- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Application of a Dual Optogenetic Silencing-Activation Protocol to Map Motor Neurons Driving Rolling Escape Behavior in Drosophila Larvae
Published: Vol 14, Iss 23, Dec 5, 2024 DOI: 10.21769/BioProtoc.5131 Views: 1504
Reviewed by: Ivan Sanchez DiazRupkatha BanerjeeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Evaluating Baseline and Sensitised Heat Nociception in Adult Drosophila
Josephine N. Massingham [...] G. Gregory Neely
Jul 5, 2021 3248 Views
Training Mice to Perform Attentional Set-Shifting Under Head Restraint
Katarina Kalajzic [...] Timothy Spellman
Sep 5, 2025 1366 Views
Mouse Vestibulo-Ocular Reflex Testing for Otolith Organs and Horizontal Semicircular Canal
Tong Zhao [...] Fangyi Chen
Nov 20, 2025 1067 Views
Abstract
Drosophila larvae exhibit rolling motor behavior as an escape response to avoid predators and painful stimuli. We introduce an accessible method for applying optogenetics to study the motor circuits driving rolling behavior. For this, we simultaneously implement the Gal4-UAS and LexA-Aop binary systems to express two distinct optogenetic channels, GtACR and Chrimson, in motor neuron (MN) subsets and rolling command neurons (Goro), respectively. Upon exposure to white LED light, Chrimson permits the influx of positive ions into Goro neurons, leading to depolarization, whereas GtACR mediates chloride influx into MNs, resulting in hyperpolarization. This method allows researchers to selectively activate certain neurons while simultaneously inhibiting others within a circuit of interest, offering a unique advantage over current optogenetic approaches, which often utilize a single type of optogenetic actuator. Here, we provide a detailed protocol for the dual silencing-activation approach using GtACR and Chrimson optogenetic channels and present a robust methodological framework for investigating the neuromuscular basis of rolling in larvae. Our cost-effective and scalable approach utilizes readily accessible equipment and can be applied to study other locomotor behaviors in Drosophila larvae, thereby enhancing our understanding of the neural circuit mechanisms underlying sensorimotor transformation.
Key features
• Enables real-time manipulation of neural activity, providing insights into the immediate effects of neuronal activation and silencing on larval behavior.
• The protocol is adaptable to different experimental setups, allowing researchers to extend its application to other sensory modalities or behavioral assays.
• Offers a standardized approach to studying nociceptive behaviors.
Keywords: DrosophilaGraphical overview
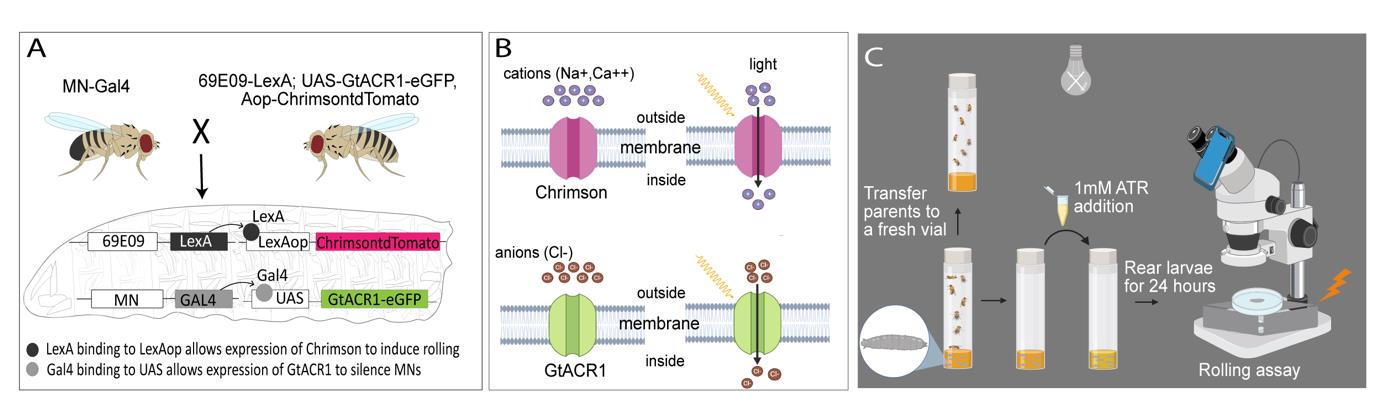
Experimental setup and procedure for the rolling assay. A) 69E06-LexA; Aop-Chrimson::tdTomato, UAS- GtACR1-eGFP female virgins are crossed with MN-Gal4 male flies, resulting in larval offspring carrying all four components essential for concurrent silencing and activation of neurons of interest. In these larvae, GtACR1-eGFP is selectively expressed in the motor neurons (MNs) of interest, whereas Aop-Chrimson::tdTomato is only expressed in Goro command neurons. B) Upon exposure to white LED light, Chrimson causes an influx of cations (Ca2+ and Na+) into Goro neurons, resulting in depolarization (activation) of these command neurons. In contrast, when exposed to white LED light, GtACR1 triggers an influx of chloride anions (Cl-), thereby hyperpolarizing (silencing) the MNs of interest. C) Both GtACR1 and Chrimson are sensitive to ambient light; therefore, the larvae should be raised in the dark until they are ready for experimentation.
Background
Noxious stimuli instinctively trigger escape behaviors in animals, prompting rapid motor responses to avoid harm. Understanding the neural mechanisms governing escape behaviors is essential for elucidating how animals respond to environmental threats. When subjected to harmful mechanical stimuli or heat, Drosophila larvae execute nocifensive escape behavior characterized by bending into a C-shape, followed by rolling and rapid forward crawling [1]. This behavior is triggered by the activation of class IV (cIV) dendritic arborization sensory neurons, which are polymodal nociceptor sensory neurons distributed throughout the body wall [2,3]. Rolling can also be induced optogenetically by stimulating nociceptive sensory neurons or interneurons within the central nervous system, including the Goro command neuron [4–6].
The body muscles of Drosophila larvae are co-innervated by type Is (small boutons, phasic firing) and a single type Ib motor neuron (MN) (big boutons, tonic firing). Type Ib MNs typically innervate one muscle, whereas Type Is MNs innervate multiple target muscles [7,8]. Here, we present a detailed protocol describing how we used a dual-optogenetic method to identify muscles and MN types essential for larval rolling escape behavior [9]. Current approaches to studying neural circuits often involve either optogenetic activation or inhibition of neural targets, limiting our ability to take full advantage of light-activated ion channels to explore circuits of interest. This protocol leverages the LexA-Aop system to express Chrimson, a cationic channelrhodopsin, in Goro neurons (69E06-LexA) to induce the larval rolling escape behavior. Concurrently, we utilize MN-Gal4 driven GtACR1, a light-sensitive chloride channel activated by the same white light as Chrimson, to acutely inhibit specific subsets of MN subsets. This dual optogenetic strategy enables the targeted and acute silencing of different MNs using different MN-Gal4 lines during the same temporal window of Goro activation. This setup, including a system to rapidly control light exposure, ensures consistent behavioral responses, thus enhancing the reproducibility and precision of behavioral studies in larvae. By implementing MN silencing in conjunction with rolling assays, we recently elucidated the neuromuscular basis of larval escape behavior in Drosophila [9].
This approach can be extended to investigate multifactorial motor neuron diseases, such as amyotrophic lateral sclerosis (ALS), which results from a combination of genetic and environmental factors that remain incompletely understood. The neural activation approach (69E06-LexA; LexA-Aop-Chrimson) can be integrated with reverse genetic screens to identify genes critical for the maintenance of motor circuit health and function, thereby elucidating novel genetic defects underlying motor neuron diseases, such as ALS. Optogenetic activation of the GORO neuron induces continuous rolling behavior in Drosophila larvae, which potentially leads to more rapid larval exhaustion than default forward crawling. The exhausted larvae may provide a more sensitive background to identify genes essential for neural function and maintenance compared to non-threatened larvae that perform forward crawling and have the opportunity to take brief pauses between crawling bouts to recover from subtle defects in motor circuit malfunction. To conduct these reverse genetic experiments, researchers can substitute UAS-GtACR1 with the UAS-RNAi line to knock down the candidate gene in the neurons of interest. Alternatively, 69E06-LexA; LexA-Aop-Chrimson activation could be employed in larvae carrying null mutant alleles for any candidate gene.
Materials and reagents
Biological materials (Drosophila stocks)
Commercially available Drosophila stocks
Type Is MN driver: w;;27E09-Gal4 (Bloomington Drosophila Stock Center, catalog number: 49227)
Type Ib MNs of ventral longitudinal (VL) and dorsal oblique (DO) muscles driver: w; exex-Gal4 (HB9-Gal4) (Bloomington Drosophila Stock Center, catalog number: 83004)
Lab-generated Drosophila stocks
Goro neuron driver: w; 69E06-LexA; Aop-Chrimson::tdTomato, UAS- GtACR1-eGFP [9]
Type Is and Ib MN1 driver: w; 27E09-Gal4; RRa-Gal4 [9]
Type Ib MNs of lateral transverse (LT) muscles driver: w; BH1-Gal4 [9]
Five type Ib MNs of dorsal longitudinal (DL) muscles driver: w; CQ-Gal4 [9]
Five type Ib MNs of DL muscles driver:: Unc4AD;vGlut DBD split Gal4 line [9]
Five Type Ib and Ib MN1 driver: w; CQ-Gal4; RRa-Gal4 [9]
Type Ib MNs of VL and DO muscles driver: w; vGlutAD;NKx6 DBD split-Gal4 [9]
Reagents
All trans-retinal (ATR) (Sigma-Aldrich, catalog number: R2500)
Agarose (Genesee Scientific, catalog number: 20-102GP)
Yellow cornmeal (Flystuff, catalog number: B078SZR5MS)
Drosophila Agar (Apex, catalog number: B078T16PNG)
Active dry yeast (SAF Instant, catalog number: B0049WLQ30)
Molasses (Oasis Supply, catalog number: B00M1ZYPXA)
Propionic acid (Genesee Scientific, catalog number: 20-271)
Tegosept (Genesee Scientific, catalog number: 20-259)
Ethanol (Sigma-Aldrich, catalog number: 64-17-5)
Solutions
Drosophila media (see Recipes)
10 mM ATR stock solution (see Recipes)
Recipes
Drosophila media
Reagent Quantity or Volume Cornmeal 67 g Drosophila agar 16 g Active dry yeast 27 g Molasses 67 mL Propionic acid 5 mL Tegosept 17 mL Distilled water 1,100 mL Begin by mixing cornmeal, agar, active dry yeast, molasses, and water in a large cooking vessel.
Place the vessel in a medium-to-high-heat setting and bring the fly food mixture to a boil.
Stir the mixture every 5–10 min to ensure even mixing.
Once boiling, reduce the heat slightly and let the mixture simmer gently for 30 min.
After 30 min of simmering, turn off the heat and allow the cooking vessel to cool until it is warm to the touch.
Carefully add the measured quantity of propionic acid and Tegosept to the warm mixture.
Transfer the well-mixed food into the Droso-Filler and pour the fly food into polypropylene vials.
Note: Any standard fly food will be suitable for this experiment; it does not need to be restricted to our recipe.
10 mM ATR stock solution
Reagent Quantity or Volume ATR 1 mg Ethanol 0.35 mL Dissolve 1 mg of ATR in 0.35 mL of ethanol to create a 10 mM stock solution.
Store the stock solution in the dark at -20 °C.
Laboratory suppliesPetri dish 10 cm Ø (e.g., Sigma-Aldrich, catalog number: CLS430167)
Spatula (Genesee Scientific, catalog number: 93-131)
Wash bottle containing distilled water (e.g., Depepe, catalog number: B07DB1HCKP)
Paintbrush (Soucolor, catalog number: B07YDDF26Y)
Wide tip forceps (DR instruments, catalog number: B008RBLO8Q)
Cellulose acetate fly plugs (VWR, catalog number: 89168-886)
Polypropylene vials (VWR, catalog number: 75813-144)
Droso-Filler (Genesee Scientific, catalog number: 59-168)
Equipment
External light source 18 W LED (remote control included) (Oeegoo, catalog number: B086JP5Y91)
Extension cord (Husky, catalog number: HD#342-576)
Microscope lens smartphone adapter (Celticbird, catalog number: B0BXWPL7H4)
Stereomicroscope (ZEISS, model: Stemi-305)
Stereomicroscope (ZEISS, model: Stemi-508)
Smartphone camera operating at 30 frames per second (fps) or more.
Note: Any smartphone brand (iPhone, Samsung, or Google Pixel) offering a high frame rate and excellent optical quality can be used. The smartphone camera lens should be aligned with the eyepiece of the microscope. A resolution of at least 12 megapixels is recommended, along with a wide-aperture lens (f/1.5 to f/1.8) to ensure optimal light capture during imaging. The default camera application on a smartphone may be used for video recording. Alternatively, camera applications such as ProCamera (iOS), Open Camera (Android), or Camera FV-5 (Android) can also be used to capture high-quality images.
Software and datasets
ImageJ (https://imagej.net/)
FFmpeg (https://ffmpeg.org/) (version 2022-08-25-git-9bf9d42d01)
Procedure
Larval preparation
Prepare the larvae by raising parent flies on Drosophila media (see Recipes) in the dark at 25 °C, ensuring optimal growth conditions.
Note: Drosophila media is compatible with ATR administration.
Prepare food supplemented with ATR for the optogenetic experiments.
Final concentration of ATR needs to be 1 mM (see Recipes).
Note: In our experiments, 100 µL of 1 mM ATR was administered to the food vial containing larvae using a pipette.
Transfer the parents to a fresh vial and thoroughly distribute the appropriate volume of ATR within the food provided to the larvae in the original vial in a dimly lit room. For the no-ATR control condition, omit the addition of ATR to the larvae's food.
Allow the larvae to incubate for 24 h in the dark after administering ATR, as it acts as the chromophore essential for light-sensitive proteins like channelrhodopsins.
Note: The ATR can be added as early as after the eggs are laid to ensure the larvae are exposed to ATR for at least 24 h. For L3 larvae, we recommend adding ATR at the early L2 stage. For L1–L2 stages, we recommend adding ATR during late embryonic stages and assay the late L1 to early L2 stages.
Imaging setup
Replace the original 10× eyepiece of the Stemi 305 scope with a microscope phone adapter featuring a 16× built-in eyepiece to attach a phone for image capture (Figure 1). For imaging of L1 stage larvae, utilize a Stemi 508 dissection scope.
Install the smartphone to the phone adapter.
Secure the smartphone vertically to the microscope using a phone adapter that firmly attaches to the microscope to ensure stability and prevent movement during imaging.
Adjust the position of the smartphone so that its camera aligns precisely with the optical output of the microscope.
Utilize the smartphone’s camera settings to fine-tune the zoom and focus, ensuring the image is sharp and the area of interest is centered in the view.
Note: The images used in our experiments were captured using the default camera application of iPhone 14 with a 12 MP camera operating at 30 fps.
Set up the external light for the experiment.
Plug in the Oeegoo 18W square LED ceiling light before the experiment and set it up to ensure that it is a white light at maximum brightness.
Turn off the light using the remote; it will retain the settings when turned on again.
Position the light beneath the imaging microscope, ensuring that it illuminates the agarose pad placed atop it within the Petri dish (Figures 1 and 2).
Note: This assay should be conducted in a room with minimal light exposure to ensure accurate results and to reduce potential interference.

Figure 1. Step-by-step demonstration of the rolling assay setup. The procedure begins with the removal of the microscope lens (2), followed by the installation of an adapter (3) to secure the smartphone. Once the smartphone is placed in position (4) for imaging, an LED light source is installed (5) to provide illumination. Finally, the setup is completed with the placement of an agarose plate (6), ready for the rolling assay.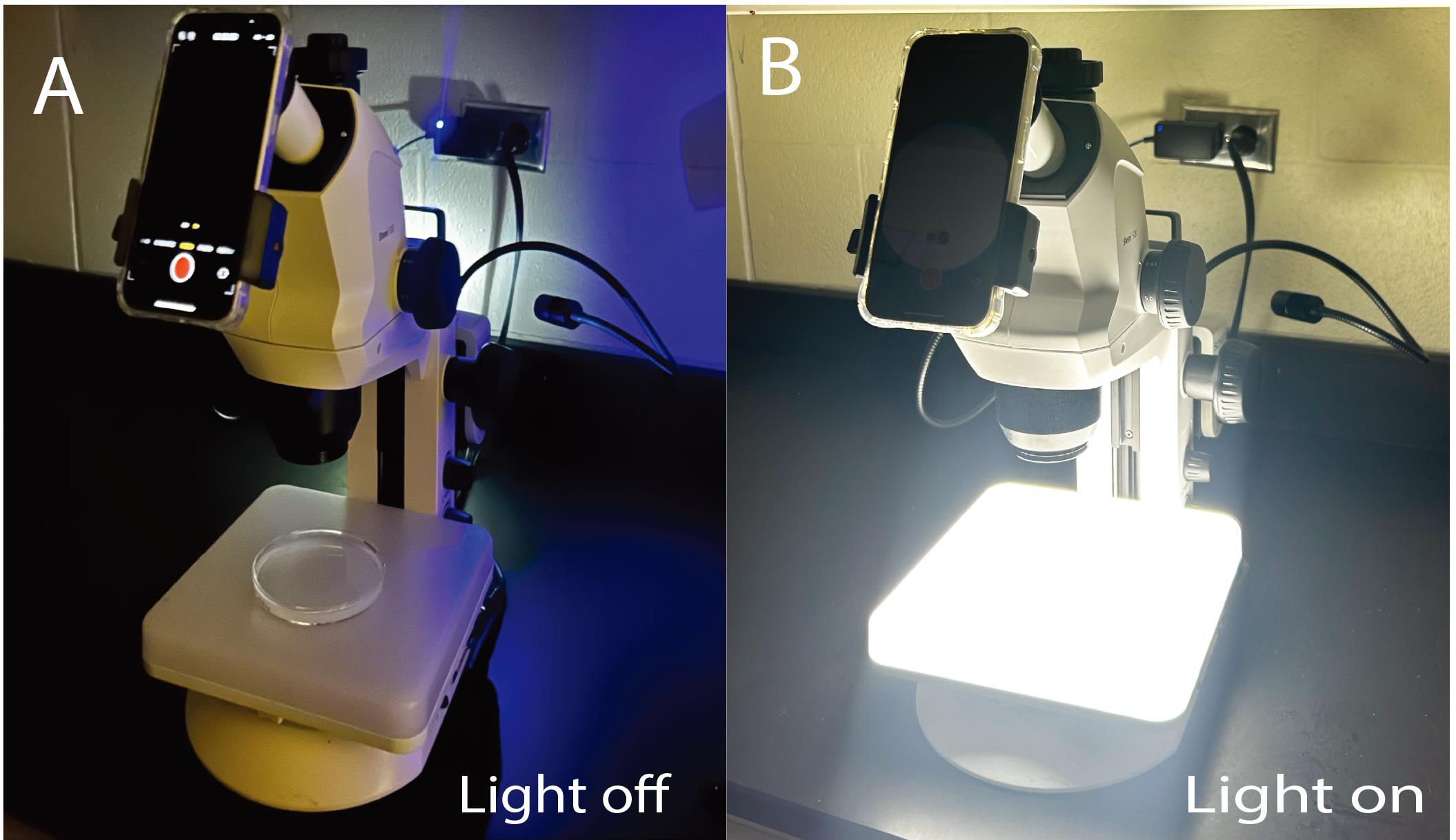
Figure 2. Experimental setup for the rolling assay. A phone camera is attached to the stereomicroscope with an agarose-filled Petri dish placed on top of the external light source (A) off and (B) on.Optogenetic induction
Using a spatula, gently collect the food or media containing the larvae (Figure 3).
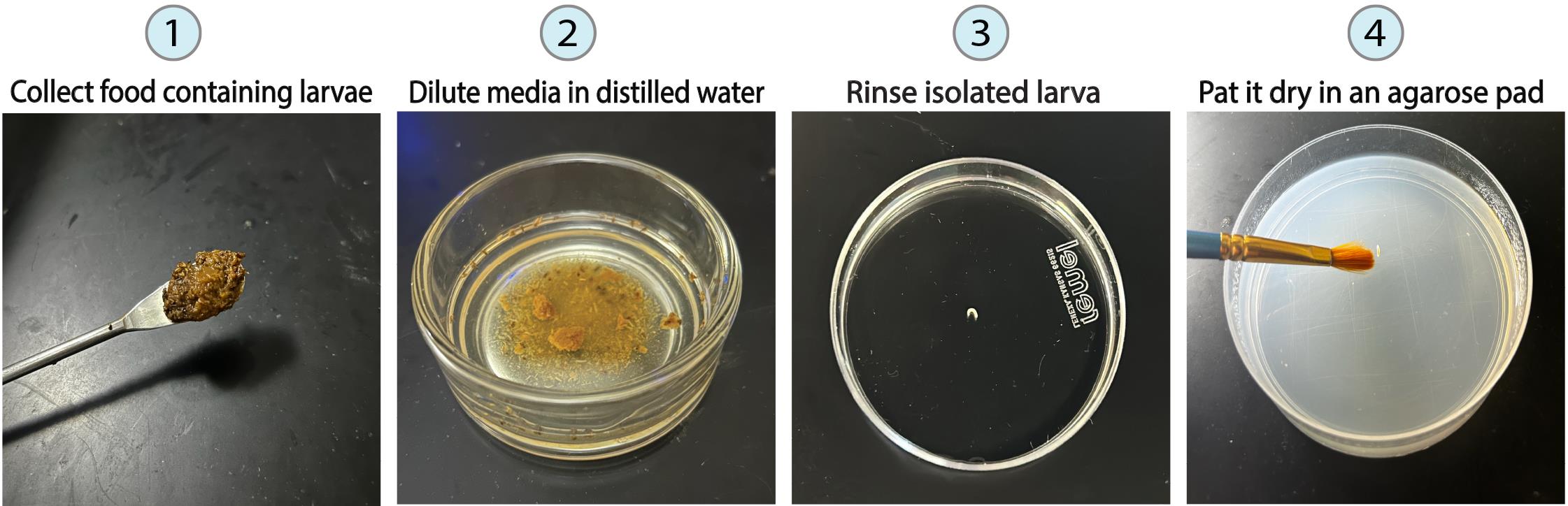
Figure 3. Step-by-step demonstration of larval preparation for optogenetic induction. Larval preparation begins by scraping the food from the vial (1) to retrieve the larvae. Next, the media is diluted in distilled water (2) to locate individual larvae. Following this, a single larva is picked and rinsed in distilled water to remove any residual food (3), placed on an agarose pad, and gently dried with a brush (4).Slightly dilute the collected media in distilled water to facilitate the identification and location of individual larvae.
Isolate each larva individually using fine-tipped forceps and transfer it into a separate Petri dish containing distilled water.
Rinse each larva in the distilled water for 1–2 s to remove any residual food or debris.
Carefully transfer it onto a 1.5% agarose pad positioned within a Petri dish to provide a stable and uniform surface for imaging purposes.
Pat dry the larva using a paintbrush and provide a 30-s period of undisturbed rest in darkness to facilitate acclimation and minimize potential influences of light exposure on their behavior.
Note: Ensure the larva is dorsal side up at the beginning of the experiment.
Begin the video recording process and ensure continuous filming for the duration of the experiment.
Induce optogenetic activation of the larva's rolling behavior by turning on Oeegoo 18W square LED ceiling light and exposing them to 30 s of intense white light.
Note: Avoid subjecting each larva to more than three repeated optogenetic inductions to avoid stress and potential fatigue, with at least 2–3 min of rest between inductions.
Transfer the recorded videos to your computer. The rolling response should be analyzed manually by researchers blinded to the experimental conditions.
Transfer the video from your phone to your computer using a USB cable or cloud service. The video file size can range from 30 to 60 MB depending on the resolution and length of the video.
Open the video using FFmpeg software to convert it to a .avi file.
Open the converted video with ImageJ software.
Note: To analyze videos in ImageJ, the computer should have at least 8 GB of RAM and 500 MB of free storage and be running Windows 10, macOS 10.13+, or Linux.
Observe larvae's movements and spot both dorsal tracheae (Figure 4 and Video 1). A full 360° roll is defined as the larva starting from the dorsal side up, rolling completely, and returning to the dorsal side up position. Identify a full roll by observing the disappearance and reappearance of both tracheae (Video 1 and Video 2).
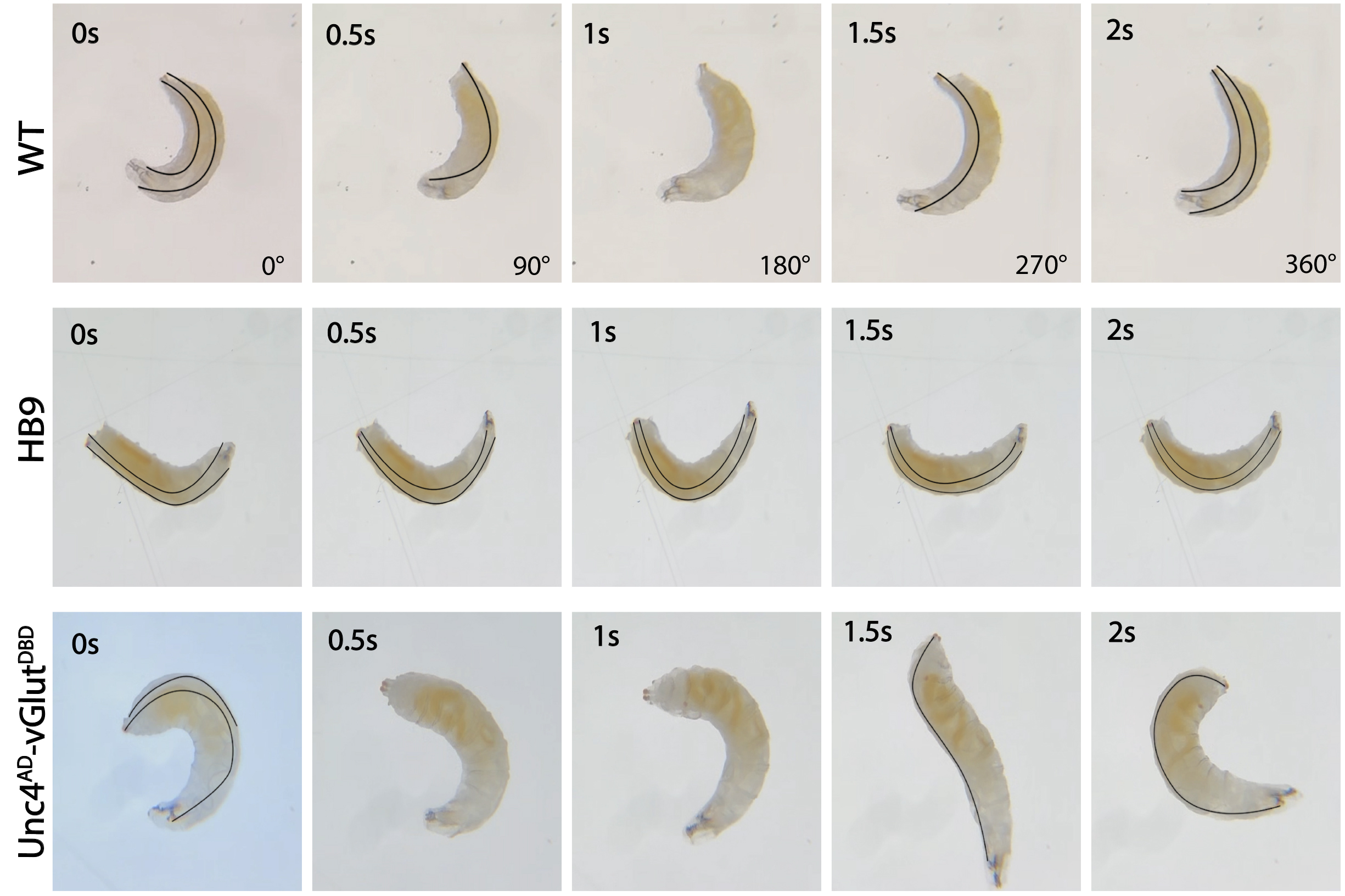
Figure 4. Sill images of L3 larvae showing 0°, 90°, 180°, 270°, and 360° of an outward roll in wild-type (GORO>Chrimson) rolling and defective rolling in HB9>UAS-GtACR1-eGFP, and Unc4AD-vGlutDBD>UAS-GtACR1-eGFP animals. Silencing Ib motor neurons (MNs) of ventral longitudinal (VL) and dorsal oblique (DO) muscles (HB9>UAS-GtACR1-eGFP) results in a significant inability to roll, approaching complete dysfunction. Silencing of dorsal longitudinal (DL) muscles (Unc4AD-vGlutDBD>UAS-GtACR1-eGFP) leads to animals being unable to complete a full 360° roll continuously. Black lines indicate dorsal tracheae. The left and right tracheae are both visible when the dorsal side is facing upward and invisible when the ventral side is facing upward.Note: The larvae in the figure appear to be at different larval stages because of varying camera zoom levels during the experiment.
Video 1. Video of a wild-type third-instar larvae showing the rolling response with a 360° rolling. Wild-type rolling is initiated by bending to one lateral side while the dorsal plane is facing upward (identified by both tracheae being visible, labeled with black lines), followed by continuous body rotation.Video 2. Examples of defective rolling. The video depicts a defective rolling in HB9>UAS-GtACR1 third-instar larva struggling to initiate rolling and Unc4AD-vGlutDBD>UAS-GtACR1-eGFP larva failing to complete a full 360° roll continuously.Consider a roll successful if the larva completes at least one 360° rotation, demonstrating the ability to rotate in a single direction from the dorsal side up back to the dorsal side up.
Criteria for failed rolling: First, confirm that the larva can crawl in an upright position (dorsal up) in the dark or with minimum light without triggering the Chrimson response. Count the rolling as a failure if, upon light stimulation, the larva responds to light but cannot bend into a C-shape, bends but cannot initiate lateral rolling, or cannot complete at least one 360° roll. A successful 360° roll contains the C-bending at a dorsal-up position (both tracheae completely visible), continuous rolling (inward or outward the curve) in a single direction without changing the direction of the curve, and the tracheae become invisible as the larva rolls to ventral up and visible again as the larva returns to dorsal up (Figure 4, Video 1, and Video 2).
Calculate the percentage of larvae that were able to perform rolling to determine the success rate for each genotype.
Calculate rolling frequency by counting the number of full 360° rolls the larva executes in 30 s (use the frame count shown by ImageJ to measure the time precisely).
Calculate the rolling duration by counting the number of frames required to complete rolling within the 30s-time frame in ImageJ. The frame count can be found in the top-left corner of the ImageJ window. Divide the number of frames by 30 to get the rolling duration in seconds as the videos are recorded at 30 fps. For instance, if a larva requires 120 frames to complete four rolls within the 30s-time frame, divide 120 by 30 to yield a rolling duration of 4 s. It is important to acknowledge that certain larvae may experience interruptions in their rolling motion, resulting in brief pauses between successive rolls. In such cases, it is recommended to exclude these interrupted frames from the calculation of rolling duration. However, these interruptions can still provide valuable insights into larvae's behavior and may be noted separately as an additional parameter for analysis.
Note: Rolling duration can be calculated using Excel by dividing the total number of frames by 30.
Import all values into a programming environment of your choice and plot bar graphs showing the percentage of animals of different genotypes that are able to complete at least one complete roll to visualize the rolling success. Also, use the scatterplot or boxplot tool to visualize the rolling duration, frequency, and number of interruptions during rolling graphically.
Statistical analyses: The distribution of rolling parameters, such as rolling frequency and duration, often does not meet normality criteria, making parametric tests like Student’s t-test unsuitable. For comparing two groups, the non-parametric Wilcoxon Rank-Sum Test is preferred. When comparing more than two genotypes, the Kruskal-Wallis test serves as a non-parametric alternative to one-way ANOVA. For multiple comparisons, corrections should be made, typically using Dunn’s method as a post-hoc test. These statistical analyses can be conducted using common software like MATLAB, R, Jupyter Notebook, or similar (Figure 5).
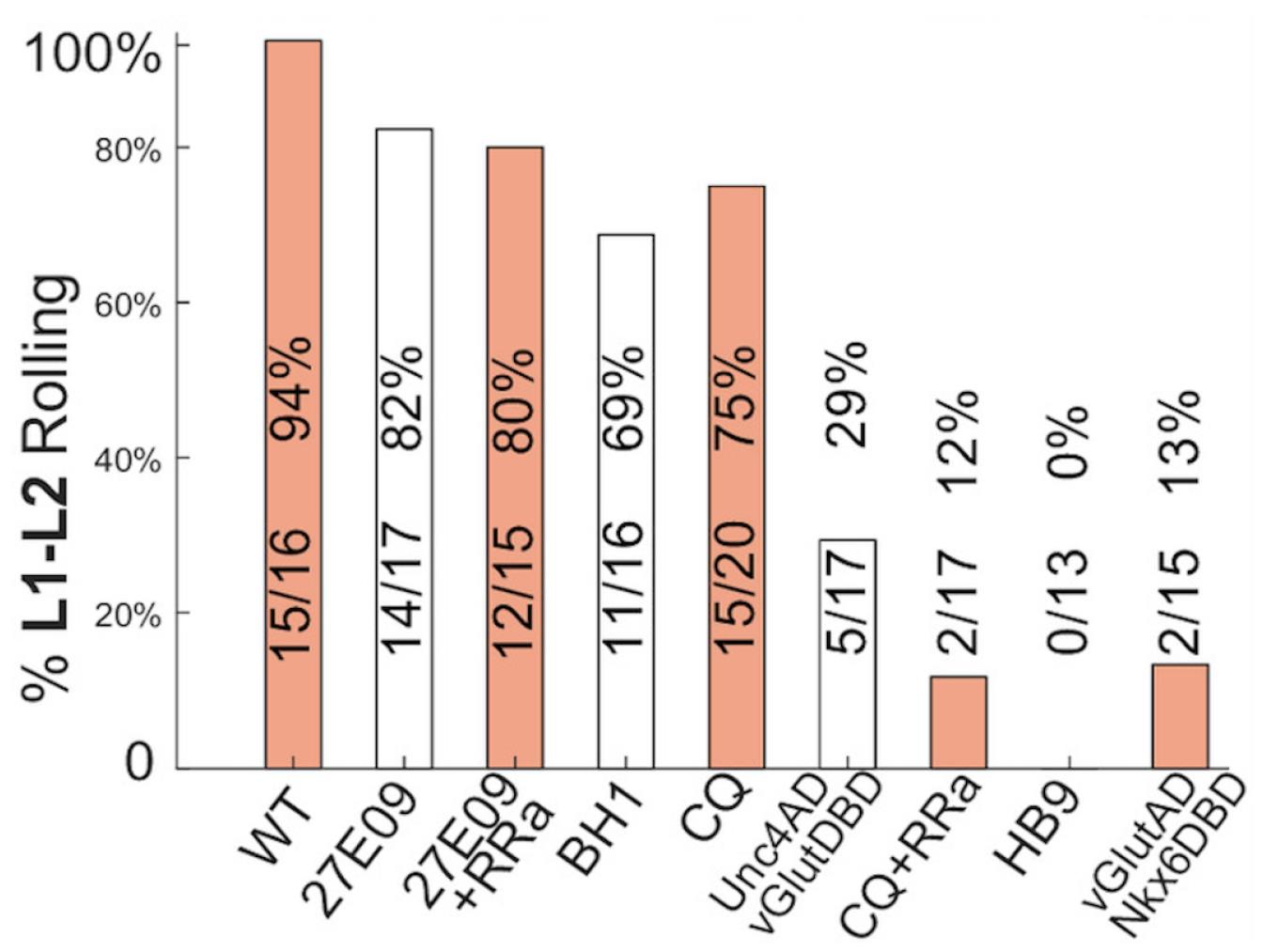
Figure 5. Experimental results showing the percentage of L1–L2 animals of different genotypes that are able to complete at least one complete roll when a different subset of motor neurons (MNs) is acutely silenced using GtACR1. Silencing MN Is (R27E09>UAS-GtACR1-eGFP) or in combination with Ib MN1 (R27E09+RRa>UAS-GtACR1-eGFP) had little or no effect on rolling performance. Silencing Ib MNs of lateral transverse (LT) muscles (BH1>UAS-GtACR1-eGFP) leads to a slightly reduced chance of successful rolling. Silencing Ib MN of dorsal longitudinal (DL) muscles caused mild (CQ>UAS-GtACR1-eGFP) to moderate (Unc4AD-vGlutDBD>UAS-GtACR1-eGFP) defect to rolling; however, silencing both Ib and Is targeting DL muscles (CQ+RRa>UAS-GtACR1-eGFP) lead to severe rolling failure. Silencing Ib MNs of ventral longitudinal (VL) and dorsal oblique (DO) muscles (HB9>UAS-GtACR1-eGFP and vGlutAD-Nkx6DBD>UAS-GtACR1-eGFP) leads to near-complete failure in rolling.While conducting optogenetics experiments, it is crucial to maintain uniform conditions across all genotypes. Variations in factors such as light luminance and ATR concentration can lead to inconsistent results. Therefore, standardizing these parameters ensures reliable and reproducible outcomes.
The dual activation and silencing technique using Chrimson and GtACR1 in this protocol can be applied to other experiments to investigate various neural circuits and behaviors. The protocol can also be expanded to include genetic tools like RNAi.
- Tracey, W., Wilson, R. I., Laurent, G. and Benzer, S. (2003). painless, a Drosophila Gene Essential for Nociception. Cell. 113(2): 261–273.
- Hwang, R. Y., Zhong, L., Xu, Y., Johnson, T., Zhang, F., Deisseroth, K. and Tracey, W. D. (2007). Nociceptive Neurons Protect Drosophila Larvae from Parasitoid Wasps. Curr Biol. 17(24): 2105–2116.
- Grueber, W. B., Jan, L. Y. and Jan, Y. N. (2002). Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129(12): 2867–2878.
- Ohyama, T., Schneider-Mizell, C. M., Fetter, R. D., Aleman, J. V., Franconville, R., Rivera-Alba, M., Mensh, B. D., Branson, K. M., Simpson, J. H., Truman, J. W., et al. (2015). A multilevel multimodal circuit enhances action selection in Drosophila. Nature. 520(7549): 633–639.
- Yoshino, J., Morikawa, R. K., Hasegawa, E. and Emoto, K. (2017). Neural Circuitry that Evokes Escape Behavior upon Activation of Nociceptive Sensory Neurons in Drosophila Larvae. Curr Biol. 27(16): 2499–2504.e3.
- Burgos, A., Honjo, K., Ohyama, T., Qian, C. S., Shin, G. e., Gohl, D. M., Silies, M., Tracey, W. D., Zlatic, M., Cardona, A., et al. (2018). Nociceptive interneurons control modular motor pathways to promote escape behavior in Drosophila. eLife. 7: e26016.
- Peron, S., Zordan, M. A., Magnabosco, A., Reggiani, C. and Megighian, A. (2009). From action potential to contraction: Neural control and excitation–contraction coupling in larval muscles of Drosophila. Comp Biochem Physiol A Mol Integr Physiol. 154(2): 173–183.
- Clark, M. Q., Zarin, A. A., Carreira-Rosario, A. and Doe, C. Q. (2018). Neural circuits driving larval locomotion in Drosophila. Neural Dev. 13(1): 6.
- Cooney, P. C., Huang, Y., Li, W., Perera, D. M., Hormigo, R., Tabachnik, T., Godage, I. S., Hillman, E. M. C., Grueber, W. B., Zarin, A. A., et al. (2023). Neuromuscular basis of Drosophila larval rolling escape behavior. Proc Natl Acad Sci USA. 120(51): e2303641120.
Data analysis
Validation of protocol
This protocol or parts of it has been used and validated in the following research article(s):
Cooney et al. [9]. Neuromuscular basis of Drosophila larval rolling escape behavior. PNAS (Figure 4, panel B, C).
General notes and troubleshooting
General notes
Troubleshooting
Problem 1: The smartphone camera cannot focus on the larva upon light stimulation.
Possible cause: The focus distance is not set up correctly in advance.
Solution: Most smartphone cameras feature auto-focusing when there is sufficient light. Before recording the experimental animals, adjust the distance between the objective and the gel pad with a wild-type larva with the LED light on until the camera stably focuses on the larva. Afterward, turn off the light to prepare the experimental larva without changing the camera settings on the phone. The camera should auto-focus on the rolling larva when the LED light is turned on.
Problem 2: The larva rolls out of the field of the recording.
Possible cause: The objective magnification level of the stereomicroscope is too high.
Solution: Reduce the zoom level. Alternatively, gently move the agarose gel pad between rolls to keep the animal in the center of the field. Do not move the gel pad before each roll is complete.
Problem 3: Difficulty locating the larva with the camera (especially for small, transparent L1 larvae).
Possible cause: The magnification level is low, or the agarose gel pad is too big for L1 larvae.
Solution: Increase the magnification level of the stereomicroscope. If there is difficulty navigating the gel when using a high magnification power, draw a small red mark with a Sharpie marker on the center of the gel. Then, put the larva near the mark, locate and focus on the red mark (and the larva) at a low magnification level, and zoom in.
Acknowledgments
Work in the Zarin lab is supported by Texas A&M grant. This protocol is derived from the original work of Cooney and Huang [9].
Competing interests
The authors declare no competing interest.
References
Article Information
Publication history
Received: Jun 21, 2024
Accepted: Oct 7, 2024
Available online: Oct 23, 2024
Published: Dec 5, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Sitaula, A., Huang, Y. and Zarin, A. (2024). Application of a Dual Optogenetic Silencing-Activation Protocol to Map Motor Neurons Driving Rolling Escape Behavior in Drosophila Larvae. Bio-protocol 14(23): e5131. DOI: 10.21769/BioProtoc.5131.
Category
Neuroscience > Basic technology > Optogenetics
Neuroscience > Behavioral neuroscience > Sensorimotor response
Neuroscience > Sensory and motor systems > Animal model
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










