- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Amylin-β-amyloid Hetero-Oligomers by Enzyme-Linked Immunosorbent Assay
Published: Vol 15, Iss 3, Feb 5, 2025 DOI: 10.21769/BioProtoc.5179 Views: 1415
Reviewed by: Olga KopachRupkatha BanerjeeAmberley D. StephensAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
A Step-By-Step Protocol for Correlative Light and Electron Microscopy Imaging of Proteinaceous Deposits in Cultured Cells and Human Brain Tissues
Peizhou Jiang and Dennis W. Dickson
Aug 5, 2025 2335 Views
Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1204 Views
Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1506 Views
Abstract
Amylin is an amyloidogenic neuroendocrine hormone co-synthesized and co-secreted with insulin from the pancreas. It readily crosses the blood–brain barrier and synergistically forms mixed amyloid plaques with β-amyloid (Aβ) in brain parenchyma. Parenchymal amylin-Aβ plaques are found in both sporadic and early-onset familial Alzheimer’s disease (AD), yet their (patho)physiological role remains elusive, particularly due to a lack of detection modalities for these mixed plaques. Previously, we developed an enzyme-linked immunosorbent assay (ELISA) capable of detecting amylin-Aβ hetero-oligomers in brain lysate and blood using a polyclonal anti-amylin antibody to capture hetero-oligomers and a monoclonal anti-Aβ mid-domain detection antibody combination. This combination allows for the recognition of distinct amylin epitopes, which remain accessible after amylin-Aβ oligomerization has begun, and precise detection of Aβ epitopes available after oligomer formation. The utility of this assay is evidenced in our previous report, wherein differences in hetero-oligomer content in brain tissue from patients with and without AD and patients with and without diabetes were distinguished. Additionally, using AD model rats, we provided evidence that our assay can be employed for the detection of amylin-Aβ in blood. This assay and protocol are important innovations in the field of AD research because they meet an unmet need to detect mixed amyloid plaques that, if targeted therapeutically, could reduce AD progression and severity.
Key features
• Detects amylin-Aβ hetero-oligomers in blood from patients with Alzheimer’s disease.
• Enables simultaneous, high-throughput analysis of hetero-oligomer content of brain and blood tissue.
• Allows exploration into the amylin-Aβ interaction during AD pathogenesis, potentially leading to novel treatment mechanisms by controlling the amylin-Aβ interaction.
Keywords: AmylinGraphical overview
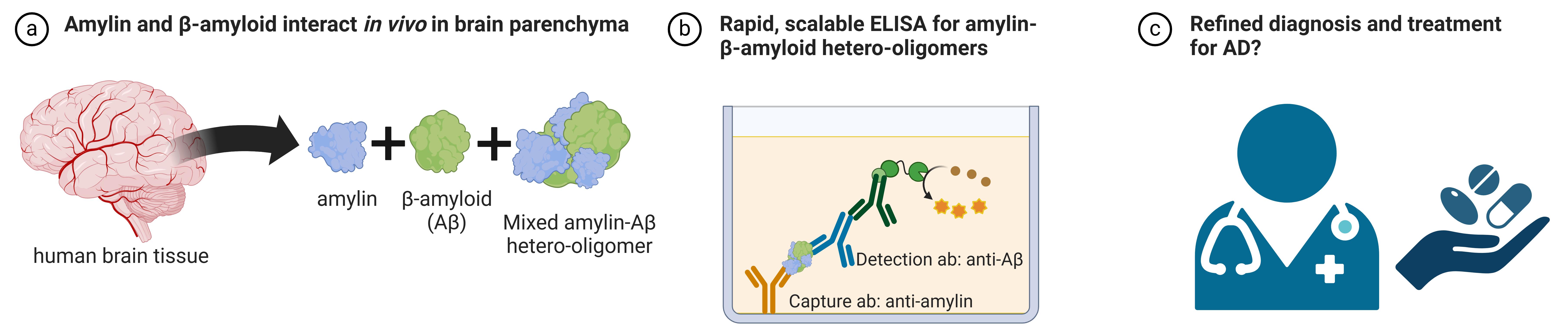
Detection of amylin-Aβ hetero-oligomers by ELISA. (A) Amyloidogenic amylin and Aβ form mixed hetero-oligomers in brain tissue. (B) The ELISA described in this protocol is capable of detecting these mixed hetero-oligomers. (C) In the future, perhaps the detection of these mixed hetero-oligomers can assist in refining diagnostic measures and treatments for Alzheimer’s disease.
Background
Alzheimer's disease (AD) has previously been linked to dysregulated pancreatic hormones, including insulin, which can exacerbate AD pathology by causing brain insulin resistance [1–2]. Amylin, a centrally acting neuroendocrine hormone co-synthesized and co-secreted with insulin from pancreatic β-cells [3], readily crosses from the blood to the brain parenchyma [4] and is reportedly involved in the central regulation of satiation [5]. In nearly all (>95% prevalence) patients with type-2 diabetes mellitus (T2DM), amylin forms cytotoxic amyloid plaques in the pancreas, which contributes to the pathogenesis of T2DM by inducing β-cell apoptosis and subsequent β-cell mass depletion [6–8]. In both sporadic and early-onset familial AD, amylin synergistically forms co-aggregates with vascular and brain parenchymal Aβ in the brain [9–14]. Consistent with findings in human AD brains, APPswe/PS1dE9 (APP/PS1) rats expressing human amylin in the pancreas (since murine amylin is non-amyloidogenic) revealed that chronic exposure to circulating amylin promoted cerebrovascular and parenchymal amylin-Aβ deposition [9–13]. These results suggest an association between Aβ pathology and amyloidogenic human amylin but the exact mechanism responsible for amylin-Aβ co-aggregation remains to be determined.
Previous research demonstrated that heterologous seeding between amylin and common Aβ fragments (e.g., Aβ42 and Aβ40) promotes amyloid formation in a manner that is comparable to that of homogenous amylin amyloid [15–17]. In vivo amylin-Aβ hetero-amyloid formation and cytotoxicity are facilitated by the co-expression of the two peptides in cells [21–22]. The hypothesis that amylin and Aβ peptides aggregate in the human AD brain to form amylin-Aβ hetero-oligomers has previously been verified by co-immunoprecipitation experiments using human brain samples from AD patients [9]. These converging data [9,15–22] support the presence of amylin-Aβ hetero-oligomers in the human brain and establish a need for their quantification. Proper detection and quantification of amylin-Aβ hetero-oligomers may further establish the connection between amylin and Aβ and their collective role in AD pathology.
While the need for a means to reliably quantify the molecular amylin-Aβ interaction in AD has been acknowledged [9–13], currently employed molecular techniques (namely immunoprecipitation, western blot, circular dichroism, and electron microscopy) limit studies to reduced sample sizes due to their inherent complexity, non-scalability, and in some cases, cost. Additionally, the denaturing conditions commonly used in methods such as western blot prove to be additional obstacles, as the size and structural stability of hetero-oligomers can be altered. Therefore, there is still an unmet need for an assay method that can detect and quantify hetero-oligomers of amylin-Aβ without interrupting molecular interactions. Previously, we reported the creation of a novel enzyme-linked immunosorbent assay (ELISA) to detect amylin-Aβ hetero-oligomers in blood and brain tissue [23] in AD model animals and human donor tissue. Here, we provide a detailed, stepwise protocol for our assay. Relative to the currently employed techniques, this assay is scalable, high-throughput, cost-effective, and non-complex, all while critically maintaining hetero-oligomer stability. The assay detailed here may prove to be a useful tool to detect, analyze, and eventually disentangle the role of the amyloidogenic amylin and Aβ proteins in AD and other pathological conditions.
Materials and reagents
Biological materials
1. Mouse anti-human-Aβ(1-16) antibody (BioLegend, catalog number: 803002)
2. Rabbit anti-amylin P2 antibody (not commercially available, see [23])
3. Mouse anti-human-total-Aβ (BioLegend, catalog number: 800720)
4. Aβ40 synthetic peptide (Anaspec, catalog number: AS-24235)
Reagents
1. ELISA assay diluent (5×) (BioLegend, catalog number: 421203)
2. Dimethyl sulfoxide (DMSO) (Sigma, catalog number: D8418)
3. Na2CO3 (Sigma, catalog number: S7795)
4. NaHCO3 (Sigma, catalog number: S5761)
5. 10× PBS (Corning, catalog number: 46-013-CM)
6. 10% Tween-20 (Bio-Rad, catalog number: 1610781)
7. Aβ40 synthetic peptide (Anaspec, catalog number: AS-24235)
8. 3,3',5,5' tetramethylbenzidine (TMB) substrate (Thermo Scientific, catalog number: 34028)
9. Stop solution (Thermo Scientific, catalog number: N600)
Solutions
1. Bicarbonate buffer (see Recipes)
2. PBST washing buffer (see Recipes)
Recipes
1. Bicarbonate buffer (250 mL)
Note: Adjust the solution’s pH to 9.6. Use distilled water as diluent.
| Reagent | Final concentration | Mass |
|---|---|---|
| Na2CO3 | 0.028 M | 0.7575 g |
| NaHCO3 | 0.071 M | 1.50 g |
2. PBST washing buffer (500 mL)
Note: Dilute 50 mL of 10× PBS to 1× using 450 mL of distilled water as diluent before preparing PBST washing buffer.
| Reagent | Final concentration | Volume |
|---|---|---|
| 1× PBS | - | 500 mL |
| 10% Tween-20 | 0.05% | 2.5 mL |
Laboratory supplies
1. 96-well clear flat-bottom polystyrene high bind microplate (Corning, catalog number: 9018)
2. Sealing film, polyester (VWR, catalog number: 89134-430)
3. Absorbent paper towels (Pacific Blue Basic, catalog number: 23504)
4. epT.I.P.S. standard 20–300 μL pipette tips (Eppendorf, catalog number: 022492047)
5. 1.5 mL standard line microcentrifuge tubes (VWR, catalog number:10025-726)
6. 50 mL reagent reservoirs (Corning, catalog number: 4870)
7. 8-channel 30–300 μL multichannel pipette (USA Scientific, catalog number: 7108-3300)
Equipment
1. xMark microplate absorbance spectrophotometer (Bio-Rad, catalog number: 1681150)
Software and datasets
1. Microsoft Excel (2019)
2. Microplate Manager Software 6 (MPM) (Bio-Rad, v6.3)
Procedure
This protocol has been validated using frozen and freshly prepared samples. The homogenization buffers and protocol used previously are described in [23]. Preparation of samples in other buffers or using other homogenization methods may be possible so long as the conditions do not disrupt amylin-Aβ binding interactions.
Unless otherwise noted, bring all reagents to room temperature prior to use. Determine ELISA plate layout/template prior to use; we recommend using the first two columns (16 wells) for the standard curve (8 standards in duplicate). Dilute 5× assay diluent to 1× final concentration using 1× PBS prior to use. A multichannel pipette is used for the addition of all solutions, except the standards/controls, which are added individually. A graphical summary of the protocol is shown in Figure 1.
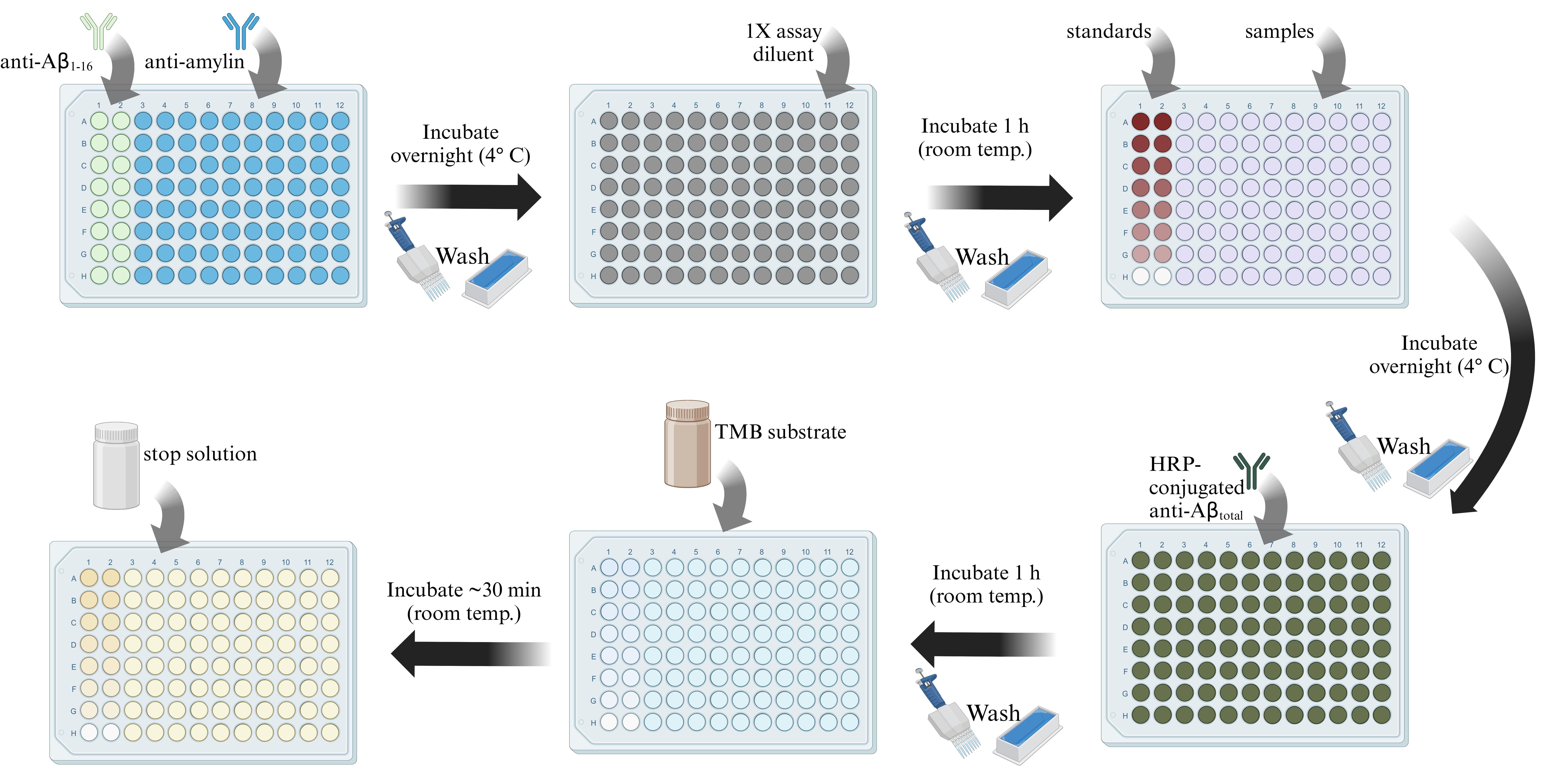
Figure 1. Graphical summary of ELISA protocol. General overview of the present ELISA protocol, highlighting key steps. Note that after the addition of the stop solution, the plate must be read in a spectrophotometer, which is not depicted in this summary figure. Data analysis steps are not shown.
A. Coating with capture antibodies
1. Dilute the mouse anti-human-Aβ(1-16) antibody to a final dilution of 1:400 using bicarbonate buffer and coat the wells of the 96-well plate to be used for the standards with 100 μL.
2. Dilute the rabbit anti-amylin P2 antibody to a final dilution of 1:400 using bicarbonate buffer and coat all sample wells of the plate with 100 μL.
3. Seal the plate with sealing film and incubate overnight at 4 °C without shaking.
B. Loading standards and samples
1. Prepare the standards.
a. Resuspend the Aβ40 peptide in a sufficient amount of DMSO to prepare the 1 ng/mL stock solution.
b. Serial dilute the stock in 1× assay diluent to create the following concentrations (in ng/mL): 10, 5, 2.5, 1.25, 0.625, 0.3125, 0.1562. Use assay diluent buffer as a blank (“0”) standard.
2. Remove the plate sealer and decant antibody solutions by gently shaking the plate (upside down) over a sink. Remove any residual solution by gently tapping the plate on an absorbent paper towel.
a. When tapping the plate, tap hard enough that any residual solution is removed from the wells and the wells are dry.
b. Repeat this method of washing/tapping for all following solution decanting steps.
3. Wash the plate with 300 μL of PBST washing buffer per well. Remove any remaining wash buffer by tapping the plate on an absorbent paper towel, as above. An automatic plate washer may also be used if available.
4. Add 300 μL of 1× assay diluent to each well and incubate for 1 h at room temperature.
5. Decant blocking solution and remove any residual solution as described above.
6. Wash the plate with 300 μL of PBST washing buffer per well 1–2 times. Decant and tap after each wash, as described above.
7. Add 100 μL of the Aβ40 peptide standard to the appropriate wells (coated with the anti-human-Aβ(1-16) antibody only). Include 100 μL of 1× assay diluent as the “0” standard/blank, as described above.
8. Dilute samples to 1:2 final dilution using 1× PBS for a final volume of 200 μL (100 μL per well, in duplicate). Plate 100 μL of diluted sample to the appropriate wells (coated with the anti-amylin P2 antibody).
9. Add 100 μL of 1× assay diluent as a blank for the samples (for wells originally coated with the P2 antibody).
10. Seal the plate and incubate overnight at 4 °C.
C. Developing the assay and data collection
1. Prepare the HRP-conjugated anti-human-total-Aβ detection antibody by diluting to a final dilution of 1:400 in 1× assay diluent.
2. Remove the plate sealer, decant the standards and samples, and remove any remaining solution, as above.
3. Wash the plate with 300 μL of PBST washing buffer per well 3 times. Decant and tap after each wash, as above.
4. Add 100 μL of prepared detection antibody per well and incubate at room temperature for 1 h.
a. It is recommended to remove the TMB from 4 °C at this time, to allow it time to acclimate to room temperature before use.
5. Decant the antibody and remove the residual solution as above.
6. Wash the plate with 300 μL of PBST washing buffer 3–4 times with decanting and tapping.
7. Add 100 μL of TMB substrate (at room temperature) per well. Incubate for approximately 30 min until standard wells show gradation and sample wells show a signal. Check the plate after 20 min of incubation and then every few minutes to prevent oversaturation of signal intensity.
8. Add 50 μL of stop solution per well in the exact same order in which the substrate was added.
9. Read the plate at 450 nm in a spectrophotometer.
Data analysis
Following data acquisition using MPM6 software, data are exported to Excel for further analysis. In each experiment, samples should be run in duplicate or triplicate for technical replicates. Average the values for all duplicate/triplicate wells. Subtract the average absorbance value of the standard blank wells (the wells with only 1× assay diluent coated with anti-human-Aβ(1-16) antibody in step A1) from all standard wells. Subtract the average absorbance value of the sample blank (the wells with only 1× assay diluent coated with anti-amylin P2 antibody in step A1) from all sample wells to remove background and non-specific signal. Create a standard curve using the absorbance values vs. standard concentrations and generate the linear regression equation. The concentration of samples can then be interpolated from the standard curve regression line. A detailed description of the data analysis method appears in our previous report [23] in the “Experimental procedures” section. An overview of the data analysis method is shown in Figure 2.
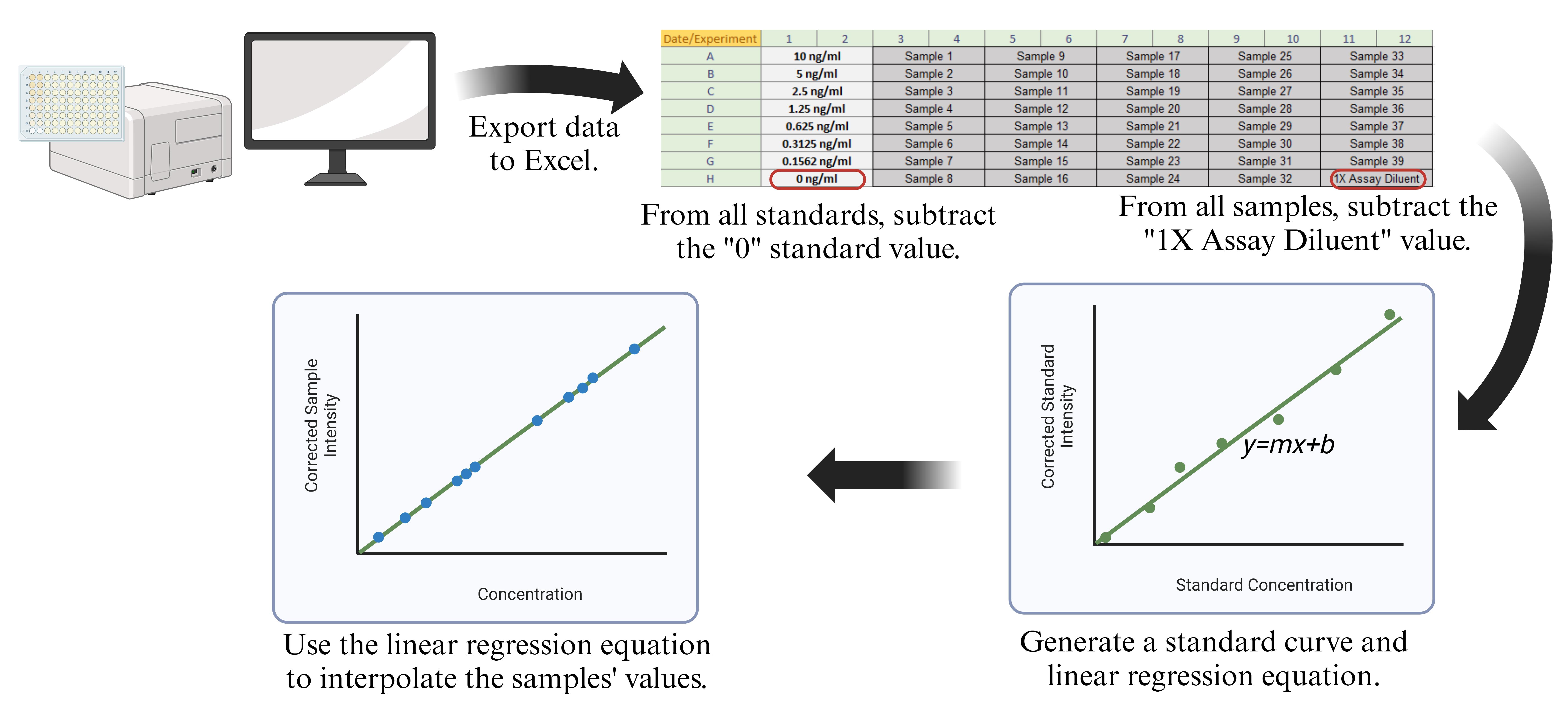
Figure 2. Data analysis overview. After exporting the values to Excel, values for duplicate wells are averaged together. For all standard values, the average of the “0” standard is subtracted. For all samples, the “1× assay diluent” average is subtracted. Then, the standard values are used to create a standard curve and generate a linear regression equation, which can be used to calculate sample values.
Validation of protocol
This protocol has been used and validated in the following research article:
• Kotiya et al. [23]. Rapid, scalable assay of amylin-β amyloid co-aggregation in brain tissue and blood. Journal of Biological Chemistry (Figure 1, panels E–F; Figure 2 panels A–E; Figure 3, panels B–E; Figure 4 panels A–C; Figure 6 panels A–E).
General notes and troubleshooting
General notes
1. For convenience, we have prepared an example plate layout (Table S1) showing standards and samples plated in duplicate. We have also included a blank plate layout, which can be filled out with sample identifiers (Table S2).
2. The inherent instability and amyloidogenicity of both amylin and β-amyloid can cause variability between measurements. We strongly recommend that all samples that are to be compared against one another be run on the same assay plate. If necessary, samples can be run as singlets in two separate plates, and the values averaged together.
3. Following the acquisition of concentrations of samples (in ng/mL), we recommend normalizing each value to the respective sample’s total protein concentration to account for individual differences and variances that occur as a result of sample preparation. We recommend reporting final values as “ng amylin/mg total protein” (or similar).
4. Though not required, we recommend wrapping the plate with aluminum foil to prevent light interference and smudging/fingerprinting on the plate’s clear bottom, which would interfere with absorbance readings.
5. Placing the plate on a plate shaker during any incubation step is not recommended; conduct all incubation and washing steps without shaking.
6. When adding solutions, we recommend always adding solutions (wash buffer, TMB, stop solution, etc.) from top to bottom, ensuring duplicate/triplicates receive each solution simultaneously.
7. We recommend initially diluting samples two-fold with 1× PBS, as described in the protocol above. Samples may need to be run with higher (if the signal is too strong) or lower (if the signal is too weak) dilution factors according to amylin-Aβ concentration in the prepared sample.
8. Although Aβ40 does not form amyloid by itself rapidly [24], we recommend aliquoting Aβ40 stock to prevent frequent freeze/thaw cycles, which can impact aggregation formation.
Troubleshooting
Problem 1: Oversaturation of standards or samples.
Possible cause: Insufficient number of washing and/or decanting steps.
Solution: Modify the number of washes during sections B and C and ensure proper decanting to remove excess/residual solution from each well before adding the next solution.
Problem 2: No (or little) gradation between standards.
Possible cause: Multiple freeze/thaw cycles of Aβ standards, incorrect preparation, or dilution.
Solution: Minimize the number of freeze/thaw cycles of the Aβ standards by aliquoting stock peptide. Only thaw the peptide standard when prepared to run the assay. Pipette the standards carefully and ensure no bubbles are present when loading the standards in the wells. If necessary, run repeat experiments with the standard curve plated in triplicate.
Supplementary information
The following supporting information can be downloaded here:
1. Table S1. Example plate layout
2. Table S2. Blank plate layout
Acknowledgments
F.D. acknowledges the following funding sources: National Institutes of Health R01 NS116058, R01 AG057290, R01 AG053999.
This protocol was adapted and modified from Kotiya et al. [23].
Competing interests
The authors declare no competing interests.
References
- Kellar, D. and Craft, S. (2020). Brain insulin resistance in Alzheimer's disease and related disorders: mechanisms and therapeutic approaches. Lancet Neurol. 19(9): 758–766.
- Biessels, G. J. and Despa, F. (2018). Cognitive decline and dementia in diabetes mellitus: mechanisms and clinical implications. Nat Rev Endocrinol. 14(10): 591–604.
- Kahn, S. E., D'Alessio, D. A., Schwartz, M. W., Fujimoto, W. Y., Ensinck, J. W., Taborsky, G. J. and Porte, D. (1990). Evidence of Cosecretion of Islet Amyloid Polypeptide and Insulin by β-Cells. Diabetes. 39(5): 634–638.
- Banks, W. A. and Kastin, A. J. (1998). Differential Permeability of the Blood–Brain Barrier to Two Pancreatic Peptides: Insulin and Amylin. Peptides. 19(5): 883–889.
- Hay, D. L., Chen, S., Lutz, T. A., Parkes, D. G. and Roth, J. D. (2015). Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Res. 67(3): 564–600.
- Westermark, P., Andersson, A. and Westermark, G. T. (2011). Islet Amyloid Polypeptide, Islet Amyloid, and Diabetes Mellitus. Physiol Rev. 91(3): 795–826.
- Jurgens, C. A., Toukatly, M. N., Fligner, C. L., Udayasankar, J., Subramanian, S. L., Zraika, S., Aston-Mourney, K., Carr, D. B., Westermark, P., Westermark, G. T., et al. (2011). β-Cell Loss and β-Cell Apoptosis in Human Type 2 Diabetes Are Related to Islet Amyloid Deposition. Am J Pathol. 178(6): 2632–2640.
- Höppener, J. W., Ahrén, B. and Lips, C. J. (2000). Islet Amyloid and Type 2 Diabetes Mellitus. N Engl J Med. 343(6): 411–419.
- Jackson, K., Barisone, G. A., Diaz, E., Jin, L. w., DeCarli, C. and Despa, F. (2013). Amylin deposition in the brain: A second amyloid in Alzheimer disease? Ann Neurol. 74(4): 517–526.
- Oskarsson, M. E., Paulsson, J. F., Schultz, S. W., Ingelsson, M., Westermark, P. and Westermark, G. T. (2015). In Vivo Seeding and Cross-Seeding of Localized Amyloidosis. Am J Pathol. 185(3): 834–846.
- Martinez‐Valbuena, I., Valenti‐Azcarate, R., Amat‐Villegas, I., Riverol, M., Marcilla, I., de Andrea, C. E., Sánchez‐Arias, J. A., del Mar Carmona‐Abellan, M., Marti, G., Erro, M., et al. (2019). Amylin as a potential link between type 2 diabetes and alzheimer disease. Ann Neurol. 86(4): 539–551.
- Ly, H., Verma, N., Sharma, S., Kotiya, D., Despa, S., Abner, E. L., Nelson, P. T., Jicha, G. A., Wilcock, D. M., Goldstein, L. B., et al. (2021). The association of circulating amylin with β‐amyloid in familial Alzheimer's disease. Alzheimer’s Dement. 7(1): e12130.
- Verma, N., Velmurugan, G. V., Winford, E., Coburn, H., Kotiya, D., Leibold, N., Radulescu, L., Despa, S., Chen, K. C., Van Eldik, L. J., et al. (2023). Aβ efflux impairment and inflammation linked to cerebrovascular accumulation of amyloid-forming amylin secreted from pancreas. Commun Biol. 6(1): 2.
- Leibold, N. S. and Despa, F. (2024). Neuroinflammation induced by amyloid-forming pancreatic amylin: Rationale for a mechanistic hypothesis. Biophys Chem. 310: 107252.
- O'Nuallain, B., Williams, A. D., Westermark, P. and Wetzel, R. (2004). Seeding Specificity in Amyloid Growth Induced by Heterologous Fibrils. J Biol Chem. 279(17): 17490–17499.
- Yan, L., Velkova, A., Tatarek‐Nossol, M., Andreetto, E. and Kapurniotu, A. (2007). IAPP Mimic Blocks Aβ Cytotoxic Self‐Assembly: Cross‐Suppression of Amyloid Toxicity of Aβ and IAPP Suggests a Molecular Link between Alzheimer's Disease and Type II Diabetes. Angew Chem Int Ed. 46(8): 1246–1252.
- Andreetto, E., Yan, L., Tatarek‐Nossol, M., Velkova, A., Frank, R. and Kapurniotu, A. (2010). Identification of Hot Regions of the Aβ–IAPP Interaction Interface as High‐Affinity Binding Sites in both Cross‐ and Self‐Association. Angew Chem Int Ed. 49(17): 3081–3085.
- Yan, L. M., Velkova, A. and Kapurniotu, A. (2014). Molecular Characterization of the Hetero-Assembly of beta-Amyloid Peptide with Islet Amyloid Polypeptide. Curr Pharm Des. 20(8): 1182–1191.
- Kapurniotu, A. (2020). Enlightening amyloid fibrils linked to type 2 diabetes and cross-interactions with Aβ. Nat Struct Mol Biol. 27(11): 1006–1008.
- Taş, K., Volta, B. D., Lindner, C., El Bounkari, O., Hille, K., Tian, Y., Puig-Bosch, X., Ballmann, M., Hornung, S., Ortner, M., et al. (2022). Designed peptides as nanomolar cross-amyloid inhibitors acting via supramolecular nanofiber co-assembly. Nat Commun. 13(1): 5004.
- Bharadwaj, P., Solomon, T., Sahoo, B. R., Ignasiak, K., Gaskin, S., Rowles, J., Verdile, G., Howard, M. J., Bond, C. S., Ramamoorthy, A., et al. (2020). Amylin and beta amyloid proteins interact to form amorphous heterocomplexes with enhanced toxicity in neuronal cells. Sci Rep. 10(1): 10356.
- Wang, Y. and Westermark, G. T. (2021). The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level. Int J Mol Sci. 22(20): 11153.
- Kotiya, D., Leibold, N., Verma, N., Jicha, G. A., Goldstein, L. B. and Despa, F. (2023). Rapid, scalable assay of amylin-β amyloid co-aggregation in brain tissue and blood. J Biol Chem. 299(5): 104682.
- Jarrett, J. T., Berger, E. P. and Lansbury, P. T. (1993). The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer's disease. Biochemistry. 32(18): 4693–4697.
Article Information
Publication history
Received: Aug 8, 2024
Accepted: Dec 5, 2024
Available online: Dec 26, 2024
Published: Feb 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Leibold, N. S., Kotiya, D., Verma, N. and Despa, F. (2025). Detection of Amylin-β-amyloid Hetero-Oligomers by Enzyme-Linked Immunosorbent Assay. Bio-protocol 15(3): e5179. DOI: 10.21769/BioProtoc.5179.
- Kotiya, D., Leibold, N., Verma, N., Jicha, G. A., Goldstein, L. B. and Despa, F. (2023). Rapid, scalable assay of amylin-β amyloid co-aggregation in brain tissue and blood. J Biol Chem. 299(5): 104682.
Category
Neuroscience > Nervous system disorders > Neurodegeneration
Neuroscience > Basic technology > High-throughput screening
Biochemistry > Protein > Immunodetection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








