- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Protocol to Purify Human Mediator Complex From Freestyle 293-F Cells
Published: Vol 15, Iss 4, Feb 20, 2025 DOI: 10.21769/BioProtoc.5185 Views: 2145
Reviewed by: Joana Alexandra Costa ReisRohini Ravindran NairAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ZnCl2 Precipitation-Assisted Sample Preparation for Proteomic Analysis
Qiqing He [...] Fuchu He
Jul 20, 2025 2721 Views
Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1806 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1208 Views
Abstract
The Mediator, a multi-subunit protein complex in all eukaryotes, comprises the core mediator (cMED) and the CDK8 kinase module (CKM). As a molecular bridge between transcription factors (TFs) and RNA polymerase II (Pol II), the Mediator plays a critical role in regulating Pol II–dependent transcription. Considering its large size and complex composition, conducting in vitro studies on the Mediator complex is challenging, especially when isolating the intact and homogeneous complex from human cells. Here, we present a method to purify the intact CKM-cMED complex from FreeStyle 293-F cells (293-F cells), which offers advantages for performing large-scale protein purification. To isolate the CKM-bound cMED without the presence of Pol II, FLAG-tagged CDK8, a subunit of the CKM complex, was expressed in 293-F cells for purification, as CKM and Pol II are mutually exclusive in their interaction with cMED. The complex is isolated from nuclear extracts through immunoaffinity purification and further purified by glycerol gradient to enhance its homogeneity. This protocol provides a time- and cost-efficient way to purify the endogenous Mediator complex for structural- and functional-based studies.
Key features
• This protocol describes a method for purifying the endogenous Mediator complex, free of Pol II, from 293-F cells.
• Does not require the use of crosslinkers, offering advantages for structural and functional studies.
Keywords: MediatorGraphical overview
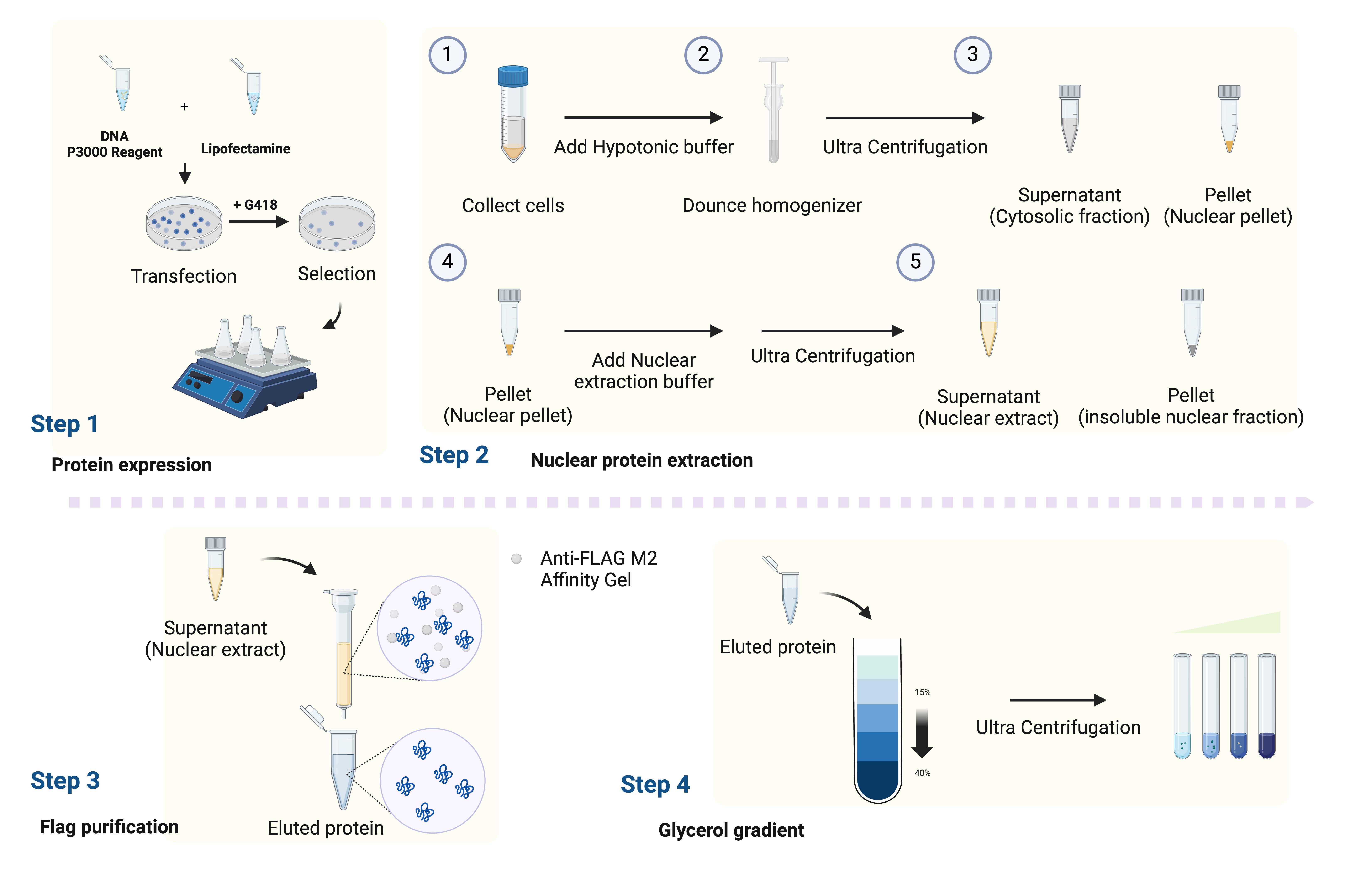
Workflow for isolating the endogenous Mediator complex. The workflow outlines the steps from generating the stable cell line to protein purification.
Background
In eukaryotes, the Mediator complex is an evolutionarily conserved transcriptional coactivator composed of 25 subunits in budding yeast and 30 subunits in humans [1]. The Mediator complex consists of a large core (cMED) with three domains: the head, middle, and tail, along with a dissociable module known as the CDK8 kinase module (CKM). The CKM contains four subunits: cyclin-dependent kinase 8 (CDK8), Cyclin C, MED12, and MED13 [2]. The primary function of cMED is to activate Pol II–dependent transcription by recruiting Pol II to promoters [3]. However, this activity is repressed by the CKM through steric hindrance mediated by MED13 [4,5]. Interestingly, recent studies suggest that CDK8 also plays a positive role in gene transcription, though the mechanisms remain poorly understood. Since the CKM serves as a key entry point for developmental and oncogenic signaling via the Mediator complex [6], isolating the CKM-cMED complex is essential for investigating its mechanisms and functions, providing deeper insights into these processes.
The human CKM-cMED complex consists of 30 subunits, and overexpressing each individually requires considerable effort. As a transcriptional coactivator, the Mediator interacts with multiple binding partners, such as RNA pol II, further complicating the process of isolating pure CKM-cMED complex. Therefore, we developed a protocol to obtain a high-quality and large quantity of CKM-cMED complex.
Previously, we established a stable HEK293 cell line expressing CDK8. However, scaling up proved challenging due to the limitations associated with adherent cells. Then, we selected the FreeStyle 293-F cells suspension cell culture system for its ease of expansion and high protein expression yields. Since CKM and Pol II are mutually exclusive for cMED binding, we overexpressed the CDK8, one of the CKM subunits, with a C-terminal FLAG tag, but not any subunits of cMED, in freestyle 293-F cells to prevent RNA Pol II contamination. The size of the FLAG tag, consisting of eight amino acids, is small and specifically recognized by the antibody conjugated to agarose beads. Additionally, the FLAG tag added to the C-terminus of CDK8 did not compromise the stability of the CKM-cMED complex and still maintained its kinase activity. By establishing stable cell lines through antibiotic selection, we could stably express FLAG-tagged recombinant CDK8 in FreeStyle 293-F cells, enabling the collection of large quantities of cells for protein purification.
For protein purification, we began by isolating nuclear extract to enrich the CKM-cMED complex as it is primarily present in the nucleus. Affinity purification was then performed using anti-FLAG M2 affinity gel to ensure sufficient purity and quality to proceed to the next step. To enhance the homogeneity of the protein complex, we further performed glycerol gradient centrifugation without a cross-linker and successfully obtained the functional endogenous CKM-cMED complex. Because the molecular weight of CKM-cMED is very large, density gradient sedimentation is an effective method to separate the complex from relatively small degradation products, disassembled components, and other impurity proteins present in the FLAG elution. This protocol provides a reliable method for isolating the homogenous endogenous CKM-cMED complex from FLAG-CDK8 expressed cells using FLAG-specific immunoprecipitation and glycerol gradient sedimentation for future investigations of structure-function relationships.
Materials and reagents
Biological materials
1. FreestyleTM 293-F cells (Thermo Fisher, catalog number: R79007), containing 1 × 10 cells suspended in 90% FreeStyleTM 293 expression medium and 10% DMSO, stored in liquid nitrogen
2. Plasmid DNA (pcDNA3.1_CDK8-F)
Reagents
1. Lipofectamine 3000 (Invitrogen, catalog number: L3000001)
2. G418 sulfate (Thermo Fisher, catalog number: 11811023)
3. ANTI-FLAG® M2 affinity gel (Sigma, catalog number: A2220)
4. cOmpleteTM, mini, EDTA-free protease inhibitor cocktail (Sigma, catalog number: 50-161-3323)
5. FreeStyleTM 293 expression medium (prewarm to 37 °C before use)
6. Phosphate buffered saline (PBS), 1×, liquid, pH 7.4 (GenDEPOT, catalog number: P2201-050)
7. HEPES (Merck, catalog number: 391338)
8. Magnesium chloride (Sigma, catalog number: M8266)
9. Potassium chloride (Sigma, catalog number: P3911)
10. Dithiothreitol (DTT) (Fisher Scientific, catalog number: BP172-5)
11. Protease inhibitor cocktail (RPI, catalog number: P50600-1)
12. Ethylenediaminetetraacetic acid (EDTA) (Sigma, catalog number: E6758)
13. NP-40 Surfact-AmpsTM detergent solution (Thermo Fisher, catalog number: 28324)
14. Glycerol (Fisher Scientific, catalog number: BP229-4)
15. Sodium chloride (Sigma, catalog number: 567440)
16. beta-mercaptoethanol (Sigma, catalog number: 444203)
17. DYKDDDDK tag Peptide (FLAG peptide) (APExBio, catalog number: A6002)
Solutions
1. Hypotonic buffer (see Recipes)
2. Nuclear extraction buffer (see Recipes)
3. Washing buffer A (see Recipes)
4. Washing buffer B (see Recipes)
5. Washing buffer C (see Recipes)
6. Elution buffer (see Recipes)
7. 15% glycerol gradient buffer (see Recipes)
8. 40% glycerol gradient buffer (see Recipes)
Recipes
1. Hypotonic buffer
10 mM HEPES pH 7.9
1.5 mM MgCl2
10 mM KCl
0.5 mM dithiothreitol (DTT)
Protease inhibitors (freshly added)
2. Nuclear extraction buffer
25 mM HEPES pH 7.9
1.5mM MgCl2
0.6 M KCl
0.5 mM DTT
0.2 mM EDTA
0.02% NP-40
20% glycerol
Protease inhibitors (freshly added)
3. Washing buffer A
25 mM HEPES pH 7.6
1.5 mM MgCl2
150 mM NaCl
10% glycerol
0.2 mM EDTA
2 mM beta-mercaptoethanol
Protease inhibitors (freshly added)
4. Washing buffer B
25 mM HEPES pH 7.6
1.5 mM MgCl2
150 mM NaCl
10% glycerol
0.2 mM EDTA
2 mM beta-mercaptoethanol
5. Washing buffer C
25 mM HEPES pH 7.6
1.5 mM MgCl2
150 mM NaCl
10% glycerol
0.2 mM EDTA
0.01% NP40
2 mM beta-mercaptoethanol
6. Elution buffer
25 mM HEPES pH 7.6
1.5 mM MgCl2
150 mM NaCl
10% glycerol
0.2 mM EDTA
0.01% NP40
2 mM beta-mercaptoethanol
200 μg/mL 1× FLAG peptide
7. 15% glycerol gradient buffer
20 mM HEPES pH 7.6
1 mM MgCl2
300 mM NaCl
0.25 mM EDTA
1 mM beta-mercaptoethanol
15% (v/v) glycerol
8. 40% glycerol gradient buffer
20 mM HEPES pH 7.6
1 mM MgCl2
300 mM NaCl
0.25 mM EDTA
1 mM beta-mercaptoethanol
40% (v/v) glycerol
Filter sterilize and store all buffers at 4 °C.
Laboratory supplies
1. Clean, sterile glass 250, 1,000, 2,000 mL flasks
2. Hemacytometer
3. 60 × 15 mm non-treated culture dish, sterile (CytoOne, catalog number: CC7672-3359)
4. 15 and 50 mL conical sterile polypropylene centrifuge tubes (Thermo Scientific, catalog number: 12-565-269, 12-565-271)
5. Dounce homogenizer
6. 1.7 mL microtubes (e.g., Axygen, catalog number: 14-222-168)
7. Centrifuge bottles with sealing closure (Thermo Scientific, catalog number: 05-564-3)
8. Polycarbonate ultracentrifuge bottles (Beckman, catalog number: 355622)
9. 4 mL ultra-clear polypropylene tubes (Beckman, catalog number: 344062)
10. Econo-Pac® chromatography columns (Bio-Rad, catalog number: 7321010)
11. NovexTM tris-glycine mini protein gels, 4%–20%, 1.0 mm, WedgeWellTM format (Invitrogen, catalog number: XP04205BOX)
Equipment
1. Standard orbital shaker (VWR, model: 1000, catalog number: 89032-088)
2. CO2 cell culture incubator
3. Inverted microscope
4. Biological safety cell culture hood
5. 37 °C water bath
6. Benchtop centrifuge (Eppendorf, catalog number: 022625501)
7. High-speed centrifuge (Thermo Sorvall, model: RC-6 Plus; PTI FiberLite F10-6x500y fixed angle rotor)
8. Ultracentrifuges (Beckman, model: Optima XPN-90; 45 Ti fixed-angle titanium rotor, SW60 Ti swinging-bucket rotor)
9. GRADIENT MASTERTM (Biocomp, Model 108)
10. Tube revolver rotator (Thermo Scientific, catalog number: 88881001)
Software and datasets
1. BioRender (https://www.biorender.com/) was used for the Graphical overview: Tang, H. (2025) https://BioRender.com/z21d813; and Figure 3: Tang, H. (2025) https://BioRender.com/t82q193
Procedure
A. Overexpression of CDK8-FLAG in FreeStyleTM 293-F cells
1. Cell culture
a. Thaw the FreeStyleTM 293-F cells quickly in a 37 °C water bath.
b. Before the vial is almost thawed, clean the vial with 70% ethanol.
c. Gently transfer the cells from the vial to the 250 mL flask containing 30 mL of prewarmed FreeStyleTM 293 expression medium inside a biosafety cabinet (cell density is around 0.33 × 106 cells/mL).
d. Place the flask on the orbital shaker (at 135 rpm) inside the incubator at 37 °C with a humidified atmosphere of 8% CO2.
e. Maintain the cells at 0.3–3 × 106 cells/mL density in a 250 mL flask.
Note: Cells should be healthy with viability greater than 90%. Low viability leads to reduced transfection efficiency.
2. Transfection and generation of stable cell line
Day 0
a. Culture 293-F cells at a density of 0.6 × 106 to 0.7 × 106 cells/mL.
Day 1
a. Measure the cell density using a hemacytometer.
b. Dilute the cells to 1 × 106 cells/mL.
c. Seed the cells in a volume of 3 mL in the 6 cm non-treated plate.
d. Dilute 3 μg of plasmid DNA and 6 μL of P3000 into 100 μL of FreeStyleTM 293 expression medium in a 1.7 mL tube.
e. Dilute 7.5 μL of lipofectamine into 100 μL of FreeStyleTM 293 expression medium in another tube.
f. Gently add the diluted lipofectamine (from step A, day 1, e) into the diluted DNA solution (from step A, day 1, d).
g. Incubate the mixture for 10–15 min at room temperature.
h. Drop the DNA–lipid complex into cells.
i. Place the 6 cm non-treated plate on the orbital shaker inside the incubator at 37 °C with 8% CO2.
Day 2
a. Change the fresh media after 8–12 h.
b. Collect the cells in a 15 mL tube.
c. Centrifuge the cells at 201× g for 3 min.
d. Discard the supernatant and resuspend the cells in 4 mL of prewarmed fresh medium.
e. Transfer the cells into the 6 cm non-treated plate.
Day 4
a. Add G418 sulfate to a final concentration of 200 μg/mL for selection.
b. Measure the cell density and viability using a hemacytometer. Change the fresh media every 3–4 days.
c. When cell density is higher than 1 × 106 cells/mL, split the cells into a 1:4 ratio in fresh medium and use a smaller amount of G418 (50 μg/mL) for maintenance.
d. Collect a small amount of cells and check protein expression using western blot (Figure 1). This is an essential checkpoint for the following steps.
e. Expand the cells to 4 L (check protein expression frequently using western blot while expanding cells).
f. Pour the medium into centrifuge bottles from glass flasks. Harvest the cells by centrifugation at 1,590× g for 10 min at 4 °C in a Thermo Sorvall RC-6 Plus Centrifuge with F10-6x500y fixed angle rotor.
g. Discard the supernatant and resuspend the cell pellet with ice-cold PBS.
h. Transfer the cells into 50 mL tubes.
i. Centrifuge at 201× g for 10 min at 4 °C.
j. Discard the supernatant and store the pellet at -80 °C.
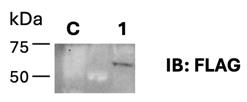
Figure 1. Expression test of hCDK8-FLAG. FreeStyleTM 293-F cells were collected after transfection. FLAG antibody was used for western blot analysis. C: FreeStyleTM 293-F cells; Lane 1: hCDK8-FLAG overexpressed in FreeStyleTM 293-F cells. Sample was collected when expanding the cell culture.
B. Purification of human cMED-CKM
1. Cell lysis and nuclear extraction
a. Defrost 40 g of 293F cell pellets stably over-expressing FLAG-CDK8 in 250 mL of pre-chilled hypotonic buffer.
b. Resuspend the cell pellet by gently pipetting.
c. Incubate on ice for 10 min.
d. Use a Dounce homogenizer (30 mL) with 20 gentle strokes to lyse the cells.
e. Aliquot the lysate into four ultracentrifuge bottles for ultracentrifugation.
f. Centrifuge (45 Ti Rotor) at 46,377.8× g for 30 min at 4 °C and discard the supernatant.
g. Resuspend the nuclear pellet with 40 mL/tube of hypotonic buffer.
h. Centrifuge (45 Ti Rotor) at 46,377.8× g for 10 min at 4 °C and discard the supernatant.
i. Resuspend the nuclear pellet with 40 mL/tube of hypotonic buffer.
j. Centrifuge (45 Ti Rotor) at 46,377.8× g for 10 min at 4 °C and discard the supernatant.
k. Resuspend all nuclear pellets with 25 mL of nuclease extraction buffer.
l. Add hypotonic buffer to the final volume of 50 mL (final salt concentration: 300 mM KCl).
m. Transfer the suspension from the 50 mL tube into the polycarbonate ultracentrifuge bottles.
n. Rotate the suspension for 1 h at 4 °C.
o. Centrifuge (45 Ti Rotor) at 126,263.7× g for 1 h at 4 °C.
2. Binding
a. Collect the supernatant and aliquot into a 50 mL tube.
b. Get 4 mL of ANTI-FLAG® M2 affinity gel in another 15 mL tube.
c. Equilibrate the resin with 10 mL of pre-chilled nuclear extraction buffer, centrifuge at 201× g for 1 min, and discard the supernatant.
d. Repeat step B2c twice.
e. Apply the resin to the nuclear extract from step B2a and incubate the sample for 4 h at 4 °C with end-over-end rotation. The incubation time is associated with the purification yield.
3. Wash
a. Load the mixture of resin and the nuclear extract into the column.
b. Wash the resin with 30 mL of washing buffer A.
c. Wash the resin with 30 mL of washing buffer B.
d. Wash the resin with 30 mL of washing buffer C.
4. cMED-CKM complex elution
a. Seal the bottom of the column and add 8 mL of elution buffer from the top.
b. Seal the top of the column and incubate the sample for 1 h at 4 °C with end-over-end rotation.
c. Collect the elution (8 mL) and run SDS-PAGE to perform a silver stain to confirm purity (Figure 2). This is a critical checkpoint for assessing the purity of the elution before further purification.
d. Concentrate the eluted solution to approximately 2.4 mL.
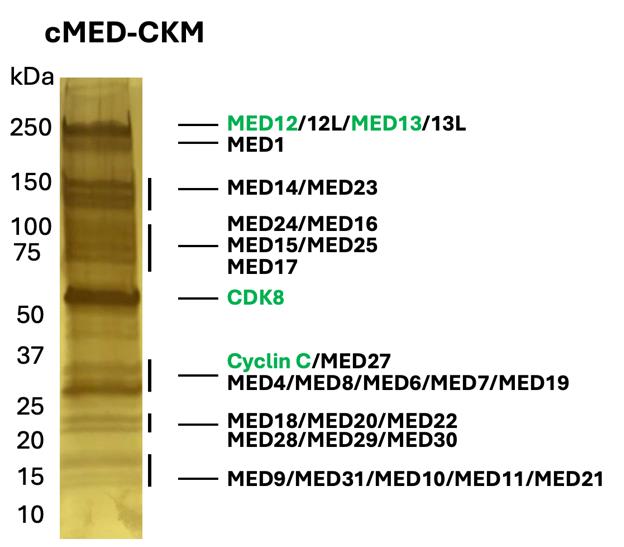
Figure 2. FLAG purification result of the Mediator complex, 4%–20% gradient gel, silver stained. The elution fraction was collected after FLAG purification. The CKM subunits, including MED12 and MED13, are indicated by green text.
5. Glycerol gradient
a. Add 1,860 μL of 40% glycerol gradient buffer (heavy solution) into ultra-clear polypropylene tubes. Using P1000 Pipetman, load 930 μL of the buffer into the tube twice.
b. Add 1,860 μL of 15% glycerol gradient buffer (light solution) gently into the same tube. Using P1000 Pipetman, load 930 μL of the buffer into the tube twice.
c. Avoid air bubble formation when adding buffer or samples. This will disturb the layers.
d. Make sure the heavy–light interface rises precisely to the middle of the tube and seal the tube.
e. Prepare six tubes. The detailed procedure is described in Figure 3.
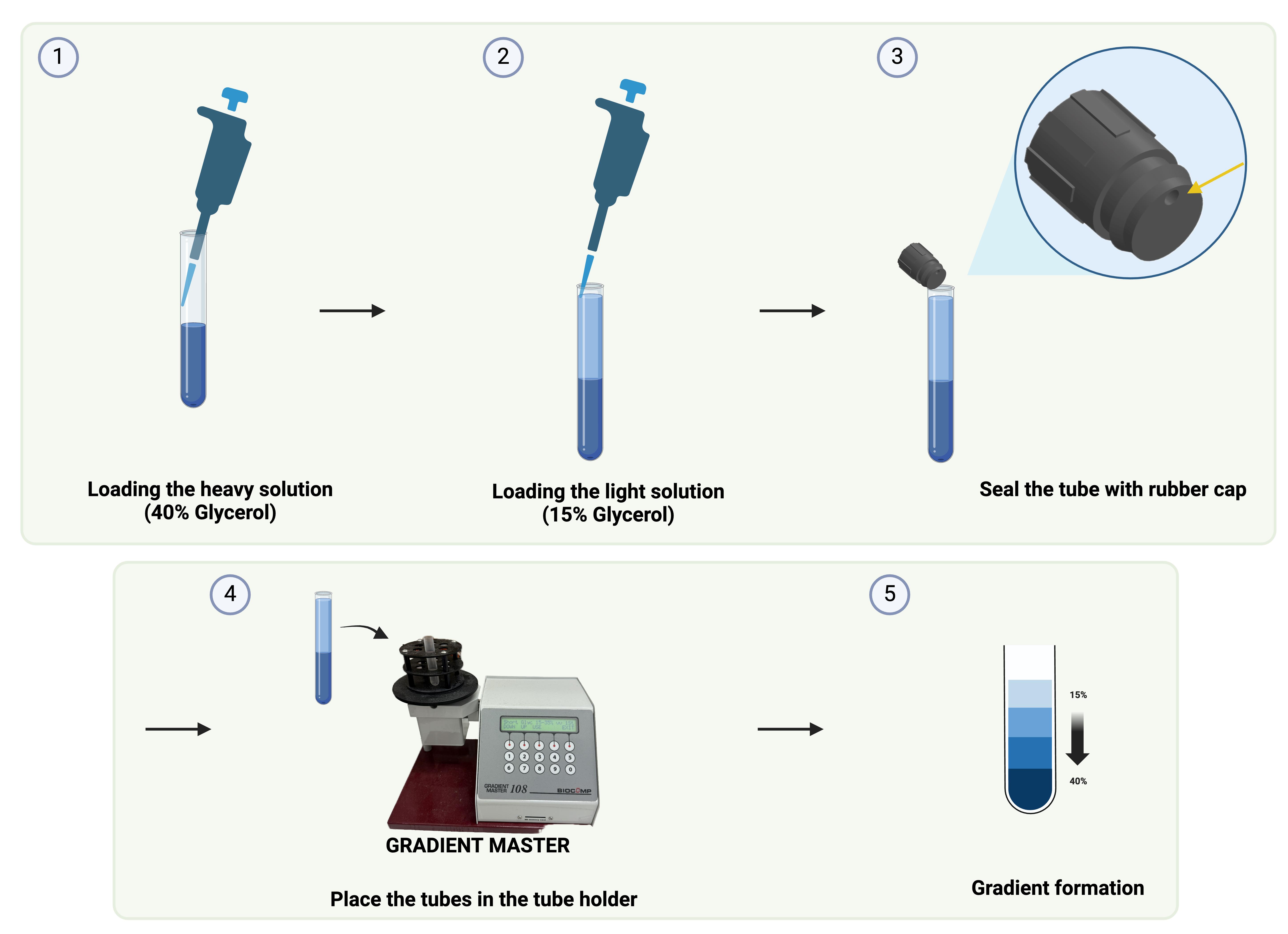
Figure 3. Flowchart of glycerol gradient
f. Use the short rubber cap to seal the tube and make sure the side with the hole is the last part to seal.
g. Place the tubes in the tube holder and use the Gradient Master instrument to rotate and mix the solutions to form the gradient. Choose the gradient 15%–40% and hit Run.
h. Remove the seal and load 400 μL of cMED-CKM complex elution by gently pipetting on top of the gradients.
i. Balance the tubes before placing them into the rotor. Caution: an improperly loaded rotor or weight difference will lead to rotor imbalance.
j. Centrifuge (SW60 Ti Rotor) at 120,937.6× g for 17 h at 4 °C.
k. Fractionate the gradients by collecting 200 μL of sample per fraction at 4 °C and analyze the fraction using SDS-PAGE (Figure 4A) and western blot (Figure 4B).
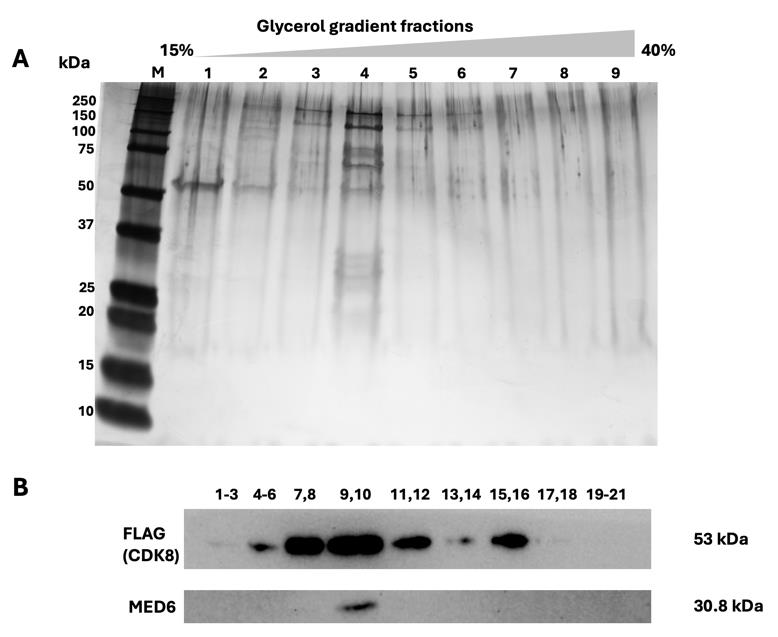
Figure 4. Overview of the glycerol gradient results. The eluted samples obtained by anti-FLAG affinity gel from Cdk8-FLAG-expressed 293-F cells were applied to glycerol gradient sedimentation. Fractions were collected from top to bottom and analyzed using a 4%–20% SDS-PAGE gel for silver stained (A) and immunoblot (B). FLAG and MED6 antibodies were used. M: Marker; 1. Fraction 1–3; 2. Fraction 4–6; 3. Fraction 7–8; 4. Fraction 9–10; 5. Fraction 11–12; 6. Fraction 13–14; 7. Fraction 15–16; 8. Fraction 17–18; 9. Fraction 19–21.
Validation of protocol
This protocol has been used in the following research article:
• Chao et al. [7]. Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module. Mol Cell. https://doi.org/10.1016/j.molcel.2024.09.001 (Figure 1B, Figure S1.B). The cryo-EM structure of cMED-CKM complex were obtained followed the purification steps from this protocol.
General notes and troubleshooting
General notes
1. In this protocol, we use 293-F cells to overexpress the CDK8-F as an example to isolate the mediator complex. It also applies to other mammalian expression strains (e.g., HEK293 cells).
2. The protocol is adaptable for expression at any scale. From 1 L of 293-F cell culture, approximately 10 g of pellet can be expected.
3. Recommended working volume for the flasks: 600 mL of cells for a 2 L shake flask and 50 mL of cells for a 250 mL flask.
Troubleshooting
A. Low transfection efficiency
1. Cell viability maintained at >90% is recommended. Use fresh cell stocks or early-passage cells.
2. Check the quality of the plasmid DNA. (Prepare the plasmid DNA with low endotoxin and contamination.)
3. Vary the amount of DNA or transfection reagent. Try 3–6 μg of plasmid DNA or 3.75–7.5 μL of lipofectamine reagent.
4. Incubation time of the mixture should be between 10 and 20 min. Longer times can lead to loss of transfection efficiency.
B. Low protein yield
1. Check the protein expression using western blot for each passage. Prolonged culture (>30 passages) may reduce the protein yield. Thaw a new batch if necessary.
2. Check the cell lysis efficiency using SDS-PAGE/western blot analysis. Increase the strokes when lysing the cells if necessary.
3. Increase the incubation time for the cell lysate with ANTI-FLAG® M2 affinity gel.
4. Verify the concentration of the FLAG peptide in the elution buffer to reach a successful protein elution.
C. Glycerol gradient shift
1. Verify the glycerol concentration using a refractometer.
2. Carefully load the gradient buffer without disturbance or air bubbles formation.
3. Ensure that a clear interface forms between the two layers before placing the tubes in the tube holder. Hold the tube steady and do not disturb the interface.
4. After adding the FLAG elution on top of the gradient, confirm a distinct interface between the FLAG elution and the gradient.
D. Poor protein purity
1. Increase the extra wash steps (e.g., 5 washes) to reduce non-specific binding if necessary.
2. Keep the samples on ice to avoid protein aggregation or degradation.
Acknowledgments
The protocol presented here is adapted from Chao et al. [7]. This work was supported by US National Institutes of Health grant R01 GM143587 (K.-L.T.).
Competing interests
The authors declare no competing interests.
References
- Verger, A., Monte, D. and Villeret, V. (2019). Twenty years of Mediator complex structural studies. Biochem Soc Trans. 47(1): 399–410.
- Tsai, K. L., Tomomori-Sato, C., Sato, S., Conaway, R. C., Conaway, J. W. and Asturias, F. J. (2014). Subunit Architecture and Functional Modular Rearrangements of the Transcriptional Mediator Complex. Cell. 158(2): 463.
- Soutourina, J. (2018). Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol. 19(4): 262–274.
- Tsai, K. L., Sato, S., Tomomori-Sato, C., Conaway, R. C., Conaway, J. W. and Asturias, F. J. (2013). A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol 20(5): 611–619.
- Elmlund, H., Baraznenok, V., Lindahl, M., Samuelsen, C. O., Koeck, P. J., Holmberg, S., Hebert, H. and Gustafsson, C. M. (2006). The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA. 103(43): 15788–15793.
- Clark, A. D., Oldenbroek, M. and Boyer, T. G. (2015). Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol. 50(5): 393–426.
- Chao, T. C., Chen, S. F., Kim, H. J., Tang, H. C., Tseng, H. C., Xu, A., Palao, L., Khadka, S., Li, T., Huang, M. F., et al. (2024). Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module. Mol Cell. 84(20): 3932–3949.e10.
Article Information
Publication history
Received: Oct 3, 2024
Accepted: Dec 3, 2024
Available online: Jan 1, 2025
Published: Feb 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Tang, H. C., Tsai, K. L. and Chao, T. C. (2025). A Protocol to Purify Human Mediator Complex From Freestyle 293-F Cells. Bio-protocol 15(4): e5185. DOI: 10.21769/BioProtoc.5185.
Category
Biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link