- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
Published: Vol 15, Iss 4, Feb 20, 2025 DOI: 10.21769/BioProtoc.5194 Views: 1911
Reviewed by: Noelia ForesiPooja SaxenaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq
Mauricio A. Reynoso [...] Kaisa Kajala
Apr 5, 2018 16198 Views

RNA Purification from the Unicellular Green Alga, Chromochloris zofingiensis
Sean D. Gallaher and Melissa S. Roth
Apr 5, 2018 8147 Views
Extraction of RNA from Recalcitrant Tree Species Paulownia elongata
Niveditha Ramadoss and Chhandak Basu
Jul 20, 2018 7711 Views
Abstract
Plant embryos are contained within seeds. Isolating them is crucial when endosperm and seed coat tissues interfere with the study of mutant genetic functions due to differing genotypes between maternal and embryonic tissues. RNA extraction from plant embryonic tissue presents particular challenges due to the high activity of RNases, the composition of the seed, and the risk of RNA degradation. The developmental stage of the embryo is a key aspect of successful isolation and RNA extraction due to the size and amount of tissue. Proper handling during RNA extraction is critical to maintain RNA integrity and prevent degradation. While commercial kits offer various methods for RNA extraction from embryos, homemade protocols provide valuable advantages, including cost-effectiveness and accessibility for labs with limited funding. Here, we present a simple and efficient protocol for extracting RNA from isolated Arabidopsis thaliana embryos at the torpedo/cotyledon stage using a homemade extraction buffer previously reported for styles of Nicotiana alata.
Key features
• This straightforward homemade protocol builds upon established methods for embryo isolation [1] and RNA extraction [2,3].
• It enables the extraction of high-quality RNA from Arabidopsis embryos at the torpedo stage.
• The protocol is designed to be both simple and cost-effective, making it an excellent option for labs with limited funding.
Keywords: Arabidopsis embryoGraphical overview
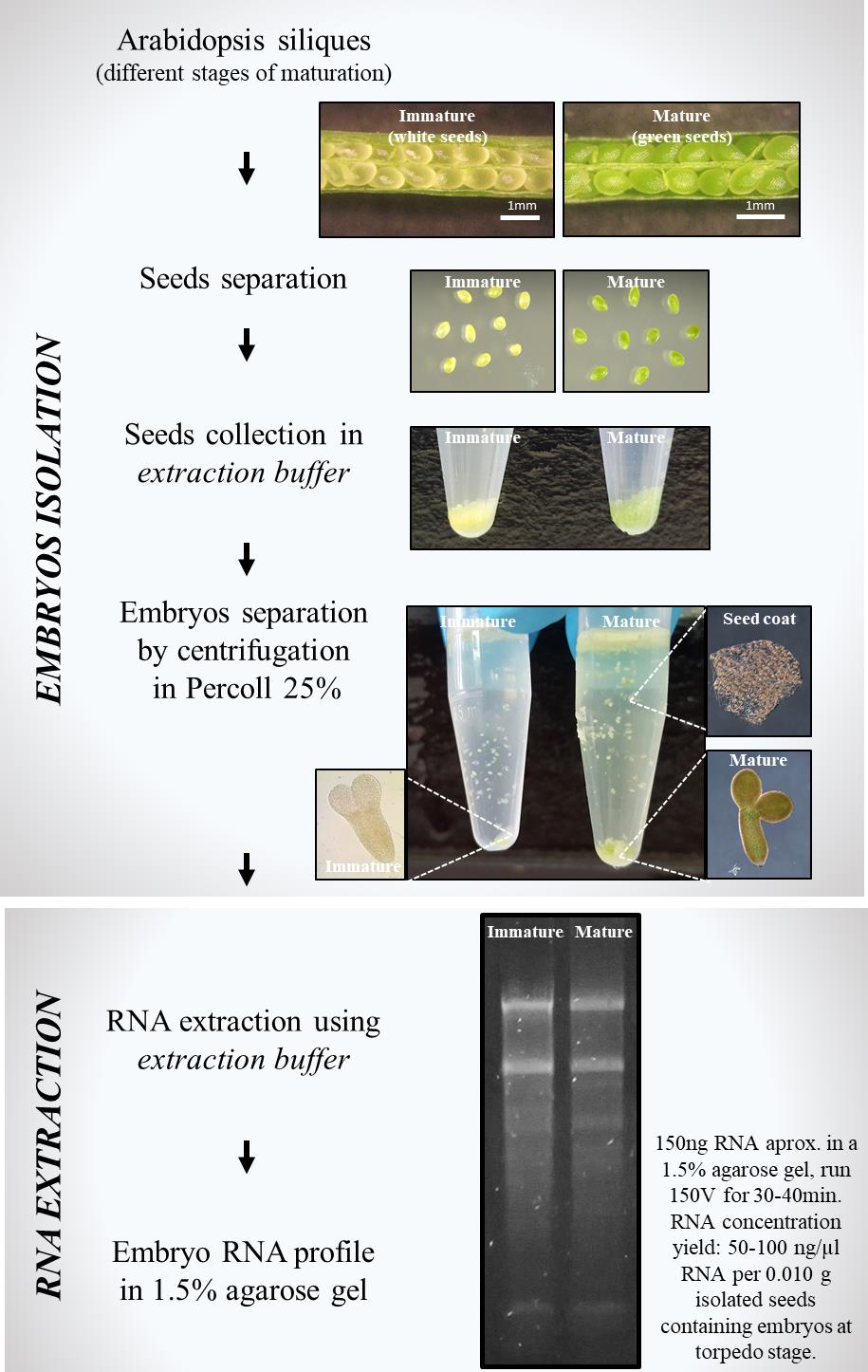
Simple method for RNA extraction from isolated Arabidopsis embryos using a homemade extraction buffer
Background
This protocol describes a straightforward two-step method for extracting RNA from isolated Arabidopsis embryos. Isolating RNA from plant tissues such as embryos, which are rich in RNases, poses significant challenges due to the risk of RNA degradation during sample preparation. To overcome these difficulties, commercial solutions such as RNAlater are commonly used to stabilize RNA by inactivating RNases, allowing for subsequent extraction using reagents like Trizol. In this protocol, we present a simple and effective homemade extraction buffer designed specifically for seed collection and RNA extraction. This buffer facilitates the isolation of high-quality RNA from Arabidopsis embryos. The method described here was successfully employed to extract RNA from developing embryos at the early torpedo and early cotyledon stages, from both immature (white) and mature (green) seeds. This approach offers an accessible and efficient alternative to commercial reagents, ensuring robust RNA recovery and preservation for downstream applications.
Materials and reagents
Biological materials
1. Arabidopsis thaliana ecotype Columbia (Col-0) wild type (WT) and embryo-lethal mutant plants from the Arabidopsis biological resource center (ARBRC, https://abrc.osu.edu/)
Reagents
1. Tris [Tri(hydroxymethyl)aminomethane] (Genbiotech, catalog number: RU2510)
2. EDTA (ethylenediamine tetraacetic acid disodium salt) (Biopack, catalog number: 1604.07)
3. Urea (Sigma, catalog number: U5378)
4. Sodium dodecyl sulfate (SDS) (Anedra, catalog number: 7211)
5. Ammonium acetate (Fluka, catalog number: 09690)
6. Lithium chloride (LiCl) (Sigma, catalog number: L9650)
7. 2-Mercaptoethanol pure (Biopack, catalog number: 9545.05)
8. Diethyl pyrocarbonate (DEPC) (Sigma, catalog number: D5758)
9. Percoll (GE Healthcare, catalog number: 17-0891-01)
10. DEPC (diethylpyrocarbonate) (Sigma, catalog number: D5758)
11. SYBRTM Safe DNA gel stain (Invitrogen, catalog number: S33102)
12. Agarose (Gene Biotech LE-Agarose 1200, catalog number: RU1010)
13. DEPC water (1 mL DEPC/L water; stirred overnight and autoclaved)
14. Phenol:chloroform:isoamyl alcohol 25:25:1 (Sigma, catalog number: P3803)
15. Chloroform (Biopack, catalog number: 1651.08)
16. Isopropanol (Mallinckrodt, catalog number: 3032-08)
Solutions
1. Buffer Tris HCl pH 8, 1 M (see Recipes)
2. EDTA pH 8, 0.5 M (see Recipes)
3. SDS 10% (see Recipes)
4. Ammonium acetate, 10 M (see Recipes)
5. Lithium chloride, 8 M (see Recipes)
6. Extraction buffer (see Recipes)
Recipes
1. Buffer Tris-HCl pH 8, 1 M
Weigh 12.11 g of Tris, add 75 mL of pure sterile water, adjust to pH 8 with HCl, and fill with pure sterile water to a final volume of 100 mL.
2. EDTA pH 8, 0.5 M
Weigh 18.6 g of EDTA, add 75 mL of pure sterile water, adjust to pH 8 using 10 M NaOH, and stir until EDTA is completely dissolved. Solubility improves while alkalinity increases; complete with pure sterile water to a 100 mL final volume.
3. SDS 10%
Dissolve 10 g of SDS in 100 mL of pure sterile water.
4. Ammonium acetate, 10 M
Dissolve 7.78 g of ammonium acetate in 10 mL of DEPC water.
5. Lithium chloride, 8 M
Dissolve 3.39 g of lithium chloride in 10 mL of DEPC water.
6. Extraction buffer (10 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Urea | 7 M | 4.2 g |
| EDTA | 10 mM | 200 μL |
| Tris-HCl (1 M, pH 8) | 100 mM | 1 mL |
| SDS 10% | 1% | 1 mL |
| 2-Mercaptoethanol | 1% | 100 μL |
| H2O | to 10 mL | *see note |
*Note: Weigh urea in a 50 mL Falcon tube and dissolve urea in half volume (5 mL for a final volume of 10 mL) of water. Vortex the solution to help it dissolve. The bottom of smaller tubes hinders the full solubility of urea. Then, add the rest of the components and complete to final volume. Keep at room temperature. Cold will precipitate SDS in the extraction buffer. No risk for later RNA extraction.
Laboratory supplies
1. Syringe, needles (13 × 0.3 mm) and pointy tips tweezers (Dumont, catalog number: 0103-2-PO)
2. Plastic grinding rod for Eppendorf tubes (SSIbio, catalog number: 1005-39)
Equipment
1. Magnifying glass (LABKLASS, model: HG468405)
2. Table centrifuge (Eppendorf, model: centrifuge 5418)
3. Vortex (VornadoTM, Benchmark Scientific, catalog number/model: BV101-B)
4. Agarose gel electrophoresis equipment (Bio-Rad, model: PowerPacTM Basic Power Supply, Horizontal Electrophoresis Systems)
5. NanoDropTM One/One (Thermo Fisher, catalog number/model: ND-ONE-W)
Procedure
A. Embryo isolation
1. Collection of seeds
a. Add 100 μL of extraction buffer (see Recipes) in a 1.5 mL Eppendorf tube. Weigh the tube.
b. Using a needle under a magnifying glass, open mature or immature siliques to collect green and/or white seeds, respectively. Place the seeds inside the Eppendorf tube containing the extraction buffer. Collect seeds from approximately 25 siliques. Weigh the tube after collecting seeds. For a satisfactory RNA extraction, the minimum recommended amount for seed collection is 0.010 g of tissue.
c. Spin down the embryos in a table centrifuge at 1,700× g for 30 s. Carefully remove the extraction buffer by pipetting. Wash the embryos three times with 1 mL of DEPC water. Spin down at 1,700× g after each washing.
2. Embryo isolation from seed coat (from Perry and Wang [2])
a. Remove 750 μL of the DEPC water per tube.
b. Shake gently the lower part of the Eppendorf tube by hand (using fingers) just to let the seeds spread in the remaining water.
c. Use a plastic grinding rod for Eppendorf tubes (plastic sticks) to press softly the sample against the tube’s wall to release embryos from seeds by applying soft pressure. Repeat three times with smooth movements.
d. Transfer the sample (250 μL) by pipetting to a new tube containing 500 μL of DEPC water and 250 μL of Percoll (25% v/v Percoll). To facilitate pipetting the sample, use a 200 μL yellow tip with the tip cut off.
e. Centrifuge at 72× g for 10 min.
f. Remove the seed coats of the upper layer together with the Percoll solution by pipetting.
g. Resuspend the embryos carefully in 250 μL of the remaining Percoll solution and transfer the sample by pipetting to a new tube containing 0.75 mL of 25% v/v Percoll solution.
h. Centrifuge at 72× g for 10 min.
i. Remove the seed coats from the upper layer and discard the remaining Percoll by pipetting.
j. Wash the embryos three times with 1 mL of DEPC water. Spin down at 72× g after each washing.
B. RNA extraction [2,3]
Before starting, prepare Eppendorf tubes containing:
I. 500 μL of phenol:chloroform:isoamyl alcohol (25:24:1) + 500 μL of extraction buffer
II. 0.5 mL of phenol:chloroform:isoamyl alcohol (25:24:1)
III. 0.5 mL of chloroform
IV. 0.1 mL of 10 M ammonium acetate
1. Remove washing water from the tube by pipetting. Add 100 μL of extraction buffer and use a plastic grinding rod to crash the tissue against the tube wall. Crash embryos completely.
2. Add the sample to tube I and vortex immediately for 2 min.
3. Centrifuge at 18,000× g for 10 min at room temperature. Take the upper phase and transfer it to tube II. Vortex vigorously for 2 min.
4. Centrifuge at 18,000× g for 10 min at room temperature. Transfer the upper phase to tube III. Vortex vigorously for 2 min.
5. Centrifuge at 18,000× g for 10 min at room temperature. Transfer the aqueous phase to tube IV. Add 1 volume of cold isopropanol. Mix by inversion. Store at -20 °C for any duration between 30 min and overnight.
6. Centrifuge at 18,000× g for 15 min at 4 °C. Discard supernatant by inverting the tube. Dry pellet at room temperature.
7. Resuspend the pellet in 300 μL of DEPC water. Maintain tubes on ice. Check that the pellet is well-solubilized. Heat sample at 55 °C for 5 min to better solubilize the pellet (optional). Add 300 μL of 8 M LiCl and mix by inversion. Place the tubes inside an ice bucket and leave on ice overnight inside the refrigerator to precipitate RNA.
8. Centrifuge at 18,000× g for 15 min at 4 °C. Discard supernatant after centrifugation.
9. Resuspend the pellet in 300 μL of DEPC water. Add 100 μL of ammonium acetate. Fill the tube with 96% cold ethanol. Precipitate at -20 °C for at least 30 min.
10. Centrifuge at 18,000× g for 15 min at 4 °C. Wash pellet with 70% ethanol. Centrifuge at 18,000× g for 15 min at 4 °C. Discard supernatant. Dry the pellet at room temperature and resuspend in 20 μL of DEPC water.
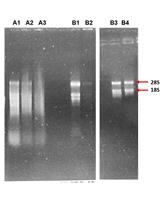
11. Check RNA profile by loading 3 μL of RNA (approximately 100–150 ng of RNA) in a 1.5% agarose gel; run at 150 V for 30–40 min. RNA concentration yield is approximately 50–100 ng/μL RNA per 0.010 g of isolated seeds containing embryos at the torpedo stage.
12. RNA can be stored at -20 °C for short-term uses. Storage at -80 °C is recommended for long-term use.
Validation of protocol
This protocol was developed by combining two existing protocols:
For RNA extraction:
McClure et al. [2]. Self incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature.
Roldán et al. [3]. Molecular and genetic characterization of novel S-RNases from a natural population of Nicotiana alata. Plant Cell Rep (using extraction buffer, no commercial kits).
For embryo isolation:
Perry, S. E. and Wang, H. [1]. Rapid isolation of Arabidopsis thaliana developing embryos. BioTechniques. (using commercial stabilization solutions).
The innovation lies in the simultaneous application of both pre-existing protocols along with the incorporation of a homemade extraction buffer for RNA extraction from tissues with high RNase activity, such as Arabidopsis embryos, without relying on commercial kits. In our hands, the homemade extraction buffer effectively prevents RNA degradation from Arabidopsis embryos. The protocol presented here represents a valuable alternative for two challenging steps when working with Arabidopsis seeds: first, the separation of embryos from seed coats (embryo isolation step), and second, RNA extraction from tissues that require RNase inhibitors and stabilization solutions to prevent RNA degradation (RNA extraction using homemade buffer step).
General notes and troubleshooting
General notes
This protocol is an alternative for RNA extraction from isolated Arabidopsis embryos using a homemade extraction buffer to prevent RNA degradation of tissues with high RNAse activity.
Embryo developmental stages play a crucial role in obtaining the necessary amount of tissue. For example, mature seeds (light green) containing embryos at advanced developmental stages, such as early cotyledon or cotyledon stages, have larger embryos and provide higher amounts of tissue when collecting from Arabidopsis siliques. In contrast, immature seeds (white seeds) contain embryos at early torpedo and torpedo developmental stages, which are smaller than embryos found in mature seeds. However, 0.010 g of tissue yield around 100–150 ng/μL RNA, suitable for cDNA synthesis. Collection of 0.010 g can be performed in approximately 30 min. The more seeds collected, the more RNA can be obtained.
This protocol can be implemented not only for immature seeds containing embryos at the early torpedo stage but also for seeds at earlier developmental stages, for example, seeds that contain embryos at the heart stage. In those cases, collecting more tissue is required because embryos are smaller in size. In our hands, this protocol works great for the isolation of embryos at early stages of development, and RNA extraction succeeds. When collecting 0.010 g of immature seeds when working with embryos at the heart stage, the tissue yield is approximately 5–10 ng/μL RNA, also suitable for cDNA synthesis. However, when working with earlier embryo developmental stages, such as globular or early heart stages in which the embryonic amount of tissue is low (in relation to the whole seeds), this protocol can be applied by skipping the embryo isolation step to work with the whole seeds (embryo, endosperm, and seed coat), performing the step of RNA extraction straightforward with the homemade extraction buffer, which will prevent RNA degradation. When collecting 0.010 g of whole immature seeds, the yield is around 60–160 ng/μL RNA, which corresponds to RNA extracted from the entire seed and not only from isolated embryos as described above but also suitable for cDNA synthesis.
Measurements of RNA concentration were done using NanoDropTM by absorbance at 260 nm and purity was estimated by the ratio 260/280 nm. RNA profile was analyzed by running (approximately) 150 ng of RNA in a 1.5% agarose gel. Note that RNA extracted from plant “white” tissue, such as embryo or root, lacks plastid RNA. In these cases, the RNA profile is composed mainly of two bands corresponding to cytoplasmic 28S and 18S rRNA, different from the RNA profile extracted from leaves in which smaller bands are present, corresponding to organellar rRNAs.
In summary, the protocol presented here is a great alternative to extract RNA from isolated Arabidopsis embryos as well as directly from seeds (when containing embryos in early developmental stages). It may be very useful when studying, for example, embryo-lethal mutants. Besides, using a homemade buffer suitable to prevent RNA degradation represents a valuable alternative when there is limited access to commercial stabilization solutions or kits.
Acknowledgments
We thank CONICET and ANPCyT for funding as well as the Institute IIB-CONICET-UNMDP. We are grateful to Dr. Roldan for suggestions when performing the RNA extraction protocol.
Competing interests
There are no competing interests.
References
- Perry, S. E. and Wang, H. (2003). Rapid isolation of Arabidopsis thaliana developing embryos. Biotechniques. 35(2): 278–282. https://doi.org/10.2144/03352bm06
- McClure, B. A., Gray, J. E., Anderson, M. A. and Clarke, A. E. (1990). Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 347(6295): 757–760. https://doi.org/10.1038/347757a0
- Roldán, J. A., Quiroga, R. and Goldraij, A. (2010). Molecular and genetic characterization of novel S-RNases from a natural population of Nicotiana alata. Plant Cell Rep. 29(7): 735–746. https://doi.org/10.1007/s00299-010-0860-6
Article Information
Publication history
Received: Sep 17, 2024
Accepted: Dec 10, 2024
Available online: Jan 5, 2025
Published: Feb 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Marchetti, F., Pagnussat, G. C. and Zabaleta, E. J. (2025). Simple Method for Efficient RNA Extraction From Arabidopsis Embryos. Bio-protocol 15(4): e5194. DOI: 10.21769/BioProtoc.5194.
Category
Plant Science > Plant molecular biology > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










