- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation and Maintenance of Patient-Derived Endometrial Cancer Organoids
(*contributed equally to this work) Published: Vol 14, Iss 20, Oct 20, 2024 DOI: 10.21769/BioProtoc.5093 Views: 2042
Reviewed by: Alessandro DidonnaRakesh Bam

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Isolation of Mammary Epithelial Cells and Fibroblasts From Mouse Tumor
Shiva Kazerounian
Feb 20, 2014 20811 Views
Isolation and Immortalization of Fibroblasts from Different Tumoral Stages
Fernando Calvo [...] Erik Sahai
Apr 5, 2014 20308 Views

In vivo Tumor Growth and Spontaneous Metastasis Assays Using A549 Lung Cancer Cells
Lei Qi [...] Min Chen
Apr 5, 2020 8819 Views
Abstract
Endometrial cancer (EC) is the leading cause of gynecologic cancer morbidity and mortality in the U.S. Despite advancements in cancer research, EC death rates are increasing, particularly high-grade endometrial cancers. The development of three-dimensional (3D) patient-derived organoid (PDO) models for EC is crucial, as they provide a more accurate representation of the biological and genetic complexity of a patient’s tumor compared to traditional 2D cell lines. Here, we describe a protocol for cultivating PDO models from normal endometrium and EC across different EC subtypes. These EC PDO models can be expanded across multiple passages and facilitate the exploration of tumor behavior and drug responses, thereby advancing our understanding of the disease and potentially leading to more effective and individualized novel therapeutic strategies.
Key features
• Establishment of patient-derived EC and normal endometrium organoids.
• Accurate replication of the various histologic and molecular subtypes of EC within the organoids.
• Our approach provides a clinically relevant platform for studying EC development, metastasis, progression, and recurrence.
• It offers potential for developing targeted therapeutic interventions tailored to specific EC subtypes.
Keywords: Endometrial cancerBackground
Endometrial cancer (EC) is the fourth most common cancer and the sixth leading cause of cancer mortality among women in the US. It is one of the few cancers experiencing a rapid rise in both incidence and mortality rates, and by 2040, it is projected to become the third most prevalent cancer and the fourth leading cause of cancer death, surpassing lung and colorectal cancers. Furthermore, EC presents a significant challenge due to its racial disparity, with the largest Black/White difference in 5-year relative survival among all cancers (63% vs. 84%) [1–4]. These concerning trends highlight the urgent need for research into the molecular and cellular basis of EC to advance our understanding and develop more effective therapeutic strategies.
Immortalized 2D cell lines and mouse models have been commonly used for EC research; however, they do not provide reliable models. 2D cell culture systems lack the critical 3D structure inherent in primary endometrial tissue, affecting glandular cell function influenced by their microenvironment and orientation. Similarly, mouse models do not accurately represent the biology of the human endometrium, highlighting the need for reliable biological models that mimic human EC biology [5,6].
Patient-derived organoids (PDOs) are 3D structures that precisely replicate the microenvironment and cellular composition of endometrial tissue, providing a clinically relevant model for studying specific histologic and molecular subtypes of EC [7–11]. This is particularly valuable for the rarer and more aggressive EC subtypes that disproportionately affect minority populations, especially African American women [2,12–14]. Additionally, the establishment of PDOs addresses the limitation of tissue samples by providing a sustainable cellular resource for ongoing research [15]. PDOs have been shown to maintain their phenotype and genotype through multiple passages, making them reliable models that preserve biological integrity over extended periods in culture [16,17]. However, there remains a need to enhance organoid culture techniques, as organoid cultures may exhibit heterogeneity and variable complexity in cellular composition, as well as poorly controlled morphogenesis during the self-assembly process [18].
Here, we present an optimized method for establishing, expanding, and banking endometrial organoids derived from patient tissue samples of normal endometrium and endometrial cancers.
Materials and reagents
Biological materials
Fresh tissue endometrial specimen in RPMI-1640 medium (Thermo Fisher Scientific, catalog number: 11879020)
Advanced DMEM/F12 (Thermo Fisher Scientific, catalog number: 12634028)
Matrigel® matrix (Corning, catalog number: 356234)
RPMI-1640 medium (Sigma-Aldrich, catalog number: R73-88)
GlutaMAX (Thermo Fisher Scientific, catalog number: 35050061)
HEPES (Sigma-Aldrich, catalog number: S7067)
R-Spondin conditioned media (homemade) [19], filter through a 0.2 μm mesh prior to use
N2 supplement (Life Technologies, catalog number: 17502048)
B27 supplement minus vitamin A (Thermo Fisher Scientific, catalog number: 12587010)
Chemically defined lipid concentrate (Thermo Fisher Scientific, catalog number: 11905031)
Recombinant human Noggin (PeproTech, catalog number: 120-10c)
Primocin (InvivoGen, catalog number: ant-pm-1)
Nicotinamide (Sigma-Aldrich, catalog number: N0636)
N-Acetyl-L-cysteine (NAC) (Sigma-Aldrich, catalog number: A9165-5G)
Y-27632 dihydrochloride (Tocris, catalog number: 1254)
A83-01 (Sigma-Aldrich, catalog number: SML0788)
Recombinant human EGF (PeproTech, catalog number: AF-100-15)
Recombinant human FGF-α (PeproTech, catalog number: 100-26)
Recombinant human FGF-4 (PeproTech, catalog number: 100-31)
Recombinant human HGF (PeproTech, catalog number: 100-39)
Recombinant human IGF-1 (PeproTech, catalog number: 100-11)
Recombinant human FGF-10 (PeproTech, catalog number: 100-26)
Recombinant human FGF-β (PeproTech, catalog number: 100-18B)
SB202190 (p38i) (Sigma-Aldrich, catalog number: S7067)
17-β Estradiol, water soluble (Sigma-Aldrich, catalog number: E4389)
Collagenase from Clostridium histolyticum (Sigma-Aldrich, catalog number: C9407)
Cell recovery solution (Corning, catalog number: 354253)
RecoveryTM cell culture freezing medium (Thermo Fisher Scientific, catalog number: 12648010)
TrypLETM express enzyme (1×), no phenol red (Thermo Fisher Scientific, catalog number: 12604103)
Phosphate buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 10010023)
ACK buffer lysis (Sigma-Aldrich, catalog number: A1049201)
Trizol reagent (Fisher Scientific, catalog number: 15-596-018)
0.4% trypan blue solution (Thermo Fisher, catalog number: 15250061)
Human tumor endometrial organoid media (see Recipes)
Human normal endometrium organoid media (see Recipes)
Human tumor endometrial organoid media (50 mL)
Reagent Final concentration Quantity or Volume Advanced DMEM/F12 1× 30 mL GlutaMAX 1% 500 μL HEPES 12.5 mM 500 μL R-Spondin conditioned media 10% 5 mL B27 2% 1 mL N2 1% 500 μL Lipid concentrate 1% 500 μL Noggin (NOG) 100 ng/mL 5 μL Primocin 100 μg /mL 100 μL Nicotinamide 2 mM 250 μL NAC 1.25 mM 102 μL Y-27632 dihydrochloride 10 μM 5 μL A83-01 0.25 μM 0.25 μL hEGF 50 ng/mL 2.5 μL hFGF-α 25 ng/mL 2.5 μL hFGF-4 50 ng/mL 5 μL hHGF 20 ng/mL 4 μL hIGF-1 40 ng/mL 2 μL SB202190 0.1 μM 0.25 μL 17-β Estradiol 10 nM 5 μL Total (optional) n/a 50 mL Store at 4 °C for a maximum of 10 days.
Human normal endometrium organoid media
Reagent Final concentration Quantity or Volume Advanced DMEM/F12 1× 30 mL GlutaMAX 1% 500 μL HEPES 12.5 mM 500 μL R-Spondin conditioned media 15% 7.5 mL B27 2% 1 mL N2 1% 500 μL Lipid concentrate 1% 500 μL Noggin (NOG) 100 μg/mL 5 μL Primocin 100 μg /mL 100 μL Nicotinamide 2 mM 250 μL NAC 1.25 mM 102 μL Y-27632 dihydrochloride 10 μM 5 μL A83-01 0.25 μM 0.25 μL hEGF 50 ng/mL 2.5 μL hFGF-10 10 ng/mL 1 μL FGF-β 2 ng/mL 2 μL SB202190 10 μM 25 μL 17-β Estradiol 1 nM 0.5 μL Total (optional) n/a 50 mL Store at 4 °C for a maximum of 10 days.
Laboratory supplies
Falcon 15 mL tube (Corning, catalog number: 352097)
Falcon 50 mL tube (Corning, catalog number: 352098)
Eppendorf 1.5 mL tube (Eppendorf, catalog number: 22364116)
Eppendorf 5.0 mL tubes (Eppendorf, catalog number: 0030119487)
CRYOVIAL® internal thread with silicone washer seal (Simport, catalog number: T311-1)
Mini-strainer 40 μm mesh (Corning, catalog number: CLS431750)
Mini-strainer 100 μm mesh (Corning, catalog number: CLS431752)
Falcon® 6-well cell culture plate (Corning, catalog number: 353046)
PYREX® 100 × 10 mm Petri dish (Corning, catalog number: 3160-100)
Pipette tips, 10, 20, 200, 1000 μL
Cell lifter (Corning, catalog number: 3008)
Forceps (Roboz Surgical Instrument, catalog number: RS-5136)
Scissors (Roboz Surgical Instrument, catalog number: RS-5910)
Equipment
Sterile tissue/cell culture hood (Labgard Class II TYPE A2)
Humidified CO2 incubator (Binder Incubator, model: CB150)
Water Bath (Sheldon Manufacturing Shel Lab H2O Bath Series)
Refrigerated centrifuge (Eppendorf, model: Centrifuge 5810R)
Light microscope (Nikon, model: Microscope Eclipse Ts2)
Portable Pipet-Aid (Thermo Scientific, model: S1 Pipet Fillers)
Cell drop automated cell counter (DeNovix, catalog number: CellDrop BF)
Fridge and freezers (4 °C, -20 °C, and -80 °C)
Software and datasets
NIS-Elements D Imaging Software
Images were acquired using the NIS-ELEMENT D_V5 software (Nikon) with a Nikon Eclipse TS2 microscope.
Procedure
Fresh tumor and normal endometrial tissue specimens were obtained from patients undergoing hysterectomy for EC. Institutional Review Board approval was obtained (study IRB #18-0897), and all patients provided informed consent prior to specimen collection. The hysterectomy specimens were transported from Northwell Health LIJ Medical Center to Cold Spring Harbor Laboratory shortly after surgery or the following morning. If tissue could not be delivered or digested the same day as the surgery, it was stored overnight at 4 °C.
Generation of organoids from endometrial tumor and normal tissue
Pipette 4.5 mL of RPMI media from the sample and transfer to a 15 mL Falcon tube.
Remove the rest of the media from the original specimen tube and wash the tissue once with 5 mL of cold 1× PBS.
Note: If the sample is bloody, add 1 mL of ACK buffer lysis to the plate and keep on ice for 10–15 min. Spin and then remove supernatant. Proceed to step A3.
Mince 5 g of tissue with shears in a Petri dish to obtain 1 mm fragments.
Transfer the tissue to a 5 mL centrifuge tube with 4.5 mL of RPMI, 500 μL of 1 mg/mL Collagenase IV, and 10 μM Y-27632 dihydrochloride.
Incubate for 1–1.5 h at 37 °C. Vigorously shake the Falcon tube approximately 10 times every 15–30 min.
Evaluate digestion for dissociated fragments under light microscopy intermittently.
Allow any remaining tissue pieces to settle to the bottom of the tube. Then, collect the supernatant and transfer to a new Falcon tube. Alternatively, filter through a 100 μm mesh. Spin down at 300× g for 5 min at 4 °C.
Remove the supernatant and resuspend the pellet in a mixed solution of 1 mL of TrypLE and 10 μM Y-27632 dihydrochloride.
Incubate for a maximum of 15 min at 37 °C. Every 5 min, pipette up and down before placing it back in the incubator at 37 °C, confirming that the larger pieces of tissue are being digested. Aim for small clumps of organoids, not single cells.
Dilute contents 1:1 with advanced DMEM to stop the reaction. Pipette up and down to mix and then spin down at 300× g for 5 min at 4 °C.
Remove supernatant and resuspend in a separate centrifuge tube in a 70:30 Matrigel and media mixture (900 μL of Matrigel and 300 μL of media for a total of 12,00 μL of volume).
Plate densely (100 cells/μL) in a 6-well plate and allow 5–10 min for the dome to polymerize in the CO2 incubator. Plate four domes (50 µL each) in each well.
Add 3 mL of culture media to each well and place in a 30 °C incubator.
On passage 0, day 1:
Check back the next day to assess organoid growth and health. The human normal crypts should have formed oval-shaped organoids. Organoids from the tumor isolation can vary greatly in size but check to see that they are growing and not dark.
On passage 0, day 2:
Freeze some normal human organoids before they turn dark (refer to Section C). If possible, take two tubes of Trizol for RNA isolation as well. Also, take two tubes of Trizol for RNA isolation of the human tumor organoids. Split (see Section B) some of the normal human organoids to see how they respond to passaging. This can be a trial run with only a few wells or more if you want to try various techniques.
On passage 0, day 3:
Freeze some human tumor organoids. Test their response to passaging on this day. Check on the normal human organoids that were split the day before and split most or all of the remaining ones this day.
On passage 0, day 4:
Check back on the tumor organoids. Split most or all of the remaining tumor organoids, if needed. Check to see if less dense normal organoids need to be combined into one well. Do not forget to exchange media with new media (2.7 mL).
Continue to assess confluency, dissociation requirements, and consolidation as your P1 organoids grow. From this point, the normal and tumor organoids can differ greatly in their needs and must be considered at different time points due to their growth rate (Figure 1).
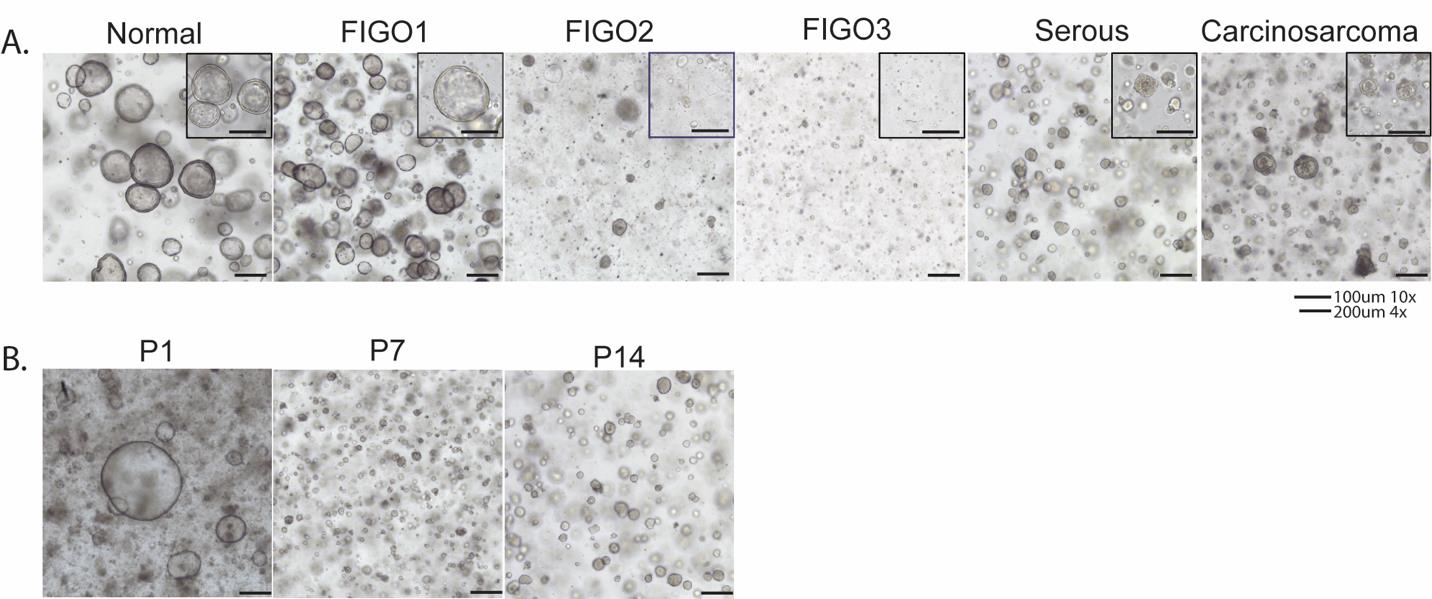
Figure 1. Patient-derived organoids from normal endometrium and cancerous tissues. A. Organoids from normal endometrium, FIGO1 (low grade), FIGO2 (intermediate grade), FIGO3 (high grade), serous cancer, and carcinosarcoma demonstrating that all histologic subtypes can be cultured and maintained through several passages. B. Serous cancer organoids demonstrating survival and maintenance over several passages in culture at passage 1 (P1), passage 7 (P7), and passage 14(P14). Scale bars: 100 μm (10×) and 200 μm (4×).Passaging of organoids
Aspirate media from desired wells and scrape off the Matrigel domes using a cell lifter.
Tilt the plate and collect all the Matrigel and organoids into the corner of the well closest to you.
Use Corning’s cell recovery solution (CRS) to collect the dome (minimum 3× volume of Matrigel domes collected) and then transfer to a new Eppendorf tube. For example, for 200 μL of Matrigel, add 600 μL of minimum CRS (for 3 wells, 1.2 mL).
Keep the tube on ice for 1 h or until organoids visibly settle. Evaluate intermittently under light microscopy for dissociation into single cells.
Spin down at 300× g for 5 min at 4 °C.
Remove supernatant and resuspend the pellet with TrypLE + 10 μM Y-27632 (≥ 500 μL of TrypLE for a 25 μL cell pellet). Lightly pipette up and down to a maximum of 10 times to break up the pellet.
Incubate at 37 °C for 10 min.
Mix the solution to break apart organoids and assess for level of dissociation under light microscopy.
Repeat steps B7 and B8 as needed until organoids are broken down to the desired level, checking every 5–10 min based on the rate of digestion. Neutralize the TrypLE with an equal volume of media.
Spin down at 300× g for 5 min at 4 °C.
Remove supernatant and resuspend with 75%/25% Matrigel and culture medium. (For 3 wells, 450:150 μL ratio; for 6 wells, 900:300 μL ratio.)
Plate and wait 10 min for the dome to polymerize at 37 °C.
Add culture medium to each well (2.7 mL/well in a 6-well plate).
Freezing organoids
Aspirate media from desired wells and scrape off the Matrigel domes using a cell lifter.
Tilt the plate and collect all the Matrigel and organoids into the corner of the well closest to you.
Use Corning’s cell recovery solution (CRS) or matrix melting solution (MMS) to collect the dome (minimum 3× volume of Matrigel domes collected), then transfer to a new Eppendorf tube.
Keep the tube on ice for 1 h.
Spin down at 300× g for 5 min at 4 °C.
Remove supernatant and suspend in 500 μL of freezing media.
Thawing frozen organoids
Remove cryovials from liquid nitrogen storage.
Warm in a 37 °C water bath for 1 min.
Add culture medium 1:1 (500 μL) and transfer to Eppendorf tube.
Spin down at 300× g for 5 min at 4 °C.
Remove supernatant and resuspend with 75%/25% Matrigel and culture medium. (For 3 wells, 450:150; for 6 wells, 900:300.)
Plate and wait 10 min for the dome to polymerize at 37 °C.
Add culture medium to each well (2.7 mL/well in a 6-well plate).
RNA isolation (low yield 50 cells/μL)
Aspirate media from desired wells.
Wash wells gently with 5 mL of 1× PBS and then remove PBS.
Add 400 μL of Trizol reagent directly to the well.
Break up Matrigel using a cell lifter and pipette up and down to mix, doing so until the solution is clear (all organoids/cells have been lysed).
Transfer to a labeled Eppendorf tube.
Vortex the tube for ~30 s.
Immediately freeze in -80°C.
RNA isolation (normal yield 100 cells/μL)
Aspirate media from desired wells and scrape off the Matrigel domes using a cell lifter.
Tilt the plate and collect all the Matrigel and organoids into the corner of the well closest to you.
Use Corning’s cell recovery solution (CRS) or matrix melting solution (MMS) to collect the dome (minimum 3× volume of Matrigel domes collected) without mixing.
Transfer the CRS with organoids to an Eppendorf tube and pipette up and down 10 times gently.
Keep on ice for 45 min.
Spin down at 300× g for 5 min at 4 °C.
Remove supernatant and resuspend the pellet with 400 μL of Trizol.
Pipette up and down until the solution is clear (all organoids/cells have been lysed).
Vortex the tube for 30 s.
Immediately freeze at -80 °C.
Quantification of organoids
Aspirate media from desired wells and scrape off the Matrigel domes using a cell lifter.
Tilt the plate and collect all the Matrigel and organoids into the corner of the well closest to you.
Use Corning’s cell recovery solution (CRS) or matrix melting solution (MMS) to collect the dome (minimum 3× volume of Matrigel domes collected), then transfer to a new Eppendorf tube.
Keep the tube on ice for 1 h.
Spin down at 300× g for 5 min at 4 °C.
Remove supernatant and suspend in 500 μL of culture media.
Add 1 μL of 0.4% trypan blue solution to 1 μL of suspension and vortex.
Place 1 μL of the mixture into a cell drop automated cell counter to determine quantification.
Validation of protocol
This protocol or parts of it has been used and validated in the following research articles:
Chung, et al. [20]. Abstract 2511: Autologous patient-derived organoid-immune cell co-culture platform for therapeutic discovery in high-grade endometrial cancer. Cancer Res.
Nizam, et al. [21]. Abstract LB236: Utilizing endometrial tumor organoids to model cancer immunomodulation. Cancer Res.
General notes and troubleshooting
General notes
Passaging should be done before organoids exhibit signs of stress or poor health. These signs are a noticeable accumulation of dead cells surrounding the organoid, loss of three-dimensional morphology, and darker color.
Collecting the dome can be done with cold 1× PBS or CRS. CRS may be helpful if you do not aim to dissociate with trypsin but just want to replate while getting rid of the Matrigel. It can also help if you notice that your organoids have not been breaking up as well as expected. To facilitate dissociation, organoids can be incubated on ice in CRS for ~20 min. When using 1× PBS or CRS, any volume from a few hundred microliters to 1 mL can be used at a time to wash the dome off.
Always orient Eppendorf tubes in the centrifuge correctly to help you predict where the pellet will form.
Organoids/cells should be pelleted properly before the supernatant is removed.
Organoid dissociation involves mechanical shearing, enzymatic, or non-enzymatic digestion. Mechanical shearing can be either with pipetting or mixing with a vortex. Enzymatic digestion utilizes trypsin or Trypl Express (TrypLE) to cleave proteins that hold the cells together. Non-enzymatic digestion via EDTA or gentle cell dissociation reagent helps break the organoids apart via calcium chelation. Normally, we will dissociate with trypsin for a few min to “loosen” up the organoid, before pipetting up and down to completely break it up. In some situations, organoids can only be mechanically sheared and then replated. This should also be considered if the organoids are very unhealthy and may not survive trypsinization. Mechanical shearing on its own also makes it extremely hard to get small organoids or single cells and is a benefit when you want to have grown organoids in only a few days. For dissociation, we use TrypLE (gentler) or Pan Trypsin (harsher). Cells will begin to lyse if left in trypsin or TrypLE for too long. Ideally, the duration of trypsinization is long enough that organoids are loose and will easily separate with a bit of pipetting. Another option is to use gentle cell dissociation reagent (GCDR) for non-enzymatic dissociation.
Collapsing domes: It is beneficial to first prewarm the plates while you are passaging, with an emphasis on 24 and 48 wells. The dome may easily collapse when plating on a room-temperature plate. Although most protocols state that a 70/30 Matrigel media ratio is best, this can vary based on your goals. For maintenance, you can go as low as 50/50, especially if the wells are large (6 wells). On the other hand, if plating in a 48-well plate, you could do 80/20 to help make sure the dome does not collapse. Pure Matrigel or a pre-made mixture can be added if working with exceedingly small amounts of cells.
Carry-over of unhealthy organoids: Healthy organoids will be bright and clear, but it is important to note when some organoids are dying but others are healthy. This can happen with carry-over from a previous passage and should be taken into consideration when deciding the next time to split. This can also occur when organoids compete for resources, i.e., they are plated densely or grow too large and take up too much room.
Acknowledgments
We thank Cold Spring Harbor Laboratory (CSHL) Cancer Center Organoid Shared Resource supported by NCI Cancer Center Support grant 5P30CA045508, Northwell Health Biospecimen Repository and Northwell Health Department of Obstetrics & Gynecology research staff for their assistance in recruiting and consenting patients and obtaining specimens for this protocol. S. B. acknowledges funding from The Mark Foundation for Cancer Research, Chan-Zuckerberg Initiative, The Oliver S. and Jennie R. Donaldson Charitable Trust and The CSHL and Northwell Health Affiliation.
Competing interests
No conflicts of interest or financial disclosures.
Ethical considerations
Informed consent was obtained from all human patients and protocol #18-0897 was approved by the Northwell IRB committee.
References
- Venkatesh, A. and Isaacs, C. (2024). Trends in Uterine Cancer Mortality in the United States: A 50-Year Population-Based Analysis. Obstet Gynecol. 143(4): e130–e131. https://doi.org/10.1097/aog.0000000000005543
- Siegel, R. L., Miller, K. D., Wagle, N. S. and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J Clin. 73(1): 17–48. https://doi.org/10.3322/caac.21763
- Desmond, D., Arter, Z., Berenberg, J. L., Killeen, J. L., Bunch, K. and Merritt, M. A. (2023). Racial and ethnic differences in tumor characteristics among endometrial cancer patients in an equal-access healthcare population. Cancer Causes Control. 34(11): 1017–1025. https://doi.org/10.1007/s10552-023-01716-9
- Park, A. B., Darcy, K. M., Tian, C., Casablanca, Y., Schinkel, J. K., Enewold, L., McGlynn, K. A., Shriver, C. D. and Zhu, K. (2021). Racial disparities in survival among women with endometrial cancer in an equal access system. Gynecol Oncol. 163(1): 125–129. https://doi.org/10.1016/j.ygyno.2021.07.022
- Gellersen, B., Brosens, I. and Brosens, J. (2007). Decidualization of the Human Endometrium: Mechanisms, Functions, and Clinical Perspectives. Semin Reprod Med. 25(6): 445–453. https://doi.org/10.1055/s-2007-991042
- Liu, T., Shi, F., Ying, Y., Chen, Q., Tang, Z. and Lin, H. (2020). Mouse model of menstruation: An indispensable tool to investigate the mechanisms of menstruation and gynaecological diseases (Review). Mol Med Rep. 22(6): 4463–4474. https://doi.org/10.3892/mmr.2020.11567
- Lancaster, M. A. and Knoblich, J. A. (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science (1979). 345(6194): e1247125. https://doi.org/10.1126/science.1247125
- Clevers H. (2016). Modeling Development and Disease with Organoids. Cell. 165(7):1586–1597. https://doi.org/10.1016/j.cell.2016.05.082
- Kim, J., Koo, B. K. and Knoblich, J. A. (2020). Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 21(10): 571–584. https://doi.org/10.1038/s41580-020-0259-3
- Boretto, M., Maenhoudt, N., Luo, X., Hennes, A., Boeckx, B., Bui, B., Heremans, R., Perneel, L., Kobayashi, H., Van Zundert, I., et al. (2019). Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol. 21(8): 1041–1051. https://doi.org/10.1038/s41556-019-0360-z
- Hibaoui, Y. and Feki, A. (2020). Organoid Models of Human Endometrial Development and Disease. Front Cell Dev Biol. 8: e00084. https://doi.org/10.3389/fcell.2020.00084
- Amant, F., Moerman, P., Neven, P., Timmerman, D., Van Limbergen, E, and Vergote, I. (2005) Endometrial cancer. Lancet. 366(9484): 491–505. https://doi.org/10.1016/s0140-6736(05)67063-8
- Bain, R. P., Greenberg, R. S. and Chung, K. C. (1987). Racial differences in survival of women with endometrial cancer. Am J Obstet Gynecol. 157(4 Pt 1):914–23. https://doi.org/10.1016/s0002-9378(87)80089-3
- Abel, M. K., Liao, C. I., Chan, C., Lee, D., Rohatgi, A., Darcy, K. M., Tian, C., Mann, A. K., Maxwell, G. L., Kapp, D. S., et al. (2021). Racial disparities in high-risk uterine cancer histologic subtypes: A United States Cancer Statistics study. Gynecol Oncol. 161(2): 470–476. https://doi.org/10.1016/j.ygyno.2021.02.037
- Fitzgerald, H. C., Dhakal, P., Behura, S. K., Schust, D. J. and Spencer, T. E. (2019). Self-renewing endometrial epithelial organoids of the human uterus. Proc Natl Acad Sci USA. 116(46): 23132–23142. https://doi.org/10.1073/pnas.1915389116
- Turco, M. Y., Gardner, L., Hughes, J., Cindrova-Davies, T., Gomez, M. J., Farrell, L., Hollinshead, M., Marsh, S. G. E., Brosens, J. J., Critchley, H. O., et al. (2017). Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol. 19(5): 568–577. https://doi.org/10.1038/ncb3516
- Katcher, A., Yueh, B., Ozler, K., Nizam, A., Kredentser, A., Chung, C., Frimer, M., Goldberg, G. L. and Beyaz, S. (2023). Establishing patient-derived organoids from human endometrial cancer and normal endometrium. Front Endocrinol (Lausanne). 14: e1059228. https://doi.org/10.3389/fendo.2023.1059228
- Zhao, Z., Chen, X., Dowbaj, A. M., et al. (2022). Organoids. Nat Rev Methods Primers. 2:94 https://doi.org/10.1038/s43586-022-00174-y
- Cantrell, M. A. and Kuo, C. J. (2015). Organoid modeling for cancer precision medicine. Genome Med. 7(1): 32. https://doi.org/10.1186/s13073-015-0158-y
- Chung, C., Nizam, A., Yueh, B., Subhash, S., Eskiocak, O., Frimer, M., Goldberg, G. L. and Beyaz, S. (2023). Abstract 2511: Autologous patient-derived organoid-immune cell co-culture platform for therapeutic discovery in high-grade endometrial cancer. Cancer Res. 83: 2511–2511. https://doi.org/10.1158/1538-7445.am2023-2511
- Nizam, A., Chung, C., Goldberg, G. L. and Beyaz, S. (2021). Abstract LB236: Utilizing endometrial tumor organoids to model cancer immunomodulation. Cancer Res. 81: LB236–LB236. https://doi.org/10.1158/1538-7445.am2021-lb236
Article Information
Publication history
Received: Jul 1, 2024
Accepted: Aug 25, 2024
Available online: Sep 29, 2024
Published: Oct 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Barbi, M., Gowthaman, D., Katcher, A., Gorman, M., Yueh, B., Nizam, A., Chung, C., Akyildiz, E. O., Frimer, M., Goldberg, G. L. and Beyaz, S. (2024). Generation and Maintenance of Patient-Derived Endometrial Cancer Organoids. Bio-protocol 14(20): e5093. DOI: 10.21769/BioProtoc.5093.
Category
Cancer Biology > General technique > Animal models > Cell isolation and culture
Biological Engineering > Biomedical engineering
Cell Biology > Cell isolation and culture > 3D cell culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









