- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Host Receptor Pili for Cryo-EM Single-Particle Reconstruction
Published: Vol 14, Iss 20, Oct 20, 2024 DOI: 10.21769/BioProtoc.5094 Views: 1437
Reviewed by: Alba BlesaCuncai GuoSrajan Kapoor

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation and Purification of β-1,3-glucan-Linked Candida glabrata Cell Wall Proteases by Ion-Exchange Chromatography, Gel Filtration, and MDPF-Gelatin-Zymography Assay
Pirjo Pärnänen [...] Pirjo Nikula-Ijäs
Mar 20, 2024 1565 Views
Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 1897 Views
Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 1951 Views
Abstract
Single-stranded RNA bacteriophages (ssRNA phages) infect their hosts by binding to the host receptor pili. Purification of pili usually involves mechanical shearing of pili from cells followed by precipitation. However, previous methods often result in low efficiency or unstable results due to pili retraction. This protocol presents an optimized method for purifying receptor type IV pili from Acinetobacter genomospecies 16 (A. gp16), incorporating enhancements in shearing and collection steps to achieve high yields. We found that repeated passage through syringe needles increases yield, and temperature control is crucial during purification. Additionally, the CsCl density gradient was optimized specifically for this specific strain. The purified type IV pili are suitable for cryogenic electron microscopy (cryo-EM) and various biochemical experiments.
Key features
• Pili purification for single-particle cryo-electron microscopy (Cryo-EM) analysis
• This protocol builds upon the F-pili purification method developed by Costa et al. [1] extending its application to the Acinetobacter genomosp. 16.
• It is optimized for higher and more stable pili yields, as well as increased reproducibility.
• The method is tested on various bacterial species and can be adapted to purify different types of pili.
Keywords: Host receptorGraphical overview
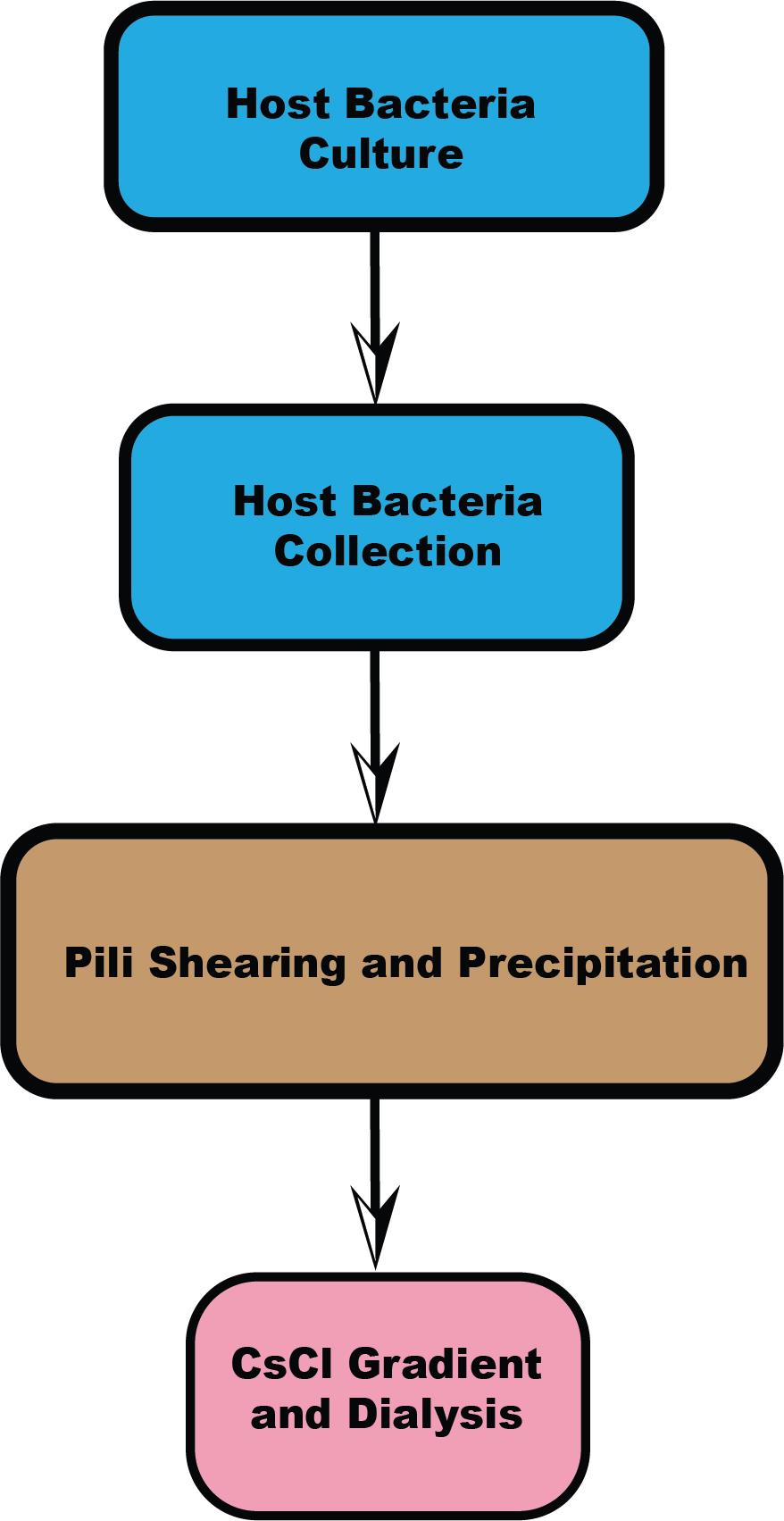
Workflow overview
Background
Acinetobacter, a Gram-negative bacterium, is an aerobic, non-flagellated, and widely distributed coccobacillus [2]. Acinetobacter baumannii, Acinetobacter calcoaceticus, Acinetobacter pittii, and Acinetobacter nosocomialis are opportunistic pathogens known for causing hospital-acquired infections. Bacteria can invade tissue and evade immune response through various mechanisms, including the production of enzymes. Adherence to the host is facilitated by structures such as pili [3–5]. Each year, U.S. hospitals report around 12,000 infections caused by multidrug-resistant Acinetobacter species. The protein secretion processes in Acinetobacter, which are critical targets for vaccine development, are not well understood. Known protein secretion systems in Acinetobacter include the Type II secretion system (T2SS), Type VI secretion system (T6SS), Type IV pilus secretion system, autotransporters, and outer membrane vesicles (OMVs) [6].
In this protocol, we focus on the type IV pili purification from Acinetobacter genomosp. 16 (A. gp16, NCBI: txid70347), which is a target host receptor of single-stranded RNA phage AP205 (ssRNA phage AP205, a virus that can infect Acinetobacter) [7]. Given the crucial role retractile pili play in the virulence of many bacteria, if infection-dependent pili detachment is a common feature of ssRNA phages, these simple viruses could potentially be used to inhibit retractile pili deployment and thereby decrease the virulence of numerous significant bacterial infections [7]. The purified pili can not only be utilized for Cryo-EM structural studies but also for biochemical or biophysical experiments to test protein binding affinity.
Type IV pili are extracellular helical appendages, typically with a diameter of 6–9 nm [8], which are composed of thousands of noncovalently linked pilin subunits. The major pilin of Acinetobacter type IV pili is the PilA monomer, which is composed of an N-termini hydrophobic α-helix, an α-β loop for connection, and the C-termini soluble β-sheet. The mature pilins assemble into type IV pilus by type IV pilus secretion system. Pilins bury the N-terminus helix inside, forming the pilus’s central axis, with the C-terminus headgroup (β-sheet) exposed outside [9].
Several protocols have been established for the purification of different types of pili [1,10,11]. While these methods share commonalities, specific steps can vary depending on the bacterial strain. We tested various pili purification techniques and found that the method developed by Costa et al. [1] for Escherichia coli (E. coli) F-pili provided higher efficiency and yield than other methods. First, we optimized and highlighted critical steps within this protocol to improve and stabilize yield, such as using a syringe to facilitate pili detachment from cells and controlling temperature to prevent pili retraction. Second, we focused on optimizing the procedure for Acinetobacter pili purification, allowing for the collection and separation of two different types of pili simultaneously. There are two types of pili outside the cell boundary: type IV pili and FilA filament. Lastly, this paper outlines a four-section process and provides two options (one for hyper-piliated strains, the other for low-yield strains), making it suitable for pili from other bacteria species as well (Graphical overview). We found that some steps could be simplified when bacterial pili yield is high. For instance, Acinetobacter pili have a relatively higher yield compared to F-pili, which have only one or two pili per cell. Microscope negative staining can be used to visually assess the number of pili per cell, determining whether the strain is hyper-piliated or has a lower yield. Based on this assessment, one can decide whether to follow the method in Sections B and C or Sections B2 and C2. Sections B and C provide a quicker method for obtaining pili from Acinetobacter and other high-yield species. Additionally, alternative steps in Sections B2 and C2, which mainly optimize the method from Costa et al. [1] are provided for strains with lower pili yields (e.g., E. coli F-pili).
In summary, this protocol has been optimized based on previous pili purification methods to enhance yield and stability, particularly for Acinetobacter pili. This method can also be generalized for the purification of pili in other species.
Materials and reagents
Biological materials
Acinetobacter genomosp. 16 (A. gp16, NCBI: txid70347)
Reagents
Difco LB broth, Miller (Miller Broth) 500 g (BD Bioscience, catalog number: 244620)
Tris (Bio-Rad, catalog number: 1610719)
Sodium chloride (NaCl) (crystalline/certified ACS) (Fisher Scientific, catalog number: 7647-14-5)
Sodium citrate dihydrate (granular/certified) (Fisher Scientific, catalog number: 6132-04-3)
(Optional) Polyethylene glycol 6000 (PEG6000) (Thermo Fisher Scientific, catalog number: 25322-68-3)
Cesium chloride (Thermo Fisher Scientific, catalog number: 7647-17-8)
Solutions
SSC buffer (see Recipes)
Dialysis buffer (see Recipes)
5% PEG6000 (see Recipes)
CsCl step gradient buffer (see Recipes)
Recipes
Note: All in sterile water.
SSC buffer
Reagent (room temperature) Final concentration (pH = 7.2) Sodium citrate 15 mM NaCl 150 mM The buffer requires autoclaving.
Dialysis buffer
Reagent (room temperature) Final concentration (pH = 8.0) Tris-HCl 50 mM NaCl 150 mM The buffer requires autoclaving.
5% PEG6000
Dissolve PEG6000 into dialysis buffer at pH = 8.0. The final volume should be 1 L of dialysis buffer with 50 g of PEG6000. The buffer requires autoclaving.
CsCl step gradient buffer
Reagent (room temperature) Final concentration (pH = 8.0) CsCl The buffer needs different densities to help form the gradient in the centrifuge tube.
Preparing density at 1.0 g/cm3, 1.1 g/cm3, 1.2 g/cm3, 1.3g/cm3, 1.4 g/cm3Dialysis buffer
Laboratory supplies
Slide-A-LyzerTM dialysis cassettes, 20 K MWCO (Thermo Fisher, catalog number: 66003)
Sterile syringes for single use, 5 mL and 3 mL (Fisher Scientific, catalog numbers: 14-955-457, 14-955-458)
Disposable hypodermic needles 25 G, 23 G, and 18 G (Fisher Scientific, catalog numbers: 26406, 26408, 26420)
BD General Use and PrecisionGlide hypodermic needles (Fisher Scientific, catalog number: 305127)
FisherbrandTM Petri dishes with clear lids (Fisher Scientific, catalog number: 0875713)
FalconTM round-bottom polypropylene test tubes with cap (Fisher Scientific, catalog number: 14-959-11B)
PierceTM Protein Concentrators PES, 30 K MWCO (different sizes) (Thermo Fisher Scientific, catalog number: 88502, 88522, 88531)
Equipment
SW 41 Ti swinging-bucket rotor (Beckman, catalog number: 331362)
13.2 mL, open-top thin-wall ultra-clear tube, 14 × 89 mm, 50 Pk (Beckman, catalog number: 344059)
Procedure
Host bacteria culture
Day 1 afternoonRecover A. gp16 (NCBI: txid70347) from -80 °C storage and place on ice.
Using a sterile inoculation loop, obtain a small amount of bacteria from the thawed stock.
Streak the loop across the surface of an LB agar plate using the quadrant streak method to gradually dilute the bacterial concentration.
Incubate the plate at 30 °C overnight to allow colony formation. After incubation, select well-isolated single colonies for further analysis or experimentation (Figure 1A).
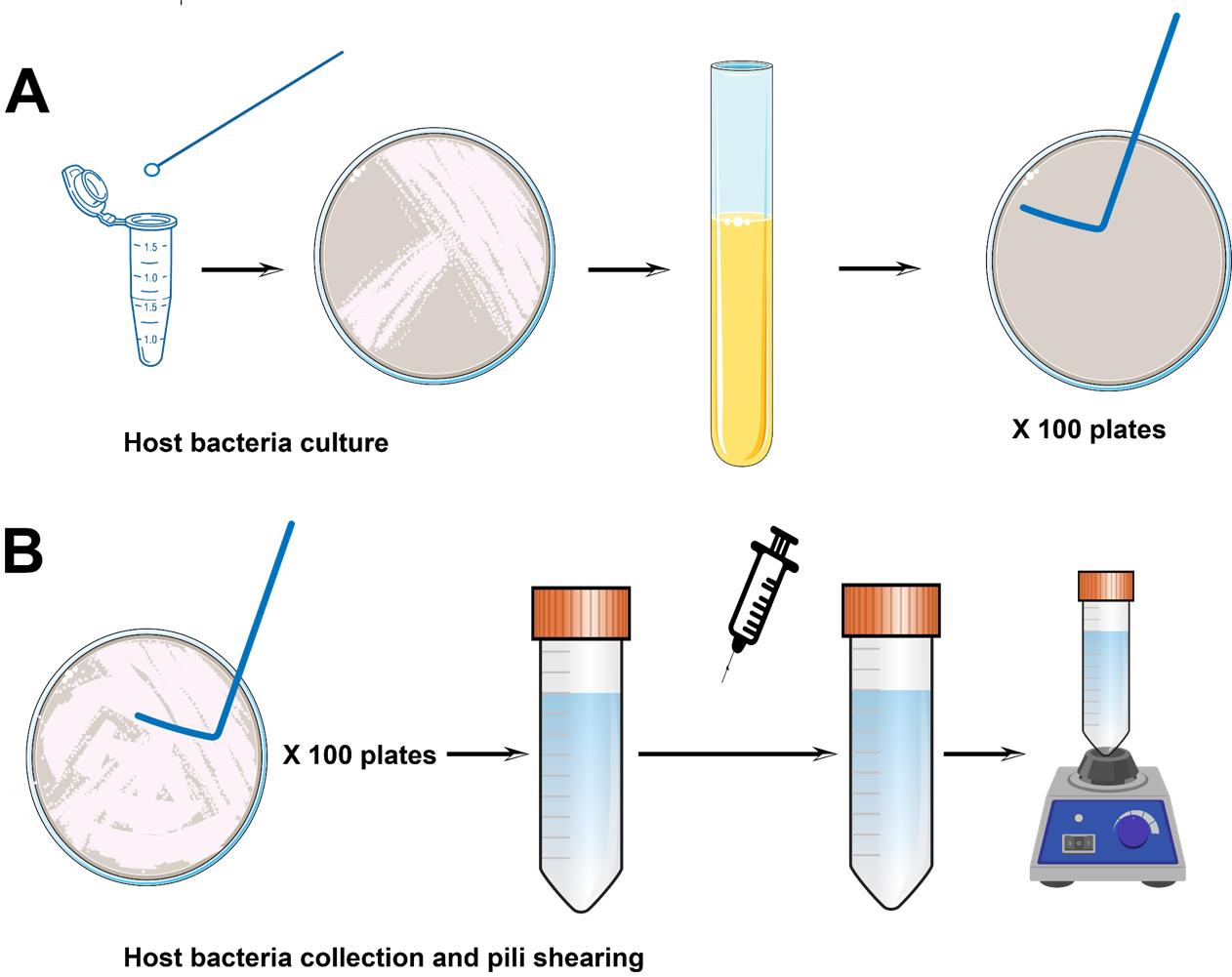
Figure 1. Host bacteria culture, collection, and pili shearing. (A) Pipeline for host bacteria inoculation and culture. (B) Pipeline for pili collection. L-shaped spreaders are used to gently scrape cells from the surface of the plate. The collected bacterial cell suspension is then passed through a syringe to further remove the pili, followed by vortexing.
Day 2 afternoon
Prepare 100 LB agar plates without antibiotics (for use on Day 3).
Pick a single colony and inoculate it into 3 mL of LB medium for overnight shaking at 30 °C.
Day 3In the morning (7 AM), take 100 µL from the 3 mL overnight culture and inoculate into 30 mL of LB medium. Incubate this A. gp16 cell culture with shaking at 200 rpm at 30 °C until the culture reaches OD600 = 0.1.
Note: For different strains, the target OD600 may vary. For example, for E. coli, OD600 = 0.6 is typical.
Add 100 µL of culture to each of the 100 prepared agar plates and spread evenly using a cell spreader.
Option 1: 100 plates. Most are for large-scale purification.
Option 2: 30 plates. For A. gp16 type IV pili, a smaller scale can also yield satisfactory results.
Option 3: Six large (25 cm × 25 cm) LB medium plates. This requires increasing the inoculum to 1 mL per plate. These larger plates reduce preparation time and simplify the procedure.
Incubate the plates at 30 °C for 30 min to allow the surface to dry slightly.
For overnight incubation, the plates should be inverted (bottom up, lid down) to prevent accumulated humidity from dripping onto the surface, which could affect pili formation. (Overnight incubation typically takes about 16 h. Deviating from this time frame may impact pili yield.)
(Option 1)
Host bacteria collection
Day 4Pre-chill the dialysis buffer (1 L) at 4 °C.
Take out the overnight plates from 30 °C and immediately place them on ice. Process five plates at a time to prevent pili retraction at room temperature.
Collect the bacterial cells from the plates using 300 µL of dialysis buffer with an L-shaped spreader, then collect the liquid from the plate surface using a 1 mL pipette into a 50 mL Falcon tube.
Note: For 25 cm × 25 cm plates, use approximately 8 mL of dialysis buffer (Figure 1B).
Pili shearing and precipitation
Day 4Pass the bacterial suspension through needles three times to shear the pili. Begin with the 18 G needle and then pass through the 25 G needle twice.
Note: Thinner needles help shear the pili but may clog.
Repeat the shearing process iteratively for all plates, processing 5–10 plates at a time. After shearing, vortex the liquid for 30 s (Figure 1B).
Centrifuge the cell solution at 3,470× g for 30 min at 4 °C to remove the cells.
Note: If the solution remains unclear, repeat the centrifugation. Higher speeds (4,000–8,000× g) can also be used as long as the cells are adequately spun down.
Collect the pili from the supernatant using a pipette. If starting with 30 plates, concentrate the pili solution to 2–3 mL using centrifugation. For larger volume purification (100 plates), concentrate the pili solution to 8–10 mL.
(Option 2)
For high-yield pili, such as A. gp16, this method is unnecessary. Sections B2 and C2 below can be used as an alternative method for pili purification of other low-yield species. It involves more steps and is relatively complicated. Refer to the general notes section for detailed explanations.
B2. Host bacteria collection (generalized method for all other kinds of pili purification)
Day 4
Pre-chill the SSC buffer at 4 °C.
Place the overnight plates on ice, processing five plates at a time to prevent pili retraction at room temperature.
Collect the bacterial cells from the plates using 500 µL of SSC buffer and an L-shaped spreader. Then, collect the liquid from the plate surface using a 1 mL pipette. For 25 cm × 25 cm plates, typically use 8 mL of SSC buffer. Slightly more buffer can be used in this step.
C2. Pili shearing and precipitation (generalized method for all other kinds of pili purification)
Day 4
Pass the bacterial suspension through needles three times to shear the pili. Begin with the 18 G needle first and then pass through the 25 G needle twice.
Note: Thinner needles help shear the pili but may clog.
Repeat the shearing process iteratively for all plates, processing 5–10 plates at a time, and vortex the liquid for 30 s.
After collecting all the plates, adjust the total volume of the solution to 1 L by adding pre-chilled SSC buffer. Incubate the liquid suspension at 4 °C for 2 h and stir gently.
Centrifuge the cell solution at 8,000× g for 30 min at 4 °C to remove any remaining cells. Centrifuge twice to make sure cells are removed, and then collect the supernatant using a pipette.
As the pili are present in large volumes, precipitate the supernatant by adding 5% PEG6000 (diluted from 50% PEG) and 500 mM NaCl. After 4 h of incubation at 4 °C, collect the precipitate by centrifuging the suspension at 15,000× g for 40–50 min, twice.
At this stage, resuspend the purified pili pellet in 2 mL of 50 mM Tris-HCl, 200 mM NaCl, pH 8.0 (F pilus).
Dialysis the 2 mL pili samples using dialysis buffer overnight.
Pili purification and dialysis
Days 5 and 6CsCl step gradient setup: The purification steps need CsCl step gradients of 1.1, 1.2, and 1.3 g/cm3 (4 mL, 3 mL, and 3 mL, respectively). If the band is formed high in the tube, consider adding 1.4 g/cm3 for 0.5 mL and reducing 1.3 g/cm3 to 2.5 mL.
When loading the CsCl gradient, ensure the needle reaches the bottom of the centrifuge tube. Different density layers are loaded onto the tube one by one. Begin by loading the low-density gradient, followed by the middle-density gradient, and finally the highest-density gradient. The needle should always reach the bottom of the centrifuge tube. Gently depress the syringe plunger to allow the lighter gradients to rise smoothly. Finally, load around 1.5 mL of the pili sample onto the top of the gradient solution (Figure 2A).
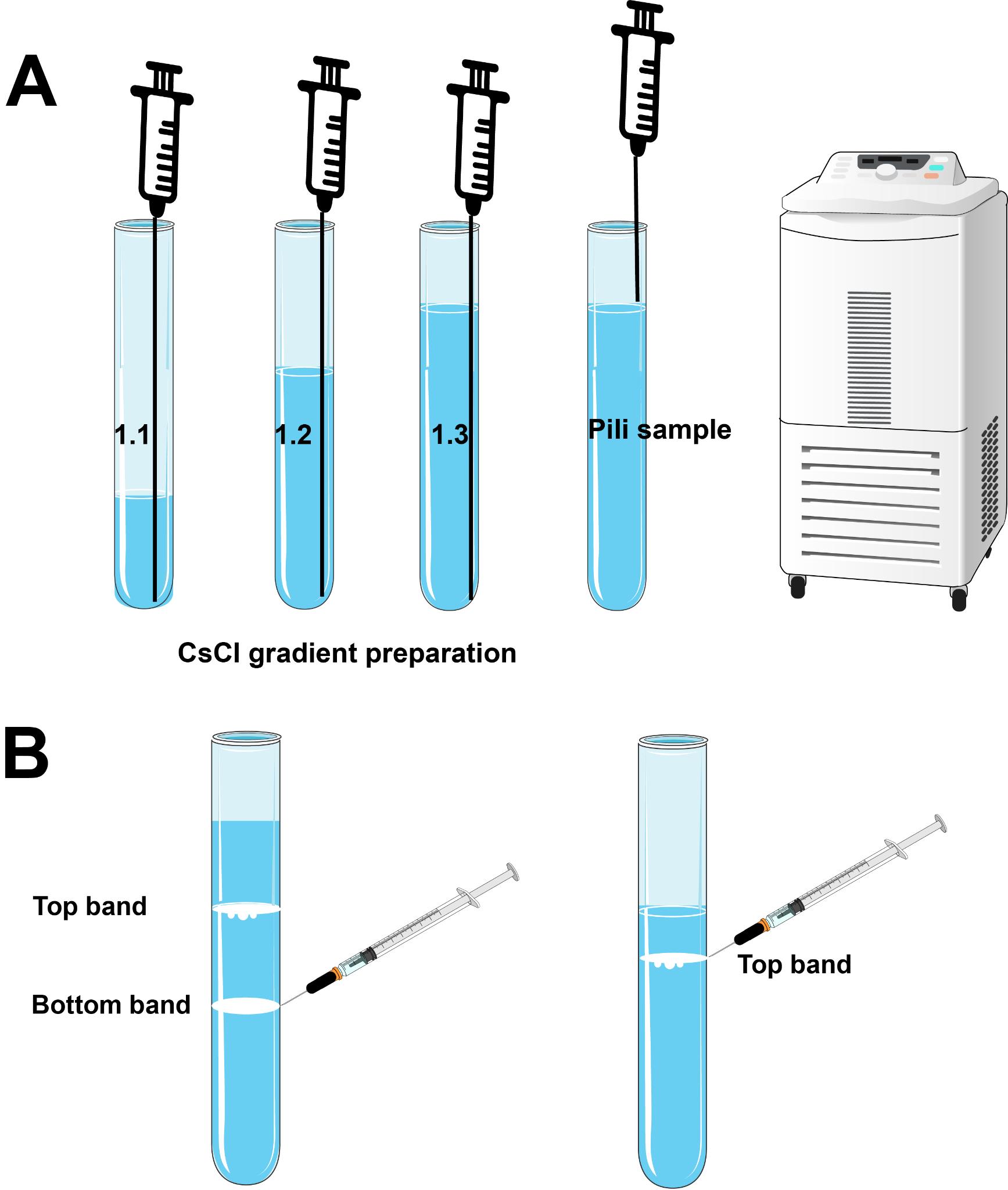
Figure 2. CsCl gradient preparation, sample loading, and sample collection. (A) CsCl gradient preparation. When loading the CsCl gradient, ensure the needle reaches the bottom of the centrifuge tube. Begin by loading the low-density gradient, followed by the middle-density gradient, and finally the highest-density gradient. The needle should always reach the bottom of the centrifuge tube. Gently depress the syringe plunger to allow the lighter gradients to rise smoothly. The pili sample is loaded onto the top of the gradient solution in the final step. (B) When extracting the bands, remove the bottom band first to avoid disturbing the gradients above it.Centrifuge the sample at 19,200× g for 19 h at 4 °C using SW 41 Ti swinging-bucket rotor. Ensure proper tube balance before starting centrifugation.
Note: For A. gp16, there will be two bands formed within the tube—the top band represents curved, thin pili, while the bottom band is composed of straight, thicker pili, which is the type IV pili. This is verified by the ssRNA phage as an antibody as well as mass spec. For F-pili, only one band forms in the tube, as strain MC4100 cells produce only one type of pili.
Use a syringe with a 23 G needle to extract the pili band from the tube.
Note: We prefer the 23 G needle because it is thin enough to precisely extract a single band while being sufficiently rigid to puncture the side of the centrifuge tube (Figure 2B).
Carefully remove the pili band and extensively dialyze it against the dialysis buffer using a dialysis cassette.
Perform initial dialysis for 4 h in 1 L of buffer, followed by overnight dialysis in 2 L of fresh buffer, and finally another 4 h of dialysis in 2 L of buffer.
Confirm the presence of pili using SDS-PAGE and verify their identity using LC-ESI MS/MS. Two types of pili are present in A. gp16. The presence and purity of type IV pili are confirmed by negative-stain electron microscopy.
Note: We used 4%–20% acrylamide gradient gels.
In Section D, the concentrated pili solution (approximately 1.5 mL) is layered on top of a CsCl gradient and centrifuged to separate into bands. For A. gp16, which yields relatively high amounts of pili, material collected from 15 plates and concentrated to 1.5 mL typically forms a visible band after CsCl gradient centrifugation. Processing pili from 100 plates yields about 9 mL of highly concentrated solution distributed into six CsCl tubes.
For low-yield strains, such as E. coli F-pili, the method described in Sections B2 and C2 should be used. This method is more complex but more broadly applicable to various pili purifications. It involves adding an SSC buffer, which aids in pili detachment. PEG6000 is used to precipitate and pellet the pili, which are then resuspended in 2 mL of dialysis buffer, resulting in a higher concentration per milliliter (containing pili from 100 plates in 2 mL). Refer to Sections B2 and C2 for details. Please note the bacterial culture temperature in Section A should be optimized if a different strain is used.
When preparing the CsCl gradient, start with 1.0, 1.2, and 1.4 g/cm3 gradients. Mix 1.0 and 1.2 to obtain a 1.1 g/cm3. Mix 1.2 and 1.4 to obtain a 1.3 g/cm3. It is better to check the density of the gradient by measuring the weight and dividing by the volume.
Different bacterial species may have multiple types of pili on the cell surface, so species other than A. gp16 might produce a different number of bands. For example, there is only one band formed for E. coli F-pili. Each layer of the band can be checked by negative stain electron microscopy (EM) to confirm its content.
We generally use 1.0, 1.1, 1.2, and 1.3 g/cm3 CsCl gradients for pili purification. If the pili band appears high in the tube, 0.5 mL of 1.4 g/cm3 CsCl can be added to slightly increase the overall density.
The CsCl gradient centrifugation should be balanced properly before loading on the high-speed centrifuge. The volume of solution in the centrifuge tube needs to be lower than the 13.2 maximum line but higher than the minimum.
Pre-check the strain: Use negative staining under a microscope to assess the number of pili outside the cell boundary.
Bacteria culture: When culturing bacteria on LB plates, invert the Petri dish to prevent moisture accumulation and dripping, which can limit pili yield.
Two critical steps: Ensure precise control of temperature to avoid pili retraction. Ensure the proper use of the syringe at least three times to remove pili from cells.
Centrifugation tips: When using a centrifuge and condenser to concentrate samples, resuspend the concentrated solution with a pipette between spins to prevent pili from sticking to the concentrator membrane.
Increase sample concentration: If the band is not visible, increase the concentration of the samples loaded into the CsCl gradient centrifugation tube. Typically, 1–1.5 mL of sample is loaded, which can contain pili from 15–20 plates or more. For F-pili, pili from 50 plates may be necessary to form a visible band.
- Costa, T. R., Ilangovan, A., Ukleja, M., Redzej, A., Santini, J. M., Smith, T. K., Egelman, E. H. and Waksman, G. (2016). Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell. 166(6): 1436–1444.e10.
- Ronish, L. A., Lillehoj, E., Fields, J. K., Sundberg, E. J. and Piepenbrink, K. H. (2019). The structure of PilA from Acinetobacter baumannii AB5075 suggests a mechanism for functional specialization in Acinetobacter type IV pili. J Biol Chem. 294(1): 218–230.
- Harding, C. M., Kinsella, R. L., Palmer, L. D., Skaar, E. P. and Feldman, M. F. (2016). Medically Relevant Acinetobacter Species Require a Type II Secretion System and Specific Membrane-Associated Chaperones for the Export of Multiple Substrates and Full Virulence. PLoS Pathog. 12(1): e1005391.
- Jones, A., Morgan, D., Walsh, A., Turton, J., Livermore, D., Pitt, T., Green, A., Gill, M. and Mortiboy, D. (2006). Importation of multidrug-resistant Acinetobacter spp infections with casualties from Iraq. Lancet Infect Dis. 6(6): 317–318.
- Dijkshoorn, L., Nemec, A. and Seifert, H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 5(12): 939–951.
- Costa, T. R. D., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M. and Waksman, G. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 13(6): 343–359.
- Meng, R., Xing, Z., Chang, J. Y., Yu, Z., Thongchol, J., Xiao, W., Wang, Y., Chamakura, K., Zeng, Z., Wang, F., et al. (2024). Structural basis of Acinetobacter type IV pili targeting by an RNA virus. Nat Commun. 15(1): 2746.
- Strom, M. S. and Lory, S. (1993). Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 47(1): 565–596.
- Mattick, J. S. (2002). Type IV Pili and Twitching Motility. Annu Rev Microbiol. 56(1): 289–314.
- Craig, L. and Altindal, T. (2019). Purification of Type IV Pili and Pilin Subunits. Methods Mol Biol.: 97–110.
- Date, T., Inuzuka, M. and Tomoeda, M. (1977). Purification and characterization of F pili from Escherichia coli. Biochemistry. 16(25): 5579–5585.
- Meng, R., Jiang, M., Cui, Z., Chang, J. Y., Yang, K., Jakana, J., Yu, X., Wang, Z., Hu, B., Zhang, J., et al. (2019). Structural basis for the adsorption of a single-stranded RNA bacteriophage. Nat Commun. 10(1): 3130.
Validation of protocol
The purification method described in this protocol has been validated in two cryo-EM structure papers published in Nature Communications [7,12]. In Figure 3A, we present bands formed during two different pili purification batches. The “Pili Only” tube (right) illustrates typical results after type IV pili purification, where Layer 1 contains curved, thin FilA pili and Layer 2 contains straight, thick type IV pili. Cryo-EM analysis (Figure 3C and 3D) confirmed the purity of these layers. In A. gp16 cells, there are more curved, thin FilA pili than the straight, thick type IV pili outside cell boundaries. This is why in the final image, curved, thin pili showed relatively higher concentration.
In Figure 3A, the “Pili-Phage” tube (left) demonstrates an experiment where phage was premixed with the pili solution before the CsCl step. The phage with RNA of higher density appears in Layer 3 as a reference. The image shown in Figure 3B shows the phage and host receptor adsorption of purified pili from Layer 1 and Layer 2. (Purified AP205 phage was added into a mixture of pili from layer 1 and layer 2.) The red arrow points to straight, thick type IV pili with phage, while the blue arrow indicates curved, thin pili, which do not act as host receptors and show no phage adsorption. The scale bar represents a length of 500 A.
For mass spec and cryo-EM results for layers 1 and 2 please see the published paper’s supplementary section [7].
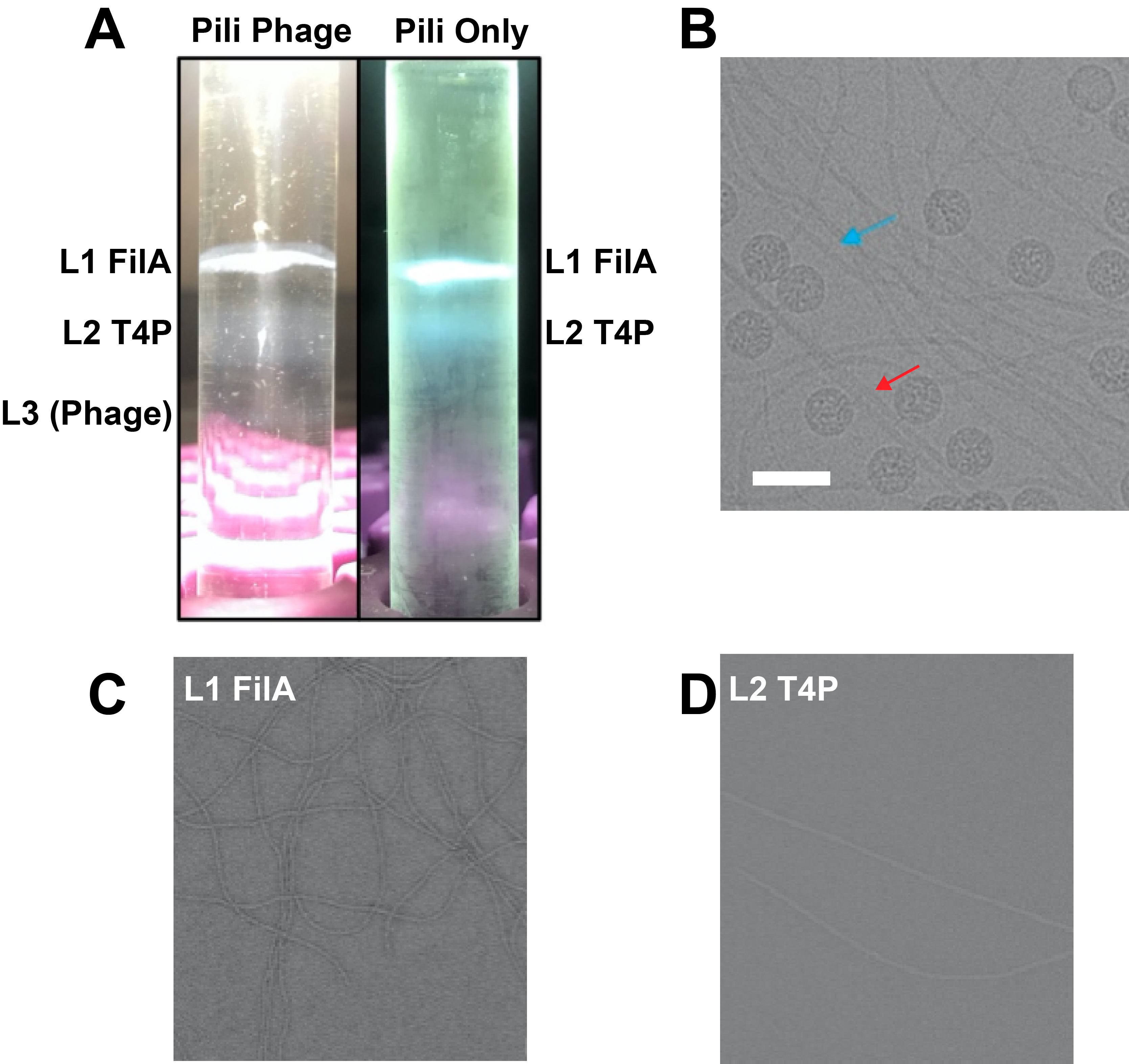
Figure 3. Validation of purified samples using EM. (A) Sample bands formed during two different purifications. The “Pili Only” tube (right) shows results after type IV pili purification, with the top band (Layer 1) containing curved, thin FilA pili, and the bottom band (Layer 2) containing straight, thicker type IV pili. The “Pili-Phage” tube (left) demonstrates an experiment where phage was premixed with the pili solution. The phage, containing RNA of higher density, appears in Layer 3 as a reference. (B) Cryo-EM image of purified pili from Layer 1 and Layer 2, along with additional purified AP205 phage. The red arrow points to straight, thick type IV pili bound with phage, while the blue arrow indicates curved, thin FilA pili, which are not host receptors and thus show no phage adsorption. The scale bar represents a length of 500 A. (C) Cryo-EM image of Layer 1 curved, thin pili. (D) Cryo-EM image of Layer 2 straight, thick pili.
General notes and troubleshooting
General notes
Troubleshooting
If no band forms at the end, it indicates either that the strain has a low yield, or a critical step was not correctly performed.
To increase reproducibility and avoid sample loss, consider the following tips:
Acknowledgments
This protocol is based on a published research paper: Meng, R., Xing, Z., Chang, J. Y., Yu, Z., Thongchol, J., Xiao, W., Wang, Y., Chamakura, K., Zeng, Z., Wang, F., et al. (2024). Structural basis of Acinetobacter type IV pili targeting by an RNA virus. Nat Commun. 15(1): 2746 [7].
Competing interests
There is no competing interest.
References
Article Information
Publication history
Received: Jun 22, 2024
Accepted: Aug 27, 2024
Available online: Sep 25, 2024
Published: Oct 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Meng, R. (2024). Host Receptor Pili for Cryo-EM Single-Particle Reconstruction. Bio-protocol 14(20): e5094. DOI: 10.21769/BioProtoc.5094.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Biophysics > Electron cryotomography
Biochemistry > Protein > Self-assembly
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








