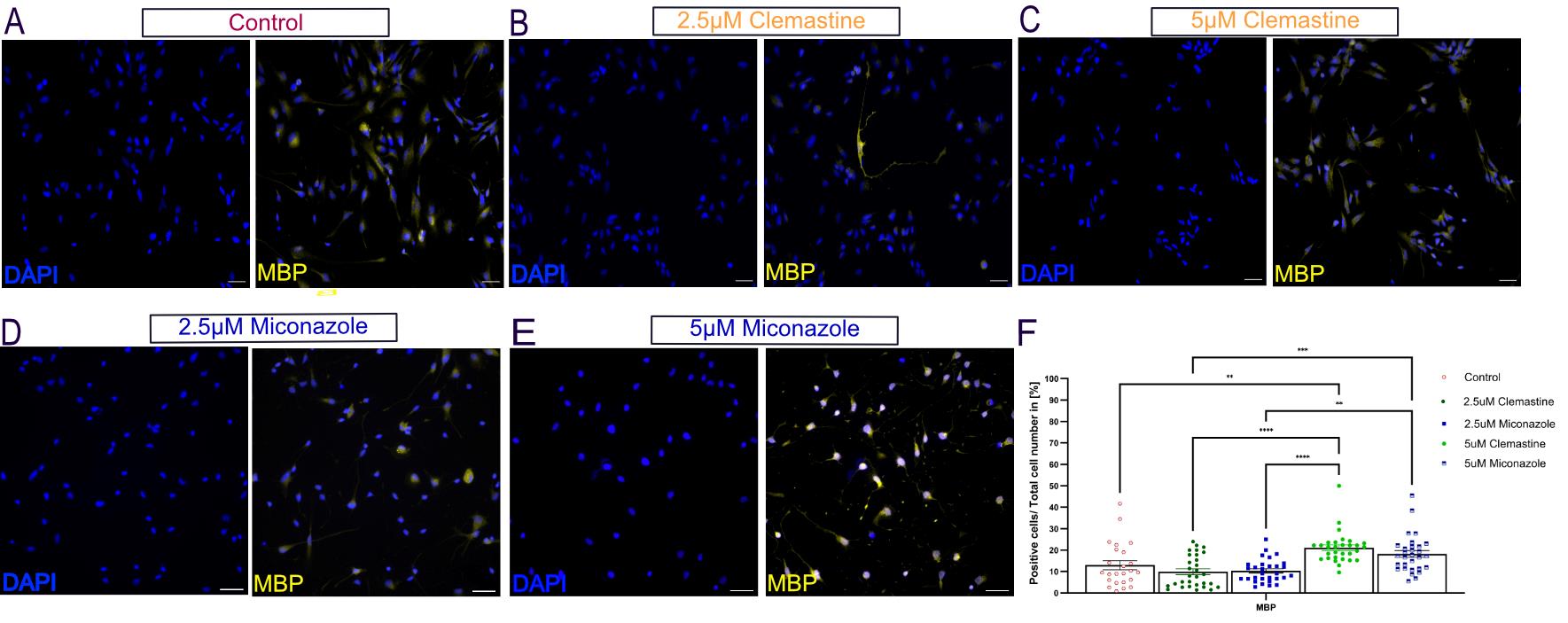
Figure 4. O4-expressing oligodendrocytes respond to pro-myelinating compounds. (A–E) O4-expressing oligodendrocytes were treated for ten days with 2.5 μM or 5 μM clemastine or miconazole, two widely validated pro-myelinating compounds [44–47]. Representative immunofluorescence images show the expression of MBP (yellow) and DAPI (blue)-counterstained nucleus. Scale bars, 50 μm. Images are representative of three biological replicates. Images were acquired and processed using identical microscope settings. (F) Corresponding quantification of MBP. The number of cells positive for MBP relative to all treated cells was determined relative to the total number of DAPI. Cell counting was performed manually by analyzing 24 randomly selected fields at 25× magnification. Data are presented as the mean ± SEM from three independent experiments; each dot = (positive cells/total cell number) * 100 from one image field. Statistical analysis was performed using two-way ANOVA followed by Tukey’s multiple comparisons test. Statistical significance was considered at ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05. DAPI: 4’,6-diamidin-2-phenylindol; MBP: myelin basic protein.
D. Neuron-OPC co-culture
1. At day 1 of co-culture, seed day-37 neurons and day-38 oligodendrocytes (OPCs) in a 5:1 ratio in poly-L ornithine (PLO)/BioLaminin (LN521)-coated coverslips in co-culture media (Recipe 4) + factors [20 ng/mL BDNF + 60 ng/mL T3 + 100 ng/ mL biotin + 1 μg/mL L2020 + 10 ng/mL NT3 + 1 μM cyclic adenosine monophosphate (cAMP) + 300 nM cholest-4-en-3-one]. The 5:1 neuron-to-OPC ratio ensures robust myelination and support in the co-culture. BDNF and NT3 support neuronal survival and growth [48]. T3 promotes oligodendrocyte maturation and myelination [49]. cAMP enhances neuron-glia communication and oligodendrocyte differentiation [50].
a. Enzymatically detach neurons from the flask with papain (135 u/vial) or Accutase as outlined in step A3.
b. Enzymatically detach OPCs from the flask with Accutase as outlined in Procedure, point 1.
c. In the same 1.5 mL Eppendorf tube, add 5,000 OPCs + 25,000 neurons.
Note: If you have more than one coverslip, each coverslip is allocated to its own Eppendorf tube, containing 5:1 neurons-to-OPCs. Do not make a master mix. It is important to get the correct ratio of cells per coverslip.
2. Seeding co-culture mixture onto coverslips.
a. Remove PLO/LN521 from coverslips.
b. Add 250 μL of co-culture media + factors + Y-27632 to the wells.
c. Gently mix cells in the Eppendorf tube and add them to the coverslips containing 250 μL of co-culture media + factors + Y-27632.
d. Place the plate into the incubator and gently move back to front and left to right to homogenously distribute the cells and avoid clumping.
Note: Instead of moving the plate as described in step D2, one can also gently pipette up and down in the well to gently mix before placing the plate into the incubator. However, when pipetting up and down, make sure the coverslip does not lift up.
3. Change media every two to three days.
Note: Cells are sensitive and might not appreciate media changes too often.
a. Prewarm co-culture media in a 37 °C water bath for a maximum of 10 min.
b. Add 20 ng/mL BDNF + 60 ng/mL T3 + 100 ng/mL biotin +1 μg/mL L2020 + 10 ng/mL NT3 + 1 μM cAMP + 300 nM cholest-4-en-3-one.
c. Gently aspirate 2/3 of the used media.
Note: Make sure to add factors for the final volume of media in the wells.
4. In our experiment, cells are fixed on day 10 of co-culturing. By day 10, signs of myelination begin to appear in the co-culture, and neurite outgrowth and initial interactions between neurons and OPCs can be observed.
a. Aspirate the used media and wash once with PBS.
b. Add 500 μL of 4% PFA and fix for 12 min at RT.
c. Wash 3× with PBS, leaving the last wash on the cells.
d. Use coverslips for immunostaining from 24 h up to two weeks (Figure 5).
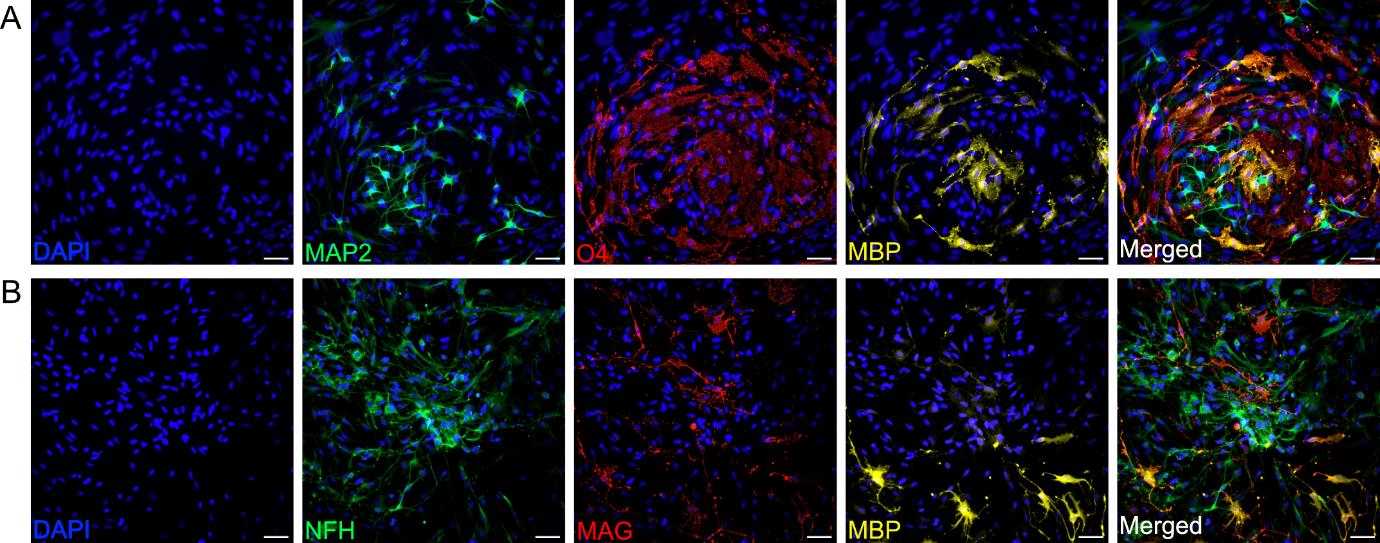
Figure 5. Characterization of neurons and oligodendrocytes grown in a co-culture for 10 days. Representative immunofluorescence images of day 10 co-cultured neurons and O4-expressing oligodendrocytes. (A) Expression of specific neuronal marker MAP2 (green), O4-expressing oligodendrocyte marker O4 (red), and mature oligodendrocyte lineage marker MBP (yellow). (B) Expression of specific neuronal marker NFH (green) and mature oligodendrocyte lineage markers MAG (red) and MBP (yellow). For all images, nuclei are counterstained with DAPI (blue). Scale bar, 50 μm. Images are representative of three biological replicates. Images were acquired and processed using identical microscope settings.
E. Immunostaining
Note: Except for changes in primary and secondary antibodies, the same protocol was used for all cell types.
1. Remove PBS.
Note: Avoid direct contact with the cells. Always add or remove liquid from the side of the well.
2. Add 300 μL of permeabilization buffer (Recipe 10) for 5 min.
3. Remove permeabilization buffer and add 300 μL of blocking buffer (Recipe 9) to the coverslips.
4. Incubate for 1 h at RT.
5. Remove blocking buffer.
6. Add primary antibodies diluted in 300 μL of blocking buffer and incubate for 3 h at RT.
7. Wash three times for 7 min with PBS.
Note: Keep protected from light from this point onward.
8. Add blocking buffer and incubate for 1 h at RT in the dark.
9. Add secondary antibodies diluted in 300 μL of blocking buffer and incubate at RT for 1.5 h in the dark.
10. Wash three times for 7 min with PBS.
11. Remove coverslips from the wells (cells face up) and dry on paper towels.
12. Allow coverslips to dry completely (white coloring) and then mount coverslips on glass slides using Fluoromount G.
Note: Remember to flip coverslips over, so cells are between the coverslip and the glass.
13. Dry at room temperature in the dark for 24 h before imaging.
Note: Once mounted and dry for 24 h, store at 4 °C protected from moisture and light.
Data analysis
Prior to experimental use, NPCs were stained at various passages (P7, P10, and P15) for the proliferative markers Pax6 and nestin. Positive cells were manually quantified using ImageJ, with NPCs typically expressing 95%–100% of Pax6 and nestin in all three passages, confirming that NPCs can be used up to passage 15 for experiments.
For neuron and oligodendrocyte differentiation, coverslips were individually stained at specific time points for lineage markers. Neuronal markers included NeuN, FoxG1, Tuj1, and MAP2, with GFAP as a negative control. Oligodendrocyte markers at various stages of differentiation included Sox10, NG2, O4, GalC, Olig2, and MBP, with MAP2 and GFAP as negative controls. Three independent experiments were analyzed, and positive cells were manually quantified using ImageJ. Typically, in a control line, approximately 25% of cells expressed early neuronal markers NeuN and FoxG1, approximately 35% expressed mature markers Tuj1 or MAP2, and less than 5% expressed GFAP. In the O4-expressing oligodendrocyte cell line on day 28, 75% of cells expressed Sox10, 30% expressed O4, 15% expressed GalC or MAP2, and less than 10% expressed NG2, MBP, or GFAP. O4-expressing oligodendrocytes were kept in culture for up to 53 days.
For the co-culture analysis, coverslips were stained to assess mature lineage markers for neurons (MAP2 and NFH) and O4-expressing oligodendrocytes, as well as more mature oligodendrocytes (O4, MAG, and MBP). Three independent experiments were conducted, combining neurons and oligodendrocytes from different experiments. Positive cells were manually quantified using ImageJ. All graphs were plotted using GraphPad Prism, and images were processed in ImageJ. Student’s T-test was used to compare two groups, while one-way or two-way ANOVA was used to compare multiple groups.
Validation of protocol
The generation of NPC was validated in Rust et al. [34] and Weber et al. [51]. The generation of oligodendrocytes overexpressing transcription factors was validated in Ehrlich et al. [25] and Chanoumidou et al. [29]. The generation of neurons was validated in Walter et al. [52].
General notes and troubleshooting
General notes
1. All cell culture work must be carried out in a sterile environment using correct BSL1 and BSL2 cell culture practices.
2. Pipette tips and coverslips are autoclaved before use.
3. Long-term storage of NPCs for up to 1–3 years requires a liquid nitrogen tank.
4. As some variation is common in NPC differentiation protocols, we recommend some quality checks before the start of a neuron-OPC co-culture.
5. Proper O4-expressing oligodendrocyte differentiation can be verified by checking for O4 expression around day 28 of the differentiation protocol, where 30%–50% of cells should express the lineage marker O4.
6. Proper neuronal differentiation can be verified by staining for MAP2/beta-III tubulin/NeuN in the cultures on day 29.
7. Antibody performance varies across suppliers. Ensure lot-to-lot consistency for primary and secondary antibodies by validating each new batch with a small-scale test before full experimental use.
Troubleshooting
Ensure even distribution of cells on coverslips to avoid clustering, which can hinder cell interactions.
Avoid frequent media changes, as cells are sensitive and may detach.
Avoid excessive pipetting during washing steps. Use a low-flow pipette tip or tilt the plate to gently remove solutions.
1. Neurons:
a. According to the manufacture protocol (CultureOneTM Supplement, Gibco), 5 million NPCs is the ideal starting cell density. Depending on the NPCs that the neurons are derived from, they might clump during differentiation stages. The clumping can be reduced by using fewer cells.
b. Neurons should not be subjected to freezing/thawing cycles, as they are prone to poor attachment.
2. Oligodendrocyte progenitor cells, O4-expressing oligodendrocytes, and oligodendrocytes:
a. To achieve optimal transduction efficiency, it is recommended to adjust the MOI, according to the specific experimental conditions.
b. OPCs might need to be split more often depending on the proliferation rate. This can also vary slightly between starting passages of NPCs.
c. OPCs can be cryopreserved at any point following the second puromycin selection. A minimum recovery period of one week is required for them to exhibit the same level of O4 lineage marker expression as day 28 O4-expressing oligodendrocytes that were not cryopreserved.
3. Co-culture:
The 5:1 ratio has been rigorously tested and found to be optimal for these cells. However, the total number of cells should be specifically adjusted for the desired experimental conditions. When testing drugs that may cause a reduction in cell numbers, it is necessary to increase the initial cell seeding density. For example, instead of using 25,000 neurons and 5,000 O4-expressing oligodendrocytes, consider starting with 30,000 neurons and 6.000 OPCs.
4. As a specific threshold for acceptable differentiation efficiency, cultures are considered ready for co-culture or downstream experiments when >30% of cells express MAP2 (neurons) or O4 (OPCs).
5. Regularly validate differentiation progress by staining a small sample of cells at intermediate stages (e.g., day 14 for OPCs, day 20 for neurons) to ensure proper lineage commitment.
Acknowledgments
We would like to thank the Larsson-Rosenquist Family Foundation for their generous support. Imaging was performed with equipment maintained by the Centre for Microscopy and Image Analysis, University of Zurich, Switzerland. The EPFL Bertarelli Platform for Gene Therapy (Lausanne, Switzerland) provided the lentiviral vector for the study.
Funding: This work was partly funded by the Jübiläumsstiftung von Swiss Life and by the Anna Mueller Grocholski-Stiftung (grant to M.C.M.). Maxi Foundation grant to MG.
Author Contributions: M.C.M. developed the concept and designed the experimental methodology. S.E.C. and S.Z. performed the experiments and analyzed the data. M.G. provided the neural progenitor cells used in the study. T.K. supplied the lentiviral vectors for oligodendrocyte differentiation. G.N. and M.C.M. supervised the work. S.E.C. and M.C.M. drafted the manuscript with contributions from all authors.
Competing interests
The authors declare no conflict of interest.
References
- Hansen, D. V., Lui, J. H., Parker, P. R. and Kriegstein, A. R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 464(7288): 554–561. https://doi.org/10.1038/nature08845
- Knowles, J. K., Batra, A., Xu, H. and Monje, M. (2022). Adaptive and maladaptive myelination in health and disease. Nat Rev Neurol. 18(12): 735–746. https://doi.org/10.1038/s41582-022-00737-3
- Kuhn, S., Gritti, L., Crooks, D. and Dombrowski, Y. (2019). Oligodendrocytes in Development, Myelin Generation and Beyond. Cells. 8(11). https://doi.org/10.3390/cells8111424
- Reillo, I., de Juan Romero, C., Garcia-Cabezas, M. A. and Borrell, V. (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 21(7): 1674–1694. https://doi.org/10.1093/cercor/bhq238
- Dean, D. C., 3rd, O'Muircheartaigh, J., Dirks, H., Waskiewicz, N., Walker, L., Doernberg, E., Piryatinsky, I. and Deoni, S. C. (2015). Characterizing longitudinal white matter development during early childhood. Brain Struct Funct. 220(4): 1921–1933. https://doi.org/10.1007/s00429-014-0763-3
- Deoni, S. C., O'Muircheartaigh, J., Elison, J. T., Walker, L., Doernberg, E., Waskiewicz, N., Dirks, H., Piryatinsky, I., Dean, D. C., 3rd and Jumbe, N. L. (2016). White matter maturation profiles through early childhood predict general cognitive ability. Brain Struct Funct. 221(2): 1189–1203. https://doi.org/10.1007/s00429-014-0947-x
- Filley, C. M. and Fields, R. D. (2016). White matter and cognition: making the connection. J Neurophysiol. 116(5): 2093–2104. https://doi.org/10.1152/jn.00221.2016
- Muetzel, R. L., Mous, S. E., van der Ende, J., Blanken, L. M., van der Lugt, A., Jaddoe, V. W., Verhulst, F. C., Tiemeier, H. and White, T. (2015). White matter integrity and cognitive performance in school-age children: A population-based neuroimaging study. Neuroimage. 119: 119–128. https://doi.org/10.1016/j.neuroimage.2015.06.014
- Petanjek, Z., Judas, M., Simic, G., Rasin, M. R., Uylings, H. B., Rakic, P. and Kostovic, I. (2011). Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci USA. 108(32): 13281–13286. https://doi.org/10.1073/pnas.1105108108
- Rabinowitch, I., Colon-Ramos, D. A. and Krieg, M. (2024). Understanding neural circuit function through synaptic engineering. Nat Rev Neurosci. 25(2): 131–139. https://doi.org/10.1038/s41583-023-00777-8
- Barres, B. A. (2008). The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 60(3): 430–440. https://doi.org/10.1016/j.neuron.2008.10.013
- Beiter, R. M., Rivet-Noor, C., Merchak, A. R., Bai, R., Johanson, D. M., Slogar, E., Sol-Church, K., Overall, C. C. and Gaultier, A. (2022). Evidence for oligodendrocyte progenitor cell heterogeneity in the adult mouse brain. Sci Rep. 12(1): 12921. https://doi.org/10.1038/s41598-022-17081-7
- Duncan, G. J., Simkins, T. J. and Emery, B. (2021). Neuron-Oligodendrocyte Interactions in the Structure and Integrity of Axons. Front Cell Dev Biol. 9: 653101. https://doi.org/10.3389/fcell.2021.653101
- Bagayogo, I. P. and Dreyfus, C. F. (2009). Regulated release of BDNF by cortical oligodendrocytes is mediated through metabotropic glutamate receptors and the PLC pathway. ASN Neuro. 1(1): https://doi.org/10.1042/AN20090006
- Jang, M., Gould, E., Xu, J., Kim, E. J. and Kim, J. H. (2019). Oligodendrocytes regulate presynaptic properties and neurotransmission through BDNF signaling in the mouse brainstem. eLife. 8: e42156. https://doi.org/10.7554/eLife.42156
- Sydnor, V. J., Larsen, B., Bassett, D. S., Alexander-Bloch, A., Fair, D. A., Liston, C., Mackey, A. P., Milham, M. P., Pines, A., Roalf, D. R., et al. (2021). Neurodevelopment of the association cortices: Patterns, mechanisms, and implications for psychopathology. Neuron. 109(18): 2820–2846. https://doi.org/10.1016/j.neuron.2021.06.016
- Thapar, A., Cooper, M. and Rutter, M. (2017). Neurodevelopmental disorders. Lancet Psychiatry. 4(4): 339–346. https://doi.org/10.1016/S2215-0366(16)30376-5
- Dong, Y. X., Zhang, H. Y., Li, H. Y., Liu, P. H., Sui, Y. and Sun, X. H. (2018). Association between Alzheimer's disease pathogenesis and early demyelination and oligodendrocyte dysfunction. Neural Regen Res. 13(5): 908–914. https://doi.org/10.4103/1673-5374.232486
- Fischer, I., Dulin, J. N. and Lane, M. A. (2020). Transplanting neural progenitor cells to restore connectivity after spinal cord injury. Nat Rev Neurosci. 21(7): 366–383. https://doi.org/10.1038/s41583-020-0314-2
- Lubetzki, C., Sol-Foulon, N. and Desmazieres, A. (2020). Nodes of Ranvier during development and repair in the CNS. Nat Rev Neurol. 16(8): 426–439. https://doi.org/10.1038/s41582-020-0375-x
- Tkachev, D., Mimmack, M. L., Ryan, M. M., Wayland, M., Freeman, T., Jones, P. B., Starkey, M., Webster, M. J., Yolken, R. H. and Bahn, S. (2003). Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 362(9386): 798–805. https://doi.org/10.1016/S0140-6736(03)14289-4
- Dincman, T. A., Beare, J. E., Ohri, S. S. and Whittemore, S. R. (2012). Isolation of cortical mouse oligodendrocyte precursor cells. J Neurosci Methods. 209(1): 219–226. https://doi.org/10.1016/j.jneumeth.2012.06.017
- O'Meara, R. W., Ryan, S. D., Colognato, H. and Kothary, R. (2011). Derivation of enriched oligodendrocyte cultures and oligodendrocyte/neuron myelinating co-cultures from post-natal murine tissues. J Vis Exp. (54). https://doi.org/10.3791/3324
- Benedetti, M. C., D'Andrea, T., Colantoni, A., Silachev, D., de Turris, V., Boussadia, Z., Babenko, V. A., Volovikov, E. A., Belikova, L., Bogomazova, A. N., et al. (2024). Cortical neurons obtained from patient-derived iPSCs with GNAO1 p.G203R variant show altered differentiation and functional properties. Heliyon. 10(5): e26656. https://doi.org/10.1016/j.heliyon.2024.e26656
- Ehrlich, M., Mozafari, S., Glatza, M., Starost, L., Velychko, S., Hallmann, A. L., Cui, Q. L., Schambach, A., Kim, K. P., Bachelin, C., et al. (2017). Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc Natl Acad Sci USA. 114(11): E2243–E2252. https://doi.org/10.1073/pnas.1614412114
- Dooves, S., Nadadhur, A. G., Gasparotto, L. and Heine, V. M. (2019). Co-culture of Human Stem Cell Derived Neurons and Oligodendrocyte Progenitor Cells. Bio Protoc. 9(17): e3350. https://doi.org/10.21769/BioProtoc.3350
- Assetta, B., Tang, C., Bian, J., O'Rourke, R., Connolly, K., Brickler, T., Chetty, S. and Huang, Y. A. (2020). Generation of Human Neurons and Oligodendrocytes from Pluripotent Stem Cells for Modeling Neuron-Oligodendrocyte Interactions. J Vis Exp. (165). https://doi.org/10.3791/61778
- von der Bey, M., De Cicco, S., Zach, S., Hengerer, B. and Ercan-Herbst, E. (2023). Three-dimensional co-culture platform of human induced pluripotent stem cell-derived oligodendrocyte lineage cells and neurons for studying myelination. STAR Protoc. 4(2): 102164. https://doi.org/10.1016/j.xpro.2023.102164
- Chanoumidou, K., Hernandez-Rodriguez, B., Windener, F., Thomas, C., Stehling, M., Mozafari, S., Albrecht, S., Ottoboni, L., Antel, J., Kim, K. P., et al. (2021). One-step Reprogramming of Human Fibroblasts into Oligodendrocyte-like Cells by SOX10, OLIG2, and NKX6.2. Stem Cell Reports. 16(4): 77–1783. https://doi.org/10.1016/j.stemcr.2021.03.001
- De Simone, U., Caloni, F., Gribaldo, L. and Coccini, T. (2017). Human Co-culture Model of Neurons and Astrocytes to Test Acute Cytotoxicity of Neurotoxic Compounds. Int J Toxicol. 36(6): 463–477. https://doi.org/10.1177/1091581817739428
- Kiray, H., Lindsay, S. L., Hosseinzadeh, S. and Barnett, S. C. (2016). The multifaceted role of astrocytes in regulating myelination. Exp Neurol. 283(Pt B): 541–549. https://doi.org/10.1016/j.expneurol.2016.03.009
- Duncan, I. D. and Radcliff, A. B. (2016). Inherited and acquired disorders of myelin: The underlying myelin pathology. Exp Neurol. 283(Pt B): 452–475. https://doi.org/10.1016/j.expneurol.2016.04.002
- Chang, E. A., Jin, S. W., Nam, M. H. and Kim, S. D. (2019). Human Induced Pluripotent Stem Cells: Clinical Significance and Applications in Neurologic Diseases. J Korean Neurosurg Soc. 62(5): 493–501. https://doi.org/10.3340/jkns.2018.0222
- Rust, R., Weber, R. Z., Generali, M., Kehl, D., Bodenmann, C., Uhr, D., Wanner, D., Zurcher, K. J., Saito, H., Hoerstrup, S. P., et al. (2022). Xeno-free induced pluripotent stem cell-derived neural progenitor cells for in vivo applications. J Transl Med. 20(1): 421. https://doi.org/10.1186/s12967-022-03610-5
- Tian, C., Liu, Q., Ma, K., Wang, Y., Chen, Q., Ambroz, R., Klinkebiel, D. L., Li, Y., Huang, Y., Ding, J., et al. (2013). Characterization of induced neural progenitors from skin fibroblasts by a novel combination of defined factors. Sci Rep. 3: 1345. https://doi.org/10.1038/srep01345
- Watanabe, K., Ueno, M., Kamiya, D., Nishiyama, A., Matsumura, M., Wataya, T., Takahashi, J. B., Nishikawa, S., Nishikawa, S., Muguruma, K., et al. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 25(6): 681–686. https://doi.org/10.1038/nbt1310
- Sautter, J., Meyer, M., Spenger, C., Seiler, R. W. and Widmer, H. R. (1998). Effects of combined BDNF and GDNF treatment on cultured dopaminergic midbrain neurons. Neuroreport. 9(6): 1093–1096. https://doi.org/10.1097/00001756-199804200-00025
- Sarnat, H. B., Nochlin, D. and Born, D. E. (1998). Neuronal nuclear antigen (NeuN): a marker of neuronal maturation in early human fetal nervous system. Brain Dev. 20(2): 88–94. https://doi.org/10.1016/s0387-7604(97)00111-3
- Wolf, H. K., Buslei, R., Schmidt-Kastner, R., Schmidt-Kastner, P. K., Pietsch, T., Wiestler, O. D. and Blumcke, I. (1996). NeuN: a useful neuronal marker for diagnostic histopathology. J Histochem Cytochem. 44(10): 1167–1171. https://doi.org/10.1177/44.10.8813082
- Cargnin, F., Kwon, J. S., Katzman, S., Chen, B., Lee, J. W. and Lee, S. K. (2018). FOXG1 Orchestrates Neocortical Organization and Cortico-Cortical Connections. Neuron. 100(5): 1083–1096 e1085. https://doi.org/10.1016/j.neuron.2018.10.016
- Soltani, M. H., Pichardo, R., Song, Z., Sangha, N., Camacho, F., Satyamoorthy, K., Sangueza, O. P. and Setaluri, V. (2005). Microtubule-associated protein 2, a marker of neuronal differentiation, induces mitotic defects, inhibits growth of melanoma cells, and predicts metastatic potential of cutaneous melanoma. Am J Pathol. 166(6): 1841–1850. https://doi.org/10.1016/S0002-9440(10)62493-5
- Wang, B., Khan, S., Wang, P., Wang, X., Liu, Y., Chen, J. and Tu, X. (2022). A Highly Selective GSK-3beta Inhibitor CHIR99021 Promotes Osteogenesis by Activating Canonical and Autophagy-Mediated Wnt Signaling. Front Endocrinol (Lausanne). 13: 926622. https://doi.org/10.3389/fendo.2022.926622
- Govier-Cole, A. E., Wood, R. J., Fletcher, J. L., Gonsalvez, D. G., Merlo, D., Cate, H. S., Murray, S. S. and Xiao, J. (2019). Inhibiting Bone Morphogenetic Protein 4 Type I Receptor Signaling Promotes Remyelination by Potentiating Oligodendrocyte Differentiation. eNeuro. 6(2). https://doi.org/10.1523/ENEURO.0399-18.2019
- Cree, B. A. C., Niu, J., Hoi, K. K., Zhao, C., Caganap, S. D., Henry, R. G., Dao, D. Q., Zollinger, D. R., Mei, F., Shen, Y. A., et al. (2018). Clemastine rescues myelination defects and promotes functional recovery in hypoxic brain injury. Brain. 141(1): 85–98. https://doi.org/10.1093/brain/awx312
- Green, A. J., Gelfand, J. M., Cree, B. A., Bevan, C., Boscardin, W. J., Mei, F., Inman, J., Arnow, S., Devereux, M., Abounasr, A., et al. (2017). Clemastine fumarate as a remyelinating therapy for multiple sclerosis (ReBUILD): a randomised, controlled, double-blind, crossover trial. Lancet. 390(10111): 2481–2489. https://doi.org/10.1016/S0140-6736(17)32346-2
- Su, X., Tang, W., Luan, Z., Yang, Y., Wang, Z., Zhang, Y., Wang, Q., Suo, L., Huang, Z., Wang, X., et al. (2018). Protective effect of miconazole on rat myelin sheaths following premature infant cerebral white matter injury. Exp Ther Med. 15(3): 2443–2449. https://doi.org/10.3892/etm.2018.5717
- Wang, P., Yang, M., Jiang, L. and Wu, Y. J. (2019). A fungicide miconazole ameliorates tri-o-cresyl phosphate-induced demyelination through inhibition of ErbB/Akt pathway. Neuropharmacology. 148: 31–39. https://doi.org/10.1016/j.neuropharm.2018.12.015
- Avila, M. A., Varela-Nieto, I., Romero, G., Mato, J. M., Giraldez, F., Van De Water, T. R. and Represa, J. (1993). Brain-derived neurotrophic factor and neurotrophin-3 support the survival and neuritogenesis response of developing cochleovestibular ganglion neurons. Dev Biol. 159(1): 266–275. https://doi.org/10.1006/dbio.1993.1239
- Dugas, J. C., Ibrahim, A. and Barres, B. A. (2012). The T3-induced gene KLF9 regulates oligodendrocyte differentiation and myelin regeneration. Mol Cell Neurosci. 50(1): 45–57. https://doi.org/10.1016/j.mcn.2012.03.007
- Shiga, H., Asou, H. and Ito, E. (2005). Advancement of differentiation of oligodendrocyte progenitor cells by a cascade including protein kinase A and cyclic AMP-response element binding protein. Neurosci Res. 53(4): 436–441. https://doi.org/10.1016/j.neures.2005.09.004
- Weber, R. Z., Achón Buil, B., Rentsch, N. H., Perron, P., Bosworth, A., Zhang, M., Kisler, K., Bodenmann, C., Zürcher, K. J., Uhr, D., et al. (2024). Human iPSC-derived cell grafts promote functional recovery by molecular interaction with stroke-injured brain. bioRxiv, 2024.2004.2003.588020. https://doi.org/10.1101/2024.04.03.588020
- Walter, N. M., Yde Ohki, C. M., Smigielski, L., Walitza, S. and Grünblatt, E. (2025). Investigating the impact of omega-3 fatty acids on oxidative stress and pro-inflammatory cytokine release in iPSC-derived forebrain cortical neurons from ADHD patients. J Psychiatr Res. 182: 257–269. https://doi.org/10.1016/j.jpsychires.2025.01.020
Article Information
Publication history
Received: Oct 29, 2024
Accepted: Jan 26, 2025
Available online: Feb 20, 2025
Published: May 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Chie, S. E., Szentpetery, Z., Generali, M., Kuhlmann, T., Natalucci, G. and Miletta, M. C. (2025). Human iPSC-Derived Neuron and Oligodendrocyte Co-culture as a Small-Molecule Screening Assay for Myelination. Bio-protocol 15(9): e5227. DOI: 10.21769/BioProtoc.5227.
Category
Stem Cell > Pluripotent stem cell > Cell differentiation
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








