- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay
Published: Vol 14, Iss 23, Dec 5, 2024 DOI: 10.21769/BioProtoc.5122 Views: 1613
Reviewed by: Emilia KrypotouChhuttan L MeenaJibin Sadasivan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
An Optimized Tat/Rev Induced Limiting Dilution Assay for the Characterization of HIV-1 Latent Reservoirs
Swarnima Mishra [...] Udaykumar Ranga
Apr 20, 2022 3014 Views
Production of Recombinant Hepatitis B virus (HBV) and Detection of HBV in Infected Human Liver Organoids
Tanvir Hossain [...] Tokameh Mahmoudi
Apr 20, 2022 3948 Views
Novel Workflows for Separate Isolation of Pathogen RNA or DNA From Wastewater: Detection by Innovative and Conventional qPCR
Kristina M. Babler [...] Ayaaz Amirali
Feb 20, 2025 3192 Views
Abstract
The quality of cellular products used in biological research can impact the accuracy of results. Epstein–Barr virus (EBV) is a latent virus that spreads extensively worldwide, and cell lines used in experiments may carry EBV and pose an infection risk. The presence of EBV in a single cell line can contaminate other cell lines used in the same laboratory, affecting experimental results. Existing tests to detect EBV can be divided into three categories: nucleic acid assays, serological assays, and in situ hybridization assays. However, most methods are time-consuming, expensive, and not conducive to high-volume clinical screening. Therefore, a simple system that allows for the rapid detection of EBV in multiple contexts, including both cell culture and tissue samples, remains necessary. In our research, we developed EBV detection systems: (1) a polymerase chain reaction (PCR)-based detection system, (2) a recombinase polymerase amplification (RPA)-based detection system, and (3) a combined RPA-lateral flow assay (LFA) detection system. The minimum EBV detection limits were 1 × 103 copy numbers for the RPA-based and RPA-LFA systems and 1 × 104 copy numbers for the PCR-based system. Both the PCR and RPA detection systems were applied to 192 cell lines, and the results were consistent with those of the assays specified in industry standards. A total of 10 EBV-positive cell lines were identified. The combined RPA-LFA system is simple to operate, allowing for rapid result visualization. This system can be implemented in laboratories and cell banks as part of a daily quality control strategy to ensure cell quality and experimental safety and may represent a potential new technique for the rapid detection of EBV in clinical samples.
Key features
• Establishes RPA and RPA-LFA detection systems for EBV.
• The RPA-LFA detection system can visualize the results in as short as 15 min.
• The specificity of the RPA and RPA-LFA assay systems for the detection of EBV is validated.
• The minimum EBV detection limit of the RPA and RPA-LFA systems is 1 × 103 copies.
Keywords: RPAGraphical overview
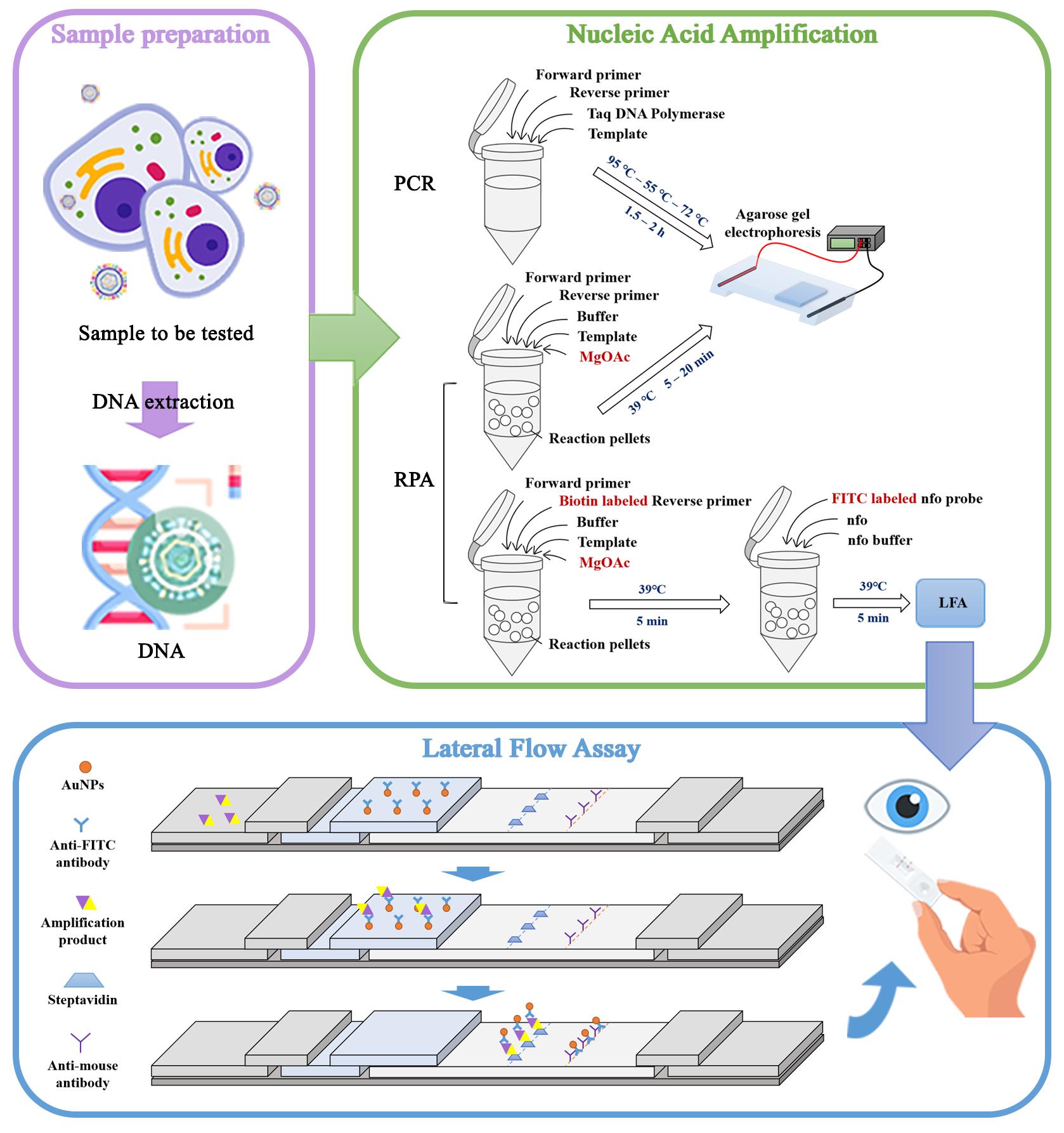
Pattern of Epstein–Barr virus (EBV) detection by the PCR, recombinase polymerase amplification (RPA), and combined RPA-lateral flow assay (LFA) systems. Reaction pellets are small particles that contain key reagents. These pellets usually contain the core components needed for the RPA reaction (recombinase, recombinase loading factor, and single-stranded binding protein). At the start of the reaction, reaction pellets dissolve these components to initiate the amplification reaction.
Background
As biological research continues to advance, cells are increasingly at risk of being misidentified or contaminated with other cell types or exogenous factors [1,2] including viruses; such contamination can be difficult to detect and remediate.
Epstein–Barr virus (EBV) is a member of the human herpesvirus family. EBV displays prolonged latency in lymphocytes, interfering with immune functions and potentially inducing cell proliferation and transformation. EBV infection involves many organ systems and is often misdiagnosed or underdiagnosed. Multiple reports in the literature have described the detection of EBV infection in various cell types maintained in cell culture banks [3,4]. These infections can be attributed to the presence of an existing EBV infection during the initial process of cell line establishment, EBV contamination of culturing materials, or improper manipulation by the experimental staff.
The most widely used of the existing EBV assays is PCR, which is a well-established and reliable molecular method. Quantitative PCR (qPCR) can be used to monitor disease progression or therapeutic efficacy by detecting changes in EBV load in the blood [5]. Iso-thermal amplification techniques, such as recombinant enzyme-mediated isothermal amplification and loop-mediated isothermal amplification (LAMP), are easy to perform, have limited equipment requirements, return rapid responses, and display high sensitivity. However, designing primers for use in the LAMP technique is complicated and can be difficult to replicate. In situ hybridization assay to detect Epstein–Barr early RNA (EBER) is the current gold standard for detecting EBV infection in clinic. However, this technique can only be applied to tissue samples and is generally limited to the clinical diagnosis of EBV-related diseases, such as cancer and lymphoma.
Recombinase polymerase amplification (RPA) is a highly sensitive and selective isothermal amplification technique that can be performed at 39 °C and can be used to amplify a large number of samples in a short period of time. RPA, first introduced in 2006 by Niall Armes of ASM Scientific Ltd., UK [6], relies on modified homologous recombination mechanisms. RPA reagents are commercially available, and the basic RPA reaction kit can be augmented by additional commercially available kits that use different probes that can be cleaved by different enzymes: exo (exonuclease III), fpg (formamidopyrimidine DNA Glycosylase), and nfo (endonuclease IV) [7]. The exo and fpg probes are typically used for real-time detection, whereas the nfo probe is typically used for detection systems that use lateral flow dipsticks.
In this study, we combined RPA technology with a lateral flow assay (LFA) to develop an RPA-LFA detection system for application to the rapid and bulk screening of EBV contamination in cell lines stored by cell banks and to conduct regular and daily inspection of cell lines used in biological experiments, ensuring the quality of cell lines and the safety of experimental personnel. This system also demonstrates high potential for clinical adaptation to improve EBV detection in blood and tissue samples.
Materials and reagents
Biological materials
B95-8 cell line (China Center for Type Culture Collection, GDC0015)
HeLa cell line (China Center for Type Culture Collection, GDC0009)
Epstein–Barr virus (China Center for Type Culture Collection, GDV132)
Reagents
Trypsin (AMEKO, catalog number: RC01145)
Ethanol (National Pharmaceutical Group Chemical Reagent Co., Ltd., catalog number: 10009228)
PBS (Gibco, catalog number: 70013032)
TIANamp Genomic DNA Kit (TIANGEN, catalog number: DP304)
E.Z.N.A Viral DNA Kit (OMEGA, catalog number: D3892)
Takara 10× loading buffer (Takara, catalog number: AA0151)
DL2,000 DNA marker (Takara, catalog number: 3427A)
TS-GelRed nucleic acid dye (TSINGKE, catalog number: TSJ003)
Agarose (TSINGKE, catalog number: TSJ001)
RNase-free water (Takara, catalog number: takara.9012)
Twist Amp Basic kit (TwistDX, catalog number: 111-035-003)
Thermo Scientific endonuclease IV(Nfo) and its buffer (FastDigest, catalog number: EN0591)
Test strip dilution (Aoke Botai Biotech)
Primer and probe:
FP: 5'-CTTGGAGACAGGCTTAACCAGACTCA-3'
RP: 5'-CCATGGCTGCACCGATGAAAGTTAT-3'
LFA-RP: 5'-[BIOTIN]CCATGGCTGCACCGATGAAAGTTAT-3'
LFA-Probe: 5'-[FITC]TGCCGGCCCCTCGAGATTCTGACCGGGGACC[THF]CTGGTTGCTCTGTTG[C3-Spacer]-3'
Laboratory supplies
Cell culture flask (NEST, catalog number: 707001)
Serological pipettes (NEST, catalog number: 327001)
Pipette tips (Biosharp, catalog numbers: BS-10-T, BS-200-T, BS-1000-T)
Micro-centrifuge tube (Biosharp, catalog number: BS-15-M)
Axygen PCR tubes (AXYGEN, catalog number: Axygen- PCR-05-A)
Lateral flow stick (Suzhou Xianda Gene Technology, catalog number: TS101)
Equipment
Biological safety cabinet (Esco, catalog number: A2AC2-S)
Refrigerator [CHANGHONG MEILING, catalog number: MCF(L)-398LDWEP]
Thermostatic water bath (Dichbio, catalog number: JB Academy)
PCR automatic serialization analyzer (SensoQuest, catalog number: Labcycler 48)
Low-speed centrifuge (DLAB, catalog number: D1008E, D1008)
Small high-speed freezer-type centrifuge (Eppendorf, model: 5420)
CO2 Incubator (Memmert, catalog number: INC)
Ultra-micro UV spectrophotometer (Thermo Fisher Scientific, model: NanoDrop2000)
Micropipettes 0.1–2.5 µL, 2–20 µL, 20–200 µL (Eppendorf, catalog number: 3123000217, 3123000233, 3123000250)
Electrophoresis apparatus (Beijing Liuyi Biotechnology Co., Ltd., catalog number: 112-0630)
Gel imaging system (Bio-Rad, catalog number: ChemiDoc XRS+)
Vortex mixer (Kylin-bell, catalog number: VORTEX-6)
Software and datasets
DNAMAN (version 8.0)
National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/)
Procedure
Primer & probe & lateral flow strip design
General primer design
Download the nucleic acid sequence of EBV from NCBI (Genebank: V01555).
Design PCR and RPA primers for conserved regions using DNAMAN 8.0 software.
Considering that the length of RPA primers is longer than that of normal PCR primers, after primer testing, we finally designed primers that can participate in both PCR and RPA as follows:
Forward primer: 5'-CTTGGAGACAGGCTTAACCAGACTCA-3'
Reverse primer: 5'-CCATGGCTGCACCGATGAAAGTTAT-3'
The position of the Epstein–Barr virus (EBV) fragment detected by primers is illustrated in Figure 1A.
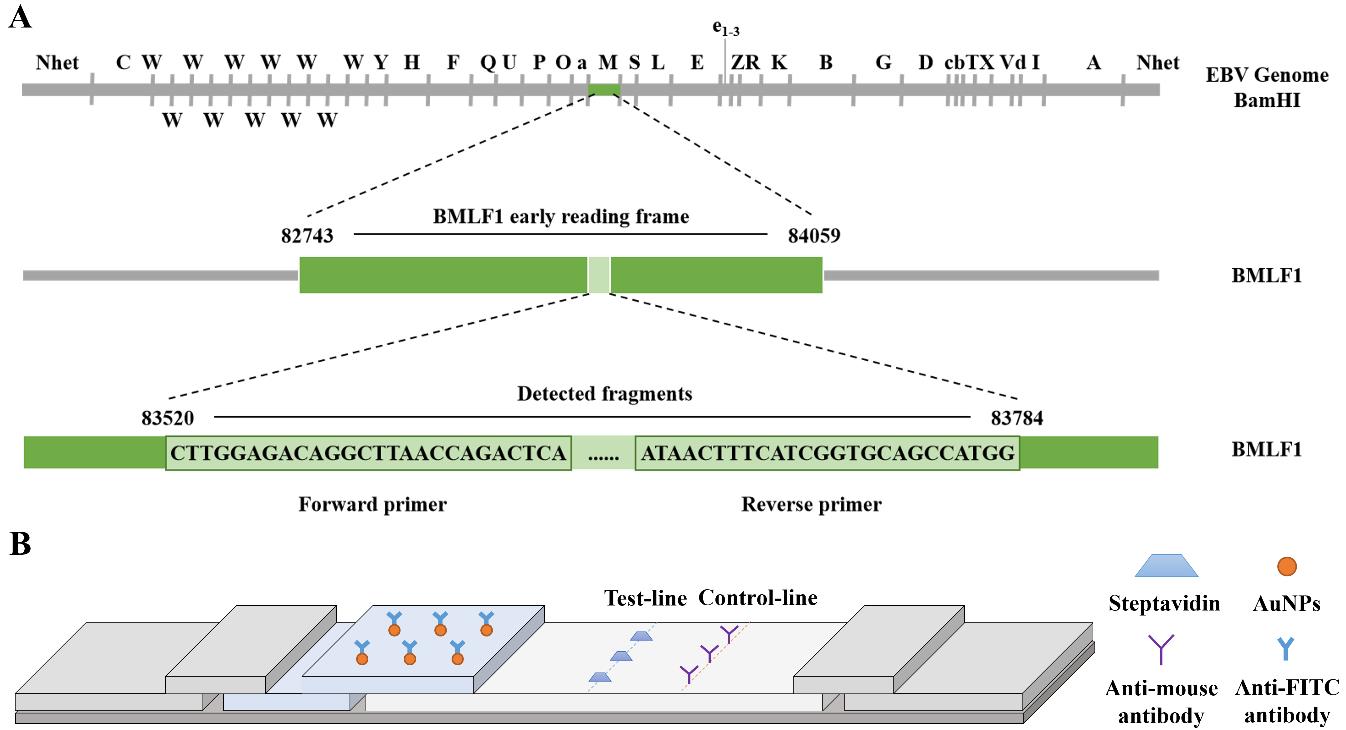
Figure 1. Schematic diagram of primer screening and lateral flow strip design. (A) Location of the Epstein–Barr virus (EBV) fragment detected by primers on the BamHI restriction map of the prototype EBV B95-8 genome. (B) Lateral flow strip design.RPA-LFA primer & probe design
The same amplified fragments as in PCR were selected but with BIOTIN modification at the 5' end of the reverse primer and a probe introduced to enhance its amplification specificity.
The probe was designed to have a FITC modification at the 5' end, a modifying group (C3-Spacer) at the 3' end, and tetrahydrofuran (THF) labeled in the middle of the 5' and 3' ends to serve as the recognition site for NFO. The final primers and probes were as follows:
Forward primer: 5'-CTTGGAGACAGGCTTAACCAGACTCA-3'
LFA-Reverse primer: 5'-[BIOTIN]CCATGGCTGCACCGATGAAAGTTAT-3'
LFA-Probe: 5'-[FITC]TGCCGGCCCCTCGAGATTCTGACCGGGGACC[THF]CTGGTTGCTCTGTTG[C3-Spacer]-3'
Lateral flow strip design
Gold particles of BIOTIN antibody and FITC antibody are immobilized on a lateral flow strip (test-line), and the target fragment will be captured to appear as a positive band. (Analysis of results: If only one control line can be observed, the result is regarded as negative; if both the control line and the test line can be observed at the same time, the result is regarded as positive; if the control line is not observed, it indicates that the test strip is invalid.) The structure of the test strip is shown in Figure 1B.
Sample preparation
DNA extraction
Collect cells in good growth condition, centrifuge with trypsin digestion, and aspirate the cell suspension. If cells are freeze-stored, directly thaw and transfer to microcentrifuge tubes and aspirate the cell suspension. The latter steps are operated according to the blood/cell/tissue genomic DNA extraction kit of Tiangen Biochemical Technology (Beijing) Co. as follows:
Place the pre-treated cell suspension (containing a cell number approximately equal to the number of cells that can be cultured with one t25) into a 1.5 mL microcentrifuge tube, centrifuge at 9,306× g for 1 min, remove the supernatant, and add 1 mL of PBS reagent to keep the cells in suspension. Resuspend to rewash the cell precipitate, centrifuge at 9,306× g for 1 min, remove the supernatant, add 200 μL of buffer solution GA, and shake the mixture until it is fully suspended.
Add 20 μL of proteinase K solution to the bottom of the micro-centrifuge tube and mix well; then, add 200 μL of buffer GB to it and mix well. Place the micro-centrifuge tube in a thermostat at 70 °C for 10 min and centrifuge briefly to remove the water droplets on the inner wall of the cap.
Add 200 μL of anhydrous ethanol to it, shake it well for 15, and centrifuge it briefly to remove the water droplets on the inner wall of the cap.
Add the liquid from this step to the adsorbent column and centrifuge at 13,400× g for 30 s. Discard the flowthrough liquid.
Add 500 μL of buffer GD with anhydrous ethanol to the column and centrifuge at 13,400× g for 30 s. Discard the waste liquid.
Add 600 μL of rinse solution PW to the column, centrifuge at 13,400× g for 30 s, and discard the waste solution. Repeat step B1g with another addition of rinse solution PW.
Place the column back into the collection tube, centrifuge at 13,400× g for 2 min, discard the waste liquid, and let dry thoroughly. Prepare a clean micro-centrifuge tube and place the column in it. Add 50 μL of ddH2O dropwise to the column and centrifuge at 13,400× g for 2 min. The resultant liquid obtained after centrifugation is the cellular DNA.
DNA dilution
Measure the DNA concentration using a NanoDrop microspectrophotometer. Calculate concentration by diluting 50 ng/μL with ddH2O and then store in the refrigerator at -20 °C.
Nucleic acid amplification
RPA-specific assay
Cellular DNA diluted with ddH2O at a concentration of 50 ng/μL is selected as the template in the system. In the reaction tube containing the RPA lyophilized powder, add 29.5 μL of buffer, 2 μL each of unlabeled 10 μM upstream and downstream primers, 1 μL of cellular extracted DNA, and 13 μL of ddH2O. Finally, add a 280 mmol/L solution of magnesium acetate in a volume of 2.5 μL to initiate the reaction. Uniformly distribute the above reaction solution and place it into the PCR instrument at 39 °C for 20 min. After the amplification reaction is completed, add 6 μL of 10× loading buffer and thoroughly mix. Then, load 10 μL of the sample into the wells of a 2% agarose gel and operate on agarose gel electrophoresis (120 V, 200 A). The amplification of the target bands is observed using a gel-image system (Figure 2A).
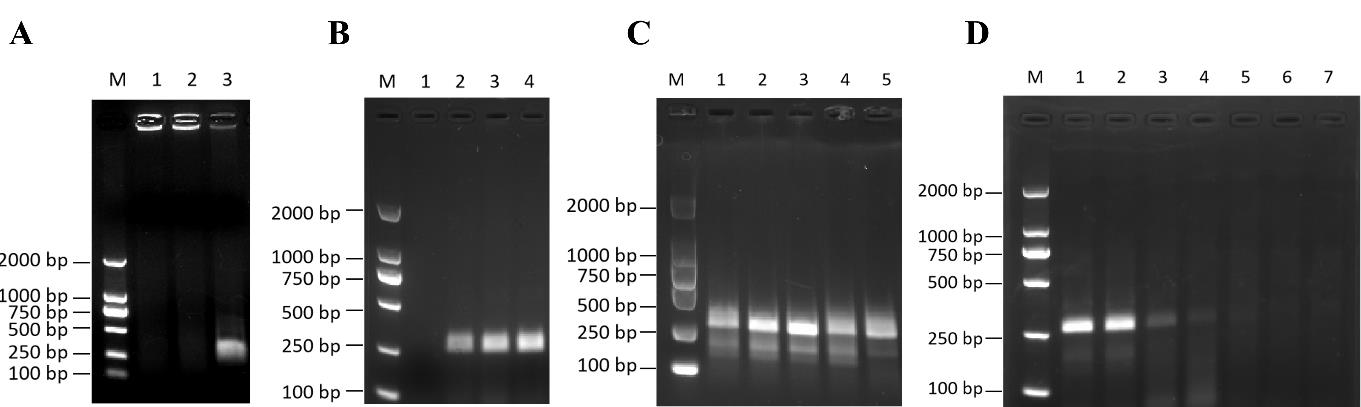
Figure 2. Establishment of recombinase polymerase amplification (RPA) testing system. (A) Validation of the RPA system. 1: ddH2O; 2: HeLa cell DNA (negative control); 3: B95-8 cell DNA (positive control); M: DL2000 marker. (B) Determination of the optimal RPA time for B95-8 cell DNA. 1: 5 min; 2: 10 min; 3: 15 min; 4: 20 min; M: DL2000 marker. (C) Determination of the optimal RPA temperature for B95-8 cell DNA. 1: 43 °C; 2: 41 °C; 3: 39 °C; 4: 37 °C; 5: 35 °C; M: DL2000 marker. (D) Sensitivity of Epstein–Barr virus (EBV) detection by the RPA method. M: DL2000 marker; 1: 1 × 106 copies/μL; 2: 1 × 105 copies/μL; 3: 1 × 104 copies/μL; 4: 1 × 103 copies/μL; 5: 1 × 102 copies/μL; 6: 1 × 101 copies/μL; 7: 1 × 100 copies/μL; M: DL2000 marker.Optimization of RPA reaction system
B95-8 cell DNA at 50 ng/μL is used as a template and optimized in terms of time and temperature, respectively. Thoroughly mix the resulting product with 6 μL of 10× loading buffer and confirm the amplification efficiency by agarose gel electrophoresis (120 V, 200 A).
Time optimization: Maintain reaction temperature at 39 while the reaction time is varied at 5, 10, 15, and 20 min to observe the amplification of different reaction times (Figure 2B).
Temperature optimization: Maintain reaction time at 15 min, and vary the reaction temperatures at 35, 37, 39, 41, and 43 °C to observe the amplification of RPA reaction at different temperatures (Figure 2C).
RPA sensitivity examination
Dilute the previously prepared EBV DNA with a ddH2O gradient to 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 1 × 101, 1 × 100 copies/μL as templates. Subsequently, use 1 μL of the diluted DNA for ordinary RPA amplification at 39 °C for 15 min. Following this amplification process, add 6 μL of 10× loading buffer to the reaction mixture. After thorough mixing, take a sample volume of 10 μL for agarose gel electrophoresis (120 V, 200 A) to verify the amplification effect (Figure 2D).
Lateral flow assay
RPA-LFA specific assay
Set up the RPA reaction with a common forward primer, a reverse primer with Biotin labeling, buffer, DNA template, ddH2O, and magnesium acetate. The reaction takes place at 39 °C for 20 min.
Add the FITC-labeled probe, nfo enzyme, and nfo enzyme buffer to the reaction product from the previous step. The specific reaction system is as follows:
Buffer 29.5 μL
10 μM FP 2 μL
10 μM LFA-RP 2 μL
10 μM probe 0.6 μL
280 mmol/L magnesium 2.5 μL
ddH2O 13 μL
RPA amplification products 50 μL
Template DNA 1 μL
NFO enzyme 1.5 μL
NFO enzyme buffer 5 μL
Dilute the final amplification product with diluent at a ratio of 1:100. Then, take 50 μL and add it to the lateral flow strip. Wait for 5 min to observe the results (Figure 3A).
Optimization of RPA-LFA reaction system
Optimize the reaction conditions of RPA-LFA, such as reaction time and temperature, using 50 ng/μL B95-8 cellular DNA as the reaction template. Dilute the amplification products at a ratio of 1:100 and then add dropwise to the test strips to wait for the detection results.
Time optimization: 1) Optimization of the first part of the reaction. The reaction time of the first step for common RPA is set to 5, 10, 15, and 20 min. After adding the probe, nfo enzyme, and its buffer, set the amplification and digestion time to 15 min. Then, test the resulting amplification product using a test strip (Figure 3B). 2) Optimization of the reaction time for the second step. The time of the first step is set at 5 min, and the time of the second step of enzymatic amplification is set at 5, 10, and 15 min. Add the amplification products obtained dropwise to the test strip to observe the results (Figure 3C).
Reaction temperature: Maintain the reaction time of both steps at 15 min and vary the reaction temperatures of the samples in both steps at 35, 37, 39, 41, and 43 °C, respectively (Figure 3D).
RPA-LFA sensitivity examination
Dilute the EBV DNA to 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 1 × 101, 1 × 100 copies/μL in a gradient, and take 1 μL of each concentration as the template for amplification. Set the temperature of the reaction to 39 °C and the amplification time for both steps to 15 min. After amplification, proportionally dilute 50 μL and add to the test strip for 5 min to observe the results (Figure 3E).
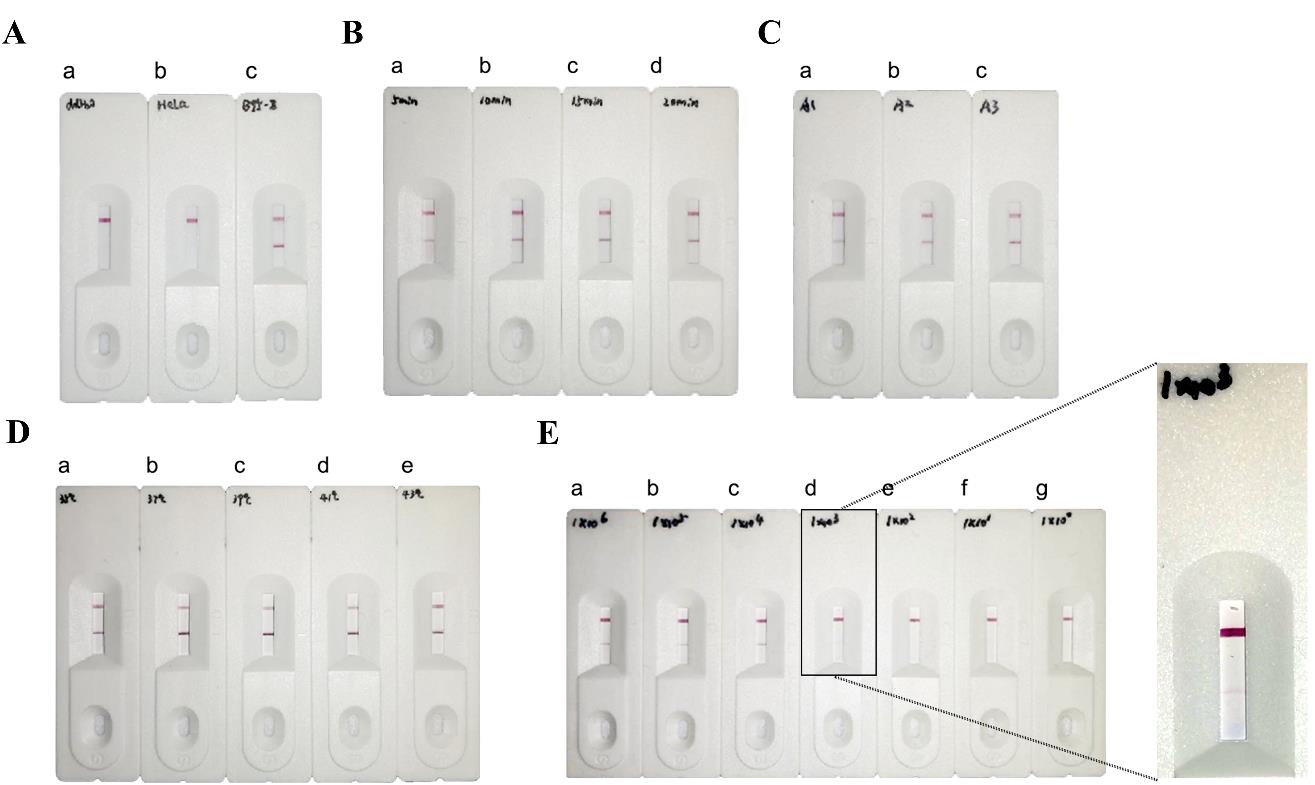
Figure 3. Establishment of combined recombinase polymerase amplification–lateral flow assay (RPA-LFA) testing system. (A) Validation of the RPA-LFA system. a: ddH2O; b: HeLa cell DNA (negative control); c: B95-8 cell DNA (positive control). (B) RPA-LFA was used to assess assays with different reaction times (first step). a: 5 min; b: 10 min; c: 15 min; d: 20 min; (C) RPA-LFA was used to assess results for assays with different reaction times (second step). a: 5 min; b: 10 min; c: 15 min. (D) Determination of the optimal RPA-LFA temperature for B95-8 cell DNA. a: 35 °C; b: 37 °C; c: 39 °C; d: 41 °C; e: 43 °C. (E) Sensitivity of Epstein–Barr virus (EBV) detection by the RPA method. M: DL2000 marker; a: 1 × 106 copies/μL; b: 1 × 105 copies/μL; c: 1 × 104 copies/μL; d: 1 × 103 copies/μL; e: 1 × 102 copies/μL; f: 1 × 101 copies/μL; g: 1 × 100 copies/μL.
Data analysis
Determination of primer and probe specificity.
The primers and probes were tested for specificity by RPA and RPA-LFA assay and the results showed good specificity of the primers and probes. To further validate their specificity, 192 cells from different species were validated using RPA. The results are shown in the Supplementary Information. In order to ensure the accuracy of the experiment, we also chose two other EBV detection methods, PCR and qPCR. The detection results of these three methods are shown in Table 1 below:
Table 1. Evaluation of EBV test results using different methods of detection
Result Method PCR qPCR RPA Positive 9 9 9 Negative 183 183 183 Total 192 192 192 To verify the specificity of the RPA-LFA system, we screened several cell lines containing endogenous viruses among the 192 tested cell lines: HeLa (containing human papillomavirus type 18), SiHa (containing human papillo-mavirus type 16), Hep-G2/2.2.15 (containing hepatitis B virus), and RK13 (containing bovine viral diarrhea virus). The RPA-LFA method was validated using these cell lines, and the results were negative for EBV (Figure 4A), indicating that the system has good specificity. In addition, we randomly selected cell lines for further validation, including three EBV-negative cell lines (A-204, H9, and HTR-8) and the remaining nine cell lines (NK-92, Daudi, ARH-77, Raji, JVM-2, A-431, Farage, MC/CAR, and CCRF-SB) that showed EBV positivity in previous tests. All cell detection results were consistent with expectations.
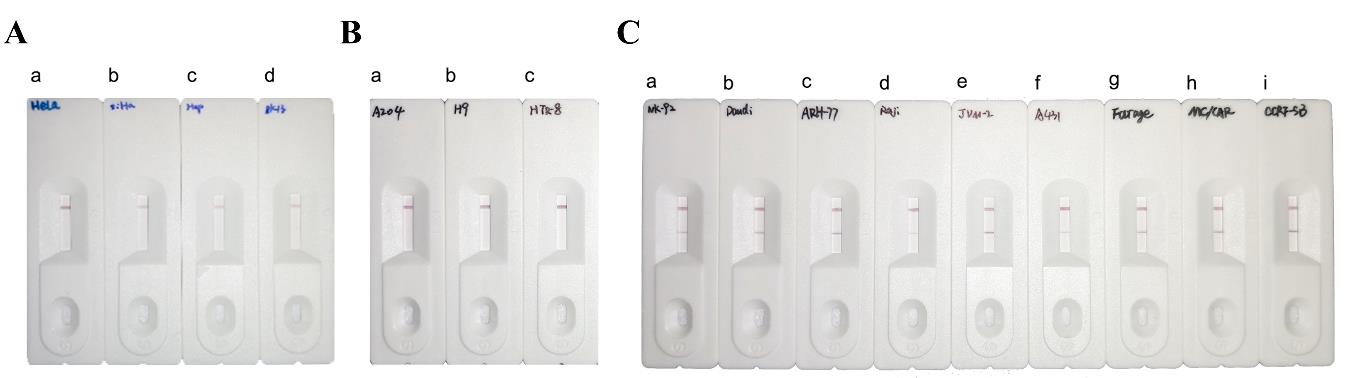
Figure 4. Combined recombinase polymerase amplification–lateral flow assay (RPA-LFA) detection of Epstein–Barr virus (EBV) system in different cells. (A) Detection of cells containing other endogenous viruses using the RPA-LFA system. a: HeLa; b: SiHa; c: Hep-G2/2.2.15; d: RK13. (B) RPA-LFA detection of EBV-negative cell lines. a: A-204; b: H9; c: HTR-8. (C) RPA-LFA detection of EBV-positive cell lines. a: NK-92; b: Daudi; c: ARH-77; d: Raji; e: JVM-2; f: A-431; g: Farage; h: MC/CAR; i: CCRF-SB.Reaction condition optimization.
Through the optimization experiments of RPA reaction conditions, it was found that the optimal reaction time is 15 min, and the optimal reaction temperature is 39 °C. In the optimization experiment of RPA-LFA reaction conditions, it was observed that positive results could still be obtained when both the first and the second steps were reacted for 5 min at a temperature of 39 °C.
Determination of detection limit.
We constructed a plasmid containing the EBV gene and used this for qPCR to plot an EBV standard curve according to the CT value (Figure 5). The CT value of 100 copies/μL was discarded because the deviation of this point was large, and a standard curve was plotted. The CT value was inversely proportional to the gene copy number, and the R2 value of the curve was 0.99. Based on this standard curve, the concentration of EBV DNA extracted from B95-8 cells was calculated, and the DNA was diluted with ddH2O to 1 × 106 copies/μL. The EBV DNA was further diluted with ddH2O to produce a gradient of 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 1 × 101, and 1 × 100 copies/μL.
The limit of detection is determined by the presence or absence of the test line. Based on the results of experiments in which viruses of different concentration gradients were used as templates, the following could be concluded: In the RPA and RPA-LFA experiments, bands were still observed on the test line when the copy number of template EBV was 1×103copies, so the limit of detection for the RPA and RPA-LFA assays were 1 × 103 copies (Figure 2D; Figure 3E).
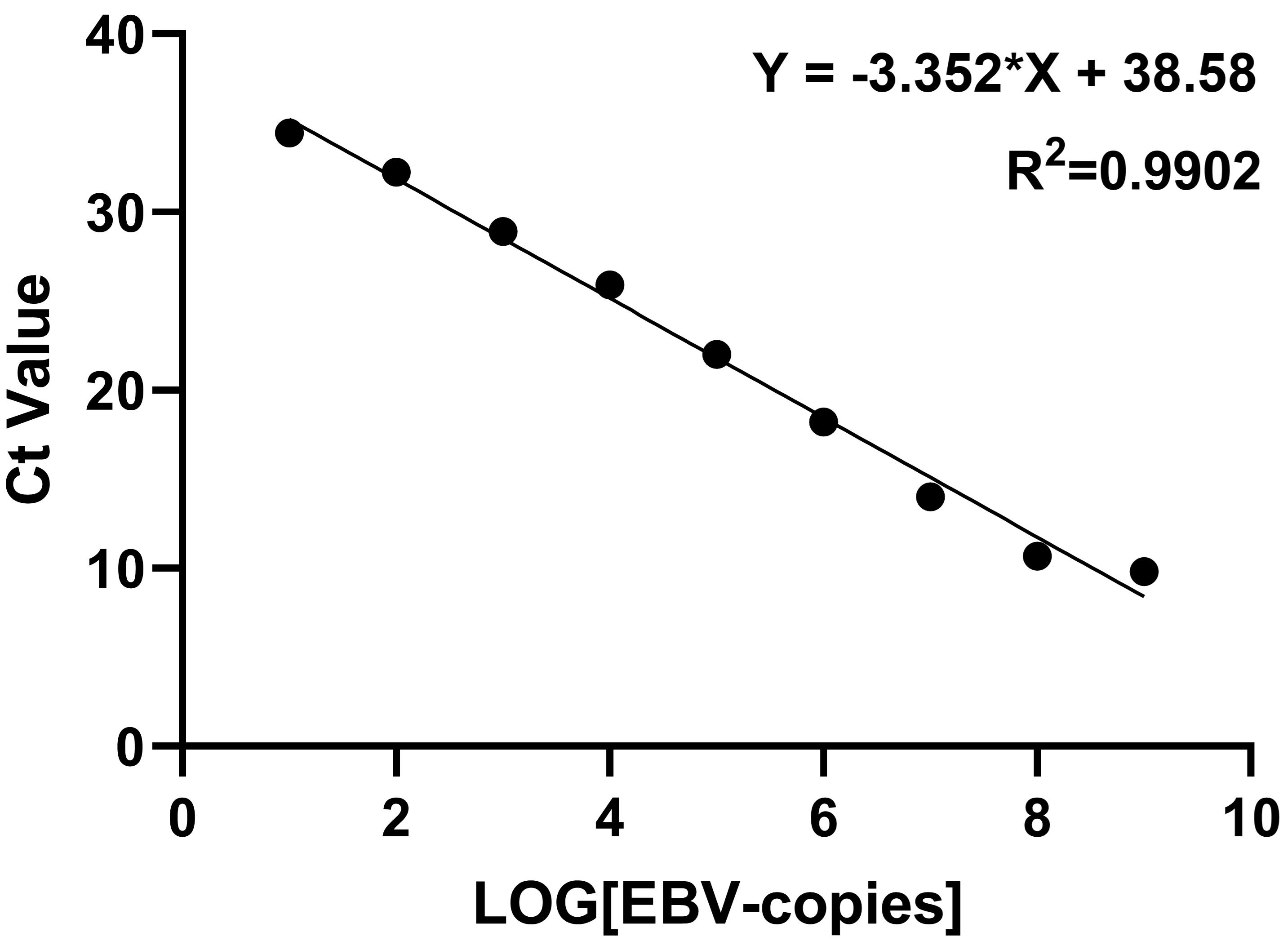
Figure 5. Epstein–Barr virus (EBV) standard curve
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Sun et al. [8] Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay.
General notes and troubleshooting
General notes
Please pay attention to cross-contamination and aerosol contamination during operation.
The length of RPA primer should be around 30 bp (a primer too short will affect the amplification speed and detection sensitivity).
When adding reaction reagents to RPA, it is recommended to add magnesium ions at the end to activate the reaction.
Troubleshooting
Problem: The drag band is obvious after agarose gel electrophoresis with RPA.
Possible cause: The reaction system is not mixed well; the agarose gel concentration is too low.
Solution: Ensure thorough mixing of the reaction system during preparation and add Mg ions to the lid at the end; centrifuge and shake down to activate the reaction; prepare an agarose gel with a concentration of 2% or higher.
Acknowledgments
This work was supported by grants from the National Science and Technology Infrastructure (NSTI-BMCR20, NSTI-BMCR21, and NSTI-BMCR22) and Science and Technology Innovation Grants of Hubei Province (2021CFB357) to Chao Shen.
The protocols described here are adapted from our previous work (Sun et al. [8]).
Competing interests
The authors declare no competing interests.
References
- Stacey, G. N. (2000). Cell contamination leads to inaccurate data: we must take action now. Nature. 403(6768): 356–356.
- Hughes, P., Marshall, D., Reid, Y., Parkes, H. and Gelber, C. (2007). The Costs of using Unauthenticated, Over-Passaged Cell Lines: How Much more Data do we Need? Biotechniques. 43(5): 575–586.
- Uphoff, C. C., Denkmann, S. A., Steube, K. G. and Drexler, H. G. (2010). Detection of EBV, HBV, HCV, HIV-1, HTLV-I and -II, and SMRV in Human and Other Primate Cell Lines. J Biomed Biotechnol. 2010: 1–23.
- Shioda, S., Kasai, F., Watanabe, K., Kawakami, K., Ohtani, A., Iemura, M., Ozawa, M., Arakawa, A., Hirayama, N., Kawaguchi, E., et al. (2018). Screening for 15 pathogenic viruses in human cell lines registered at the JCRB Cell Bank: characterization of in vitro human cells by viral infection. R Soc Open Sci. 5(5): 172472.
- Hübner, M., Bozic, M., Konrad, P. M., Grohs, K., Santner, B. I. and Kessler, H. H. (2015). Analytical and clinical performance of a new molecular assay for Epstein-Barr virus DNA quantitation. J Virol Methods. 212: 39–43.
- Piepenburg, O., Williams, C. H., Stemple, D. L. and Armes, N. A. (2006). DNA Detection Using Recombination Proteins. PLoS Biol. 4(7): e204.
- Lobato, I. M. and O'Sullivan, C. K. (2018). Recombinase polymerase amplification: Basics, applications and recent advances. TrAC, Trends Anal Chem. 98: 19–35.
- Sun, Y., Tang, D., Li, N., Wang, Y., Yang, M. and Shen, C. (2024). Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay. Viruses. 16(1): 106.
Supplementary information
The following supporting information can be downloaded here:
- Figure S1. RPA detection of EBV system in different cells.
Article Information
Publication history
Received: Aug 1, 2024
Accepted: Oct 4, 2024
Available online: Oct 16, 2024
Published: Dec 5, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Sun, Y., Tang, D., Li, N., Wang, Y., Yang, M. and Shen, C. (2024). Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay. Bio-protocol 14(23): e5122. DOI: 10.21769/BioProtoc.5122.
Category
Microbiology > Pathogen detection > PCR
Molecular Biology > DNA > PCR
Medicine
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link