- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Native Acetyl CoA Carboxylase From Mammalian Cells
Published: Vol 15, Iss 4, Feb 20, 2025 DOI: 10.21769/BioProtoc.5221 Views: 2190
Reviewed by: Komuraiah MyakalaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ZnCl2 Precipitation-Assisted Sample Preparation for Proteomic Analysis
Qiqing He [...] Fuchu He
Jul 20, 2025 2721 Views
Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1806 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1208 Views
Abstract
Fatty acid (FA) biosynthesis is a crucial cellular process that converts nutrients into metabolic intermediates necessary for membrane biosynthesis, energy storage, and the production of signaling molecules. Acetyl-CoA carboxylase (ACACA) plays a pivotal catalytic role in both fatty acid synthesis and oxidation. This cytosolic enzyme catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, which represents the first and rate-limiting step in de novo fatty acid biosynthesis. In this study, we developed a rapid and effective purification scheme for separating human ACACA without any exogenous affinity tags, providing researchers with a novel method to obtain human ACACA in its native form.
Key features
• Detailed protocol for the purification of native ACACA.
• ACACA is biotinylated in mammalian cells.
Keywords: ACACABackground
Acetyl-CoA carboxylase (ACACA) plays a pivotal catalytic role in both fatty acid synthesis and oxidation [1]. This cytosolic enzyme catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, which is the first and rate-limiting step of de novo fatty acid biosynthesis. This process involves two steps: first, the ATP-dependent carboxylation of biotin, carried out by the biotin carboxylase (BC) domain; second, the transfer of the carboxyl group from carboxylated biotin to acetyl-CoA [2–4]. Mammals possess two acetyl CoA carboxylase (ACC) isoforms: ACACA, located in the cytosol of the liver and adipose tissues, and ACACB, associated with the mitochondrial outer membrane in heart and muscle tissues [5,6].
Recent developments in tumor fatty acid metabolism have provided new insights into potential therapeutic strategies for cancer. Studies have shown that tumor cells exhibit abnormal lipid metabolism reprogramming [7]. ACACA is often overexpressed in various tumor types, and elevated levels of its expression correlate with poor prognosis in cancer patients [8]. Further investigations have revealed that metabolic reprogramming can enhance the malignant progression of tumor cells by altering their ability to sense essential nutrients [9]. These findings underscore the critical role of metabolic reprogramming in tumorigenesis and development, highlighting it as a hallmark of malignant cells [10]. Currently, preclinical and clinical trials targeting lipid metabolism are ongoing [11–13]. In gastric cancer, the phosphorylated ACACA is closely associated with tumor cell behavior [14].
ACACA functions as a homodimer with an approximate molecular weight of 510 kDa, resulting from the dimerization of its carboxyl-terminal (CT) domain [4]. Eukaryotic ACACA comprises several structural domains: biotin carboxylase (BC), biotin carboxyl carrier protein (BCCP), and carboxytransferase (CT), along with the interacting structural domain (BT) and the non-catalytic central structural domain (CD), which connects the BC and CT domains. The CD contains four structural elements: the N-terminal CDN, the linking CDL, and the tandem C-terminal cell cycle protein-dependent kinases CDC1 and CDC2 [4].
ACACA represents a promising target for drug development aimed at treating tumor growth [4,15] and obesity-related diseases [16–18]. It also serves as a target site for various commercial herbicides [19]. In this study, we developed an efficient method to purify the recombinantly expressed human ACACA without affinity tags. Using this method to purify ACACA allows the isolation of endogenous proteins in a state closest to their natural conformation. This is particularly advantageous for structural and enzyme activity analysis of human ACACA proteins in their native states since a tag on a protein’s C or N terminus can sometimes bring side effects. Notably, this protocol eliminates the need for enzymatic cleavage of purification tags, thereby avoiding concerns about potential interference from exogenous labels on the folding and stability of the target protein. This protocol enables the purification of ACACA without the use of exogenous labels, preserving the protein’s natural structure. By avoiding the influence of labels on protein stability and folding, the method is highly advantageous for conducting functional experiments and structural analyses. Its simplicity and efficiency make it a valuable tool for researchers studying the behavior of ACACA in a native-like environment. This method offers several advantages. On one hand, it enables the purification of endogenous ACACA without the need for purification labels, preserving the protein in a state closest to its natural environment while eliminating the complex enzymatic tag-cleavage step that is typically required in labeled purification methods. On the other hand, by avoiding the use of affinity tags, researchers can directly utilize native tissues or cells to efficiently purify endogenous ACACA. This approach significantly saves time and energy, making it a streamlined and resource-efficient solution. However, we acknowledge that a limitation of purifying ACACA without exogenous labels is the difficulty in estimating the yield of ACACA from native tissues, as its expression levels can vary significantly across different tissues. This variability introduces uncertainty regarding the amount of tissue required to purify sufficient quantities of endogenous ACACA. In general, it is feasible to harness our method to purify endogenous ACACA.
Materials and reagents
Biological materials
1. DH10Bac competent cells (Shanghai WEIDI Biology, catalog number: DL1071S)
2. pBMCL1 plasmid (Addgene, catalog number: 178203) [20]
3. 293F cells (bio-83951)
4. Sf9 cells (bio-68378)
5. DH5α (Tian Gen Biology, catalog number: CB101)
6. 293 expression medium FreeStyleTM (Gibco, catalog number: 12338026)
7. Sf-900TM II SFM (Gibco, catalog number: 10902088)
8. Forward primer (Qing Ke Biology):
tgcctttctctccacaggtgtaaggaattcATGGATGAACCATCTCCCTTGGCCCAACC
Reverse primer:
ggctagcggccgcccgggatccTCACGTGGAAGGGGAATCCATTGT
Reagents
1. Tris (hydroxymethyl) aminomethane (Tris base) (VWR Life Science, catalog number: 0497-5KG)
2. StrepTactin beads 4FF (Smart-Lifesciences, catalog number: SA053100)
3. Yeast extract (Thermo Fisher, catalog number: LP0021B)
4. Tryptone (Thermo Fisher, catalog number: LP0042B)
5. NaCl (VWR Life Science, catalog number: 0241-10KG)
6. SDS (Sodium dodecyl sulfate) (VWR Life Science, catalog number: SA053100)
7. APS (Ammonium persulfate) (Biorigin, catalog number: BN20179)
8. TEMED (N,N,N',N'-Tetramethylethylenediamine) (Yuan Ye, catalog number: R21208)
9. 30% Acrylamide (NOVON, catalog number: SS0826)
10. Agar powder (NOVON, catalog number: zz14022)
11. Ampicillin (Lablead, catalog number: ST008)
12. Fetal bovine serum (FBS) (Gibco, catalog number: 26010074)
13. D-biotin (NOVON, catalog number: ST2051)
14. Sodium butyrate (Macklin, catalog number: S817488)
15. Protein ladder (Thermo Fisher, catalog number: 26619)
16. X-Gal (MedChemExpress, catalog number: HY-15934)
17. Coomassie Brilliant Blue (BioDee, catalog number: BN20720)
18. IPL-41 medium (Thermo Fisher, catalog number: 11405081)
19. Cellfectin 2 reagent (Thermo Fisher, catalog number: 10362-100)
20. HCl (Nan Jing Reagent, catalog number: C0680110228)
21. Protease inhibitors (Thermo Scientific, catalog number: A32965)
22. Na2ATP (NOVON, catalog number: MC0709)
23. MgCl2 (Macklin, catalog number: 7791-18-6)
24. Protein loading buffer (Beyotime, catalog number: P0015)
25. Sf9 medium (Gibco, catalog number: 10902088)
26. 293F FreeStyle medium (Gibco, catalog number: C12338018)
27. Kanamycin (Lablead, catalog number: K9316)
28. Gentamicin (MedChemExpress, catalog number: HY-A0276A)
29. Tetracycline (Lablead, catalog number: T9302)
30. Isopropyl-β-D-thiogalactopyranoside (IPTG) (Beyotime, catalog number: ST098)
31. Agarose (Yeasen, catalog number: CB005)
32. Glycerol (VWR, catalog number: 0167)
33. Agarose Gel Recovery kit (TIANGEN, catalog number: DP209)
34. P1, P2, P3 (TIANGEN, catalog number: DP105)
35. BamH1-HF (New England Biolabs, catalog number: R3136)
36. EcoR1-HF (New England Biolabs, catalog number: R3101)
37. Isopropanol (Macklin, catalog number: 67-63-0)
38. 75% ethanol (Macklin, catalog number: 64-17-5)
39. Phospho-ACC(Ser79) (Cell Signaling, catalog number: D7D11)
40. Horseradish peroxidase-labeled goat anti-rabbit IgG (H+L) (Beyotime, catalog number: A0208)
41. Methanol (Macklin, catalog number: 67-56-1)
42. Tween-20 (Merck, catalog number: P1379)
43. Skimmed milk powder (YiLi)
44. PVDF (Beyotime, catalog number: FFP80)
45. Chemiluminescent substrate (Thermo Scientific, catalog number: YK378690)
46. Acetyl CoA Carboxylase Activity Detection kit (Solarbio, catalog number: BC6020)
47. Seamless Cloning kit (Beyotime, catalog number: D7010M)
48. β-mercaptoethanol (β-ME) (Merck, catalog number: M3148)
49. n-Dodecyl-beta-D-maltoside (DDM) (Thermo Fisher, catalog number: 329370250)
50. CHS (Anatrace, catalog number: CH210)
51. Water (Hangzhou Wahaha Group)
52. Glycine (VWR Life Science, catalog number: 0167-5KG)
53. Na2EDTA·2H2O (Thermo Fisher, catalog number: 15576028)
Solutions
1. Lysis buffer (see Recipes)
2. Wash buffer (see Recipes)
3. Separation gel 8% (see Recipes)
4. Separation gel 10% (see Recipes)
5. Concentrated gel 5% (see Recipes)
6. LB medium (see Recipes)
7. Ampicillin LB agar plates (see Recipes)
8. Blue and white spot screening plates (see Recipes)
9. TBST (see Recipes)
10. 10× SDS-PAGE running buffer (see Recipes)
11. 50× TAE (see Recipes)
Recipes
1. Lysis buffer
50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5% glycerol, 5 mM β-M), 1% DDM/CHS.
2. Wash buffer
50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 5 mM β-M), 5 mM Na2ATP.
3. Separation gel 8%
7,050 μL of double-distilled water, 4,000 μL of 30% acrylamide, 3,800 μL of 1.5 M Tris-HCl (pH 8.8), 150 μL of 10% SDS, 150 μL of 10% APS, 9 μL of TEMED.
4. Separation gel 10%
6,000 μL of double-distilled water, 4,950 μL of 30% acrylamide, 3,750 μL of 1.5 M Tris-HCl (pH 8.8), 150 μL of 10% SDS, 150 μL of 10% APS, 15 μL of TEMED.
5. Concentrated gel 5%
3,400 μL of double-steamed water, 830 μL of 30% acrylamide, 630 μL of 1.0 M Tris-HCl (pH 6.8), 50 μL of 10% SDS, 50 μL of 10% APS, 5 μL of TEMED.
6. LB medium
Dissolve 10 g of NaCl, 10 g of tryptone, and 5 g of yeast extract in 600 mL of water. Add water to 1 L followed by sterilization under high pressure at 121 °C for 20 min.
7. Ampicillin LB agar plates
Dissolve 10 g of NaCl, 10 g of tryptone, 10 g of agar powder, and 5 g of yeast extract in 600 mL of water. Add water to 1 L followed by autoclaving at 121 °C for 20 min. After sterilization, allow the temperature to drop to 55 °C, then add ampicillin antibiotics to a final concentration of 100 mg/mL. Pour the agar onto plates in a sterile environment and allow it to solidify at room temperature. Store LB agar plates at 4 °C for future use.
8. Blue and white spot screening plates
Dissolve 10 g of NaCl, 10 g of peptone, 10 g of agar powder, and 5 g of yeast extract in 600 mL of water, dilute to 1 L, and sterilize under high pressure at 121 °C for 20 min. After sterilization, let the temperature drop to 55 °C. Add 7 μg/mL gentamicin, 10 μg/mL tetracycline, 100 μg/mL X-gal, 40 μg/mL IPTG, and 50 μg/mL kanamycin antibiotics. Place the plates in a hood until they solidify. Store them at 4 °C in the dark for future use.
9. TBST
20 mM Tris, 150 mM NaCl, and 0.1% Tween-20.
10. 10× SDS-PAGE running buffer
Add 60.4 g of Tris, 144.4 g of glycine, 20 g of SDS, and distilled water to a volume of 2 L.
11. 50× TAE
Add 484 g of Tris, 74.4 g of Na2EDTA·2H2O of SDS, and distilled water to a volume of 2 L.
Laboratory supplies
1. 6-well plate (NEST, catalog number: 703001)
2. SF9 cell culture flask (NEST, catalog number: 781011)
3. 293 cell culture flask (NEST, catalog number: 785111)
4. Plates (Corning, catalog number: CLS430167)
Equipment
1. Ultrasonic disruptor (Scientz, model: JY92-IIDN)
2. Centrifuge (RWD, model: M1324)
3. Gravity column empty column (Beyotime, catalog number: FCL12)
4. Centrifuge (Beckman, model: Avanti JXN-26)
5. Hirascan ACD spectrometer (Applied Photophysics, model: Chirascan V100)
Software and datasets
1. SnapGene Viewer v.6.0.2
Procedure
A. Transformation of DH10Bac competent cells (to obtain the bacmids to produce the baculovirus)
1. Amplify the ACACA gene by PCR (pre-denaturation: 95 °C for 3 min; denaturation: 95 °C for 30 s; annealing: 62 °C for 30 s; extension: 72 °C for 40 s; cycle number 30× between the denaturation and extension steps; maintaining temperature at 4 °C) using the primers mentioned in the biological materials list, performed in 1× TAE buffer, recover the corresponding bands using the Agarose Gel Recovery kit at a concentration of 56 ng/μL.
2. Insert the target gene at the 2,292nd nucleic acid position of the pBMCL1 vector using the BamH1 and EcoR1 enzyme cleavage sites. The length of the inserted target fragment is 7,042 bp. Recycle the linearized plasmids using an Agarose Gel Recovery kit with a final concentration of 13 ng/μL.
3. Then, clone the coding sequence for full-length ACACA into the plasmid pBMCL1 followed by the verification of DNA sequencing.
4. Recombine fragments and vectors in a 1:1 molar ratio. Add 4 μL of the linearized vector and 1 μL of the target fragment to 5 μL of 2× cloning mix and place at 50 °C for 15 min. After ligation, incubate the mixture at 4 °C. Then, add the mixture to 100 μL of DH5α competent cells, incubate on ice for 30 min, and heat-shock at 42 °C for 60 s. Further incubate the mixture on ice for 2 min. Add 200 μL of antibiotic-free LB medium and incubate at 37 °C at 220 rpm for 45 min. Then, coat the cells on an ampicillin-resistant plate and incubate at 37 °C overnight.
5. Mix 1 μL (10 ng) of pBMCL1-ACACA plasmid (Figure 1A) with 100 μL of DH10Bac competent cells in a microcentrifuge tube. Incubate on ice for 30 min. Heat-shock the mixture at 42 °C for 45 s, then incubate on ice for 2.5 min. Add 900 μL of LB medium (without antibiotics) and incubate at 37 °C in a shaking incubator for 5 h.
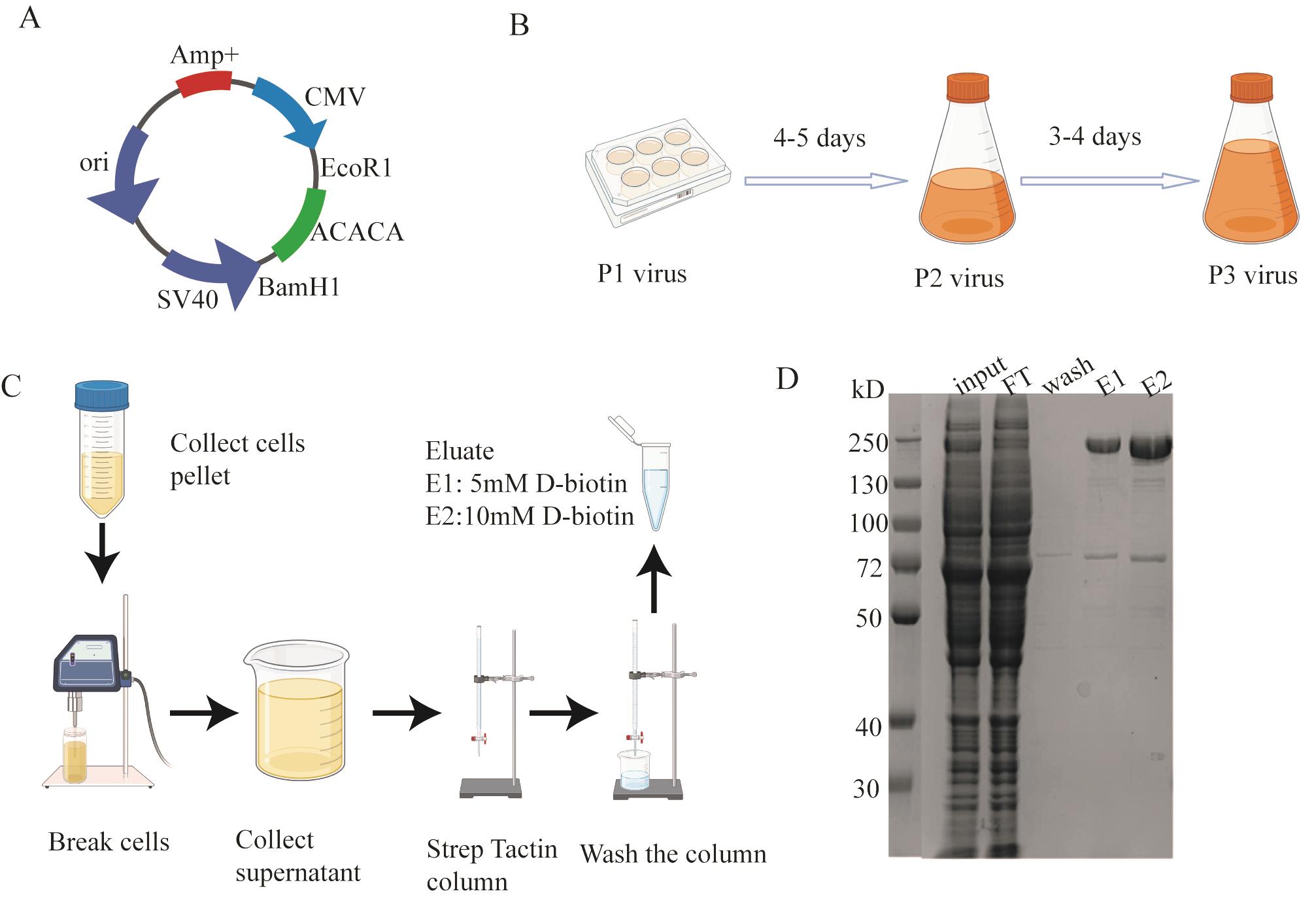
Figure 1. Cloning and protein expression and purification. A. ACACA recombinant plasmid map. B. Illustration for virus passaging. C. Diagram for purifying ACACA protein. D. SDS-PAGE for ACACA purification.
6. Plate 100 μL of the transformation mixture onto a blue and white spot screening plate containing kanamycin (50 μg/mL), gentamicin (7 μg/mL), tetracycline (10 μg/mL), X-Gal (100 μg/mL), and IPTG (40 μg/mL).
7. Incubate the plate at 37 °C for 48 h.
8. Select white colonies and streak for isolation of single transformed colonies. Incubate at 37 °C for 12 h.
9. Inoculate a single transformed colony into 4 mL of LB medium containing 50 μg/mL kanamycin, 7 μg/mL gentamicin, and 10 μg/mL tetracycline. Incubate overnight at 37 °C with shaking at 220 rpm.
10. Centrifuge the overnight culture at 1,600× g for 10 min at room temperature. Discard the supernatant.
11. Purify the bacmid using P1, P2, and P3 solution from a Plasmid Mini Prep kit and centrifuge at 17,805× g for 2 min.
12. Mix the supernatant with 800 μL of isopropanol for 1.5 h at -20 °C and centrifuge at 17,805× g for 15 min at 4 °C.
13. Wash the pellet with 750 μL of 75% ethanol at 17,805× g for 5 min at 4 °C. Repeat twice. Dry and allow the ethanol to evaporate completely in a hood.
14. Dissolve the bacmids in 30 μL of ddH2O for immediate use.
B. Transfection of ACACA-pBMCL1 bacmid into Sf9 Cells (to obtain baculoviruses to infect HEK293 cells)
1. Seed Sf9 cells (0.8 × 106) in a 6-well plate and incubate at room temperature for 1 h to allow the cells to adhere.
2. Add 5 μL of bacmids and 6 μL of Cellfectin transfection reagent in 100 μL of serum-free IPL-41 medium. Incubate the mixture at room temperature for 20 min.
3. Add the transfection mixture gently to the adherent cells using a 200 μL pipette followed by gentle rotation before incubation at 27 °C for 5 h. Replace with fresh IPL-41 medium and incubate for 5 days to obtain the P1 virus.
4. Amplify virus in Sf9 cells (Figure 1B).
a. Add 50 μL of P1 baculovirus to 50 mL of Sf9 cells at the density of 1 million per milliliter for 3–4 days. Collect P2 virus supernatant by pelleting the cells down using a centrifuge at 2,500× g for 10 min.
b. To produce P3 virus, add 200 μL of P2 baculovirus to 200 mL of Sf9 cells at the density of 1 million per milliliter for 3–4 days. Collect P3 virus supernatant by pelleting the cells down using a centrifuge at 2,500× g for 10 min. Store P3 virus in the dark at 4 °C for future use.
C. Expression of ACACA in 293F cells (to express the target protein)
1. Infect 800 mL of 293F cells in 293F FreeStyle medium with 1% FBS at the density of 2.0 × 106 cells/mL at a 1:10 ratio using the P3 virus. After 8 h, add 10 mM sodium butyrate to the cells to boost the expression of ACACA. Harvest the cells 48 h post-infection by centrifugation using a Beckman Avanti JXN-26 centrifuge with a JLA-8.1000 rotor at 8,983× g for 10 min at 4 °C. Then, freeze cells in liquid nitrogen for future use.
D. Purification of ACACA protein using StrepTactin beads
1. Resuspend the 293F cells in 30 mL of lysis buffer. Perform all steps at 4 °C.
2. Lyse the cells using sonication: break for 1 s, stop for 3 s, for a total of 5 min, centrifuge at 208,888× g for 60 min 4 °C, and load the supernatant onto a Strep Trap column pre-equilibrated with lysis buffer.
3. After loading the sample to 3 mL StrepTactin beads, wash the column with 20 times the bed volume of wash buffer.
4. Elute endogenous ACACA using lysis buffer supplemented with 15 mL of 5 mM and 15 mL of 10 mM D-biotin.
5. Concentrate the eluted ACACA at 2,500 rpm every 5 min at 4 °C to 250 μL using an Amicon ultra-centrifugal filter with 100 kDa MW cutoff.
6. Use 5% concentrated gel and 10% separation gel to check the eluate. Add 2.5 µL of protein loading buffer to 10 µL of sample. Perform electrophoresis in the SDS-PAGE buffer at a constant voltage of 220 V for 36 min. Afterward, stain the gel with Coomassie Brilliant Blue for 15 min. To decolorize, add water and microwave the gel on high heat for 15 min.
7. Load the concentrated sample into a 24 mL Superdex200 column to further purify the sample. Concentrate the sample to 250 μL. After running the size exclusion column to further purify ACACA, collect a total of four 500 μL sample fractions. In order to estimate the fraction of ACACA, besides performing the SDS-PAGE experiments, we also calculated the areas of peak1, peak2, and peak3. The area of peak1 accounts for about 91.2% of the total area, which means the amount of ACACA accounted for approximately 91.2% of the total protein. The concentration of ACACA was 0.8 mg/mL, and the total volume collected was 2 mL, yielding 2 mg of protein per liter. We used mass spectrometry to further identify that the protein is ACACA.
8. Circular dichroism determination: First, equilibrate the machine (Hirascan ACD spectrometer) using eluting buffer, using 200 μL of sample with the protein concentration of 0.2 mg/mL (Figure 2D, E).
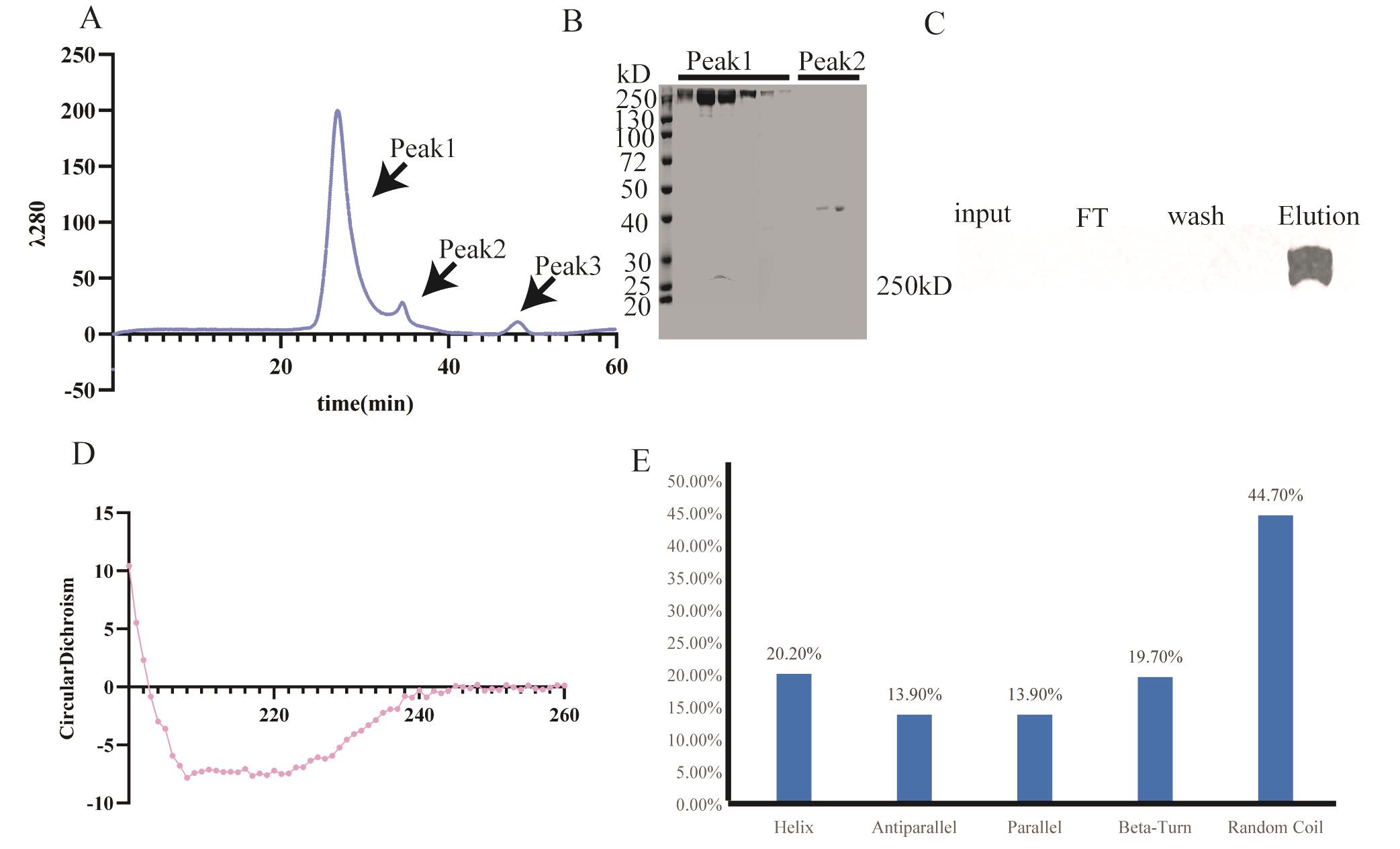
Figure 2. Validation for ACACA. A. Purification of ACACA by size exclusion chromatography. B. SDS-PAGE for size exclusion chromatography. C. Western blot result for purification of ACACA from mouse tumor tissue. D. Result for ACACA circular dichroism experiment. E. Protein secondary structure composition of ACACA.
9. Enzyme activity measurement: Preheat the spectrophotometer for 30 min and set the wavelength to 340 nm. Calibrate the instrument to zero using distilled water. Add the following reagents sequentially to a 1 mL cuvette: 250 μL of reagent 1 (see Reagent no. 46), 50 μL of sample, 500 μL of working solution (see Reagent no. 46), and 200 μL of reagent 2 (see Reagent no. 46) for the measurement. For the blank, add 250 μL of reagent 1, 50 μL of distilled water, 500 μL of working solution, and 200 μL of reagent 2. First, measure the absorbance (A1) at 340 nm for 10 s. Immediately place the cuvette at 37 °C to incubate for 10 min. After incubation, measure absorbance (A2) at 340 nm after 10 s of mixing. Calculate enzyme activity as 73.95 U/mL. [Enzyme activity = (A1M - A2M - (A1B - A2B)) × 321.5; M for materials, B for blank].
E. Purification of endogenous ACACA in mouse tumors using StrepTactin beads
1. First, lyse the tissues using sonication followed by centrifugation at 208,888× g for 60 min to remove the large debris. Then, load the supernatant onto a Strep Trap column pre-equilibrated with lysis buffer.
2. After loading the sample to 1 mL strep beads, wash the column with 20 times the bed volume of wash buffer.
3. Elute endogenous ACACA with 5 mL of lysis buffer supplemented with 10 mM D-biotin.
4. Concentrate the eluted ACACA at 1,460× g at 4 °C using an Amicon ultra-centrifugal filter with 100 kDa MW cutoff. The final concentrated volume is 250 μL.
5. Perform western blot analysis with a 5% concentrated gel and 8% separation gel. Run the gel at a constant voltage of 80 V for 120 min. Transfer the proteins at 300 mA for 2 h to a PVDF membrane (activated with 100% methanol for 2 min). Block the membrane with 7% milk powder for 1 h. Wash the membrane three times with 5 mL of TBST for 10 min each. Incubate the membrane with the primary antibody (see Reagent no. 39) overnight at 4 °C. Wash the membrane three times with 5 mL of TBST for 10 min each. Incubate with the secondary antibody (see Reagent no. 40) at room temperature for 1 h. Wash the membrane three times with 5 mL of TBST for 10 min each. Add 1 mL of chemiluminescent substrate to visualize the results.
Data analysis
Purification result
The purification of ACACA was performed following the protocol illustrated (Figure 1C). We used 293F FreeStyle medium to cultivate 293F cells. After cell lysis, the resulting supernatant was subjected to StrepTactin bead chromatography. The nonspecific proteins bound to the beads were removed using wash buffer. The target protein was subsequently eluted using elution buffer containing 10 mM D-biotin. SDS-PAGE analysis (Figure 1D) revealed that the purified eluent predominantly contained two proteins: the target protein ACACA and the heat shock protein HS71A. HS71A is a heat-shock protein that acts as a molecular chaperone to assist in the correct folding of proteins. We successfully removed through size-exclusion chromatography (Figure 2B), yielding a highly purified ACACA sample suitable for downstream applications.
To evaluate the purity, after running the size exclusion column, a total of four 500 μL sample fractions were collected. To analyze the purity, we calculated the areas of peak1, peak2, and peak3. The area of peak 1 accounts for about 91.2% of the total area, which means the amount of ACACA accounted for approximately 91.2% of the total protein. We extracted ACACA from the mouse tumor tissues using the following procedure: The tissue was homogenized and subjected to ultrasonic treatment, employing a cycle of 1-s bursts followed by 3-s pauses for a total duration of 5 min. The sonicated sample was then centrifuged at 308,444× g for 60 min. The resulting supernatant was collected and incubated with beads pre-equilibrated with lysis buffer. Following incubation, the beads were washed to remove contaminated proteins, and the target protein was then eluted. To verify the success of the protein purification, we further conducted western blot analysis. Specifically, we analyzed the following fractions: (1) the initial supernatant after lysis, (2) the flowthrough (FT) after incubation with the beads, (3) the wash buffer containing nonspecifically bound proteins removed from the beads, and (4) the elution buffer containing the target protein. The results shown below confirmed the presence of ACACA in the elution buffer, validating this extraction protocol for mouse tumor tissues. However, ACACA in the input failed to be detected, probably due to the low amount of ACACA in the input (Figure 2C). We next performed circular dichroism analysis (Figure 2D). The results indicate that the helix accounted for 20.20%, the β sheet parallel and antiparallel accounted for 13.9%, the β turn accounted for 19.7%, and the random coiled coil accounted for 44.7% (Figure 2E).
Validation of protocol
To validate this protocol, we performed liquid chromatography–mass spectrometry and circular dichroism. Mass spectrometry demonstrated ACACA’s identity. Circular dichroism proved that the purified ACACA retains its secondary structures (Figure 2D). Also, we performed the enzyme activity experiment; the calculated enzyme activity was 73.95U/mL. Importantly, we also successfully isolated the native ACACA from the tumor tissues following the established protocol (Figure 2C).
General notes and troubleshooting
Troubleshooting
1. The cell sonication time must be long enough; otherwise, it will lead to incomplete cell lysis.
2. After the protein sample is loaded onto the beads, the flow rate must be slow; otherwise, it will result in less protein collection.
3. During the concentration process, attention should be paid to protein aggregation, and it is best to concentrate at the speed of 2,500 rpm and use a pipette to gently mix the solution every 5 min.
Acknowledgments
We thank Ding X. and Niu L. from the Mass Spectrometry Technology Center of the Institute of Biophysics, Chinese Academy of Sciences, and other lab members in the Institute of Physics, Chinese Academy of Sciences (IOP, CAS) for their support in protein purification. This work was supported by the National Natural Science Foundation of China (32471015), Chinese Academy of Sciences (E2VK311) to H. Zhu.
Competing interests
The authors declare no competing interests.
References
- Cronan, J. E. and Waldrop, G. L. (2002). Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 41(5): 407–435.
- Kim, C. W., Moon, Y. A., Park, S. W., Cheng, D., Kwon, H. J. and Horton, J. D. (2010). Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc Natl Acad Sci USA. 107(21): 9626–9631.
- Colbert, C. L., Kim, C. W., Moon, Y. A., Henry, L., Palnitkar, M., McKean, W. B., Fitzgerald, K., Deisenhofer, J., Horton, J. D., Kwon, H. J., et al. (2010). Crystal structure of Spot 14, a modulator of fatty acid synthesis. Proc Natl Acad Sci USA. 107(44): 18820–18825.
- Hunkeler, M., Hagmann, A., Stuttfeld, E., Chami, M., Guri, Y., Stahlberg, H. and Maier, T. (2018). Structural basis for regulation of human acetyl-CoA carboxylase. Nature. 558(7710): 470–474.
- Bianchi, A., Evans, J. L., Iverson, A. J., Nordlund, A. C., Watts, T. D. and Witters, L. A. (1990). Identification of an isozymic form of acetyl-CoA carboxylase. J Biol Chem. 265(3): 1502–1509.
- Abu-Elheiga, L., Brinkley, W. R., Zhong, L., Chirala, S. S., Woldegiorgis, G. and Wakil, S. J. (2000). The subcellular localization of acetyl-CoA carboxylase 2. Proc Natl Acad Sci USA. 97(4): 1444–1449.
- Liberti, M. V. and Locasale, J. W. (2016). The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 41(3): 211–218.
- Wang, C., Rajput, S., Watabe, K., Liao, D. F. and Cao, D. (2010). Acetyl-CoA carboxylase-a as a novel target for cancer therapy. Front Biosci (Sch Ed). 2(2): 515–26.
- Chantranupong, L., Wolfson, R. L. and Sabatini, D. M. (2015). Nutrient-Sensing Mechanisms across Evolution. Cell. 161(1): 67–83.
- Schvartzman, J. M., Thompson, C. B. and Finley, L. W. (2018). Metabolic regulation of chromatin modifications and gene expression. J Cell Biol. 217(7): 2247–2259.
- Koundouros, N. and Poulogiannis, G. (2019). Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 122(1): 4–22.
- Batchuluun, B., Pinkosky, S. L. and Steinberg, G. R. (2022). Lipogenesis inhibitors: therapeutic opportunities and challenges. Nat Rev Drug Discovery. 21(4): 283–305.
- Bueno, M. J., Jimenez-Renard, V., Samino, S., Capellades, J., Junza, A., López-Rodríguez, M. L., Garcia-Carceles, J., Lopez-Fabuel, I., Bolaños, J. P., Chandel, N. S., et al. (2019). Essentiality of fatty acid synthase in the 2D to anchorage-independent growth transition in transforming cells. Nat Commun. 10(1): 5011.
- Fang, W., Cui, H., Yu, D., Chen, Y., Wang, J. and Yu, G. (2014). Increased expression of phospho-acetyl-CoA carboxylase protein is an independent prognostic factor for human gastric cancer without lymph node metastasis. Med Oncol. 31(7): 15.
- Abramson, H. N. (2011). The Lipogenesis Pathway as a Cancer Target. J Med Chem. 54(16): 5615–5638.
- Harwood, H., Petras, S. F., Shelly, L. D., Zaccaro, L. M., Perry, D. A., Makowski, M. R., Hargrove, D. M., Martin, K. A., Tracey, W., Chapman, J. G., et al. (2003). Isozyme-nonselective N-Substituted Bipiperidylcarboxamide Acetyl-CoA Carboxylase Inhibitors Reduce Tissue Malonyl-CoA Concentrations, Inhibit Fatty Acid Synthesis, and Increase Fatty Acid Oxidation in Cultured Cells and in Experimental Animals. J Biol Chem. 278(39): 37099–37111.
- Zhang, H., Tweel, B., Li, J. and Tong, L. (2004). Crystal Structure of the Carboxyltransferase Domain of Acetyl-Coenzyme A Carboxylase in Complex with CP-640186. Structure. 12(9): 1683–1691.
- Wakil, S. J. and Abu-Elheiga, L. A. (2009). Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. 50: S138–S143.
- Rendina, A. R., Craig-Kennard, A. C., Beaudoin, J. D. and Breen, M. K. (1990). Inhibition of acetyl-coenzyme A carboxylase by two classes of grass-selective herbicides. J Agric Food Chem. 38(5): 1282–1287.
- Guo, W., Wang, M. and Chen, L. (2022). A co-expression vector for baculovirus-mediated protein expression in mammalian cells. Biochem Biophys Res Commun. 594: 69–73.
Article Information
Publication history
Received: Oct 2, 2024
Accepted: Jan 15, 2025
Available online: Feb 9, 2025
Published: Feb 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Sun, Y., Li, J., Zhao, L. and Zhu, H. (2025). Purification of Native Acetyl CoA Carboxylase From Mammalian Cells. Bio-protocol 15(4): e5221. DOI: 10.21769/BioProtoc.5221.
Category
Biochemistry > Protein > Isolation and purification
Cancer Biology > Cancer biochemistry > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link







