- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Improved Focus-Forming Assay for Determination of the Dengue Virus Titer
Published: Vol 14, Iss 20, Oct 20, 2024 DOI: 10.21769/BioProtoc.5084 Views: 2391
Reviewed by: Luis Alberto Sánchez VargasRan ChenDebashis Dutta

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
On-demand Labeling of SNAP-tagged Viral Protein for Pulse-Chase Imaging, Quench-Pulse-Chase Imaging, and Nanoscopy-based Inspection of Cell Lysates
Roland Remenyi [...] Mark Harris
Feb 20, 2019 6977 Views
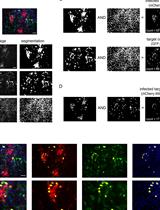
Image-based Quantification of Direct Cell-to-cell Transmission of Bovine Viral Diarrhea Virus
Fernando Merwaiss and Diego E. Alvarez
Aug 5, 2019 5318 Views
Primer ID Next-Generation Sequencing for the Analysis of a Broad Spectrum Antiviral Induced Transition Mutations and Errors Rates in a Coronavirus Genome
Shuntai Zhou [...] Ronald Swanstrom
Mar 5, 2021 7450 Views
Abstract
Dengue virus (DENV), a common and prevalent mosquito-borne endemic disease, is caused by four serotypes (DENV-1–4) and has spread rapidly on a global scale over the past decade. A crucial step in the development of antiviral therapeutics requires the utilization of in vitro cell-based techniques, such as plaque assays and focus-forming assays (FFA) for virus quantification. Vero cells have been widely used for FFA and plaque assay; however, there are instances when their efficacy and efficiency in the detection of certain clinical DENV isolates are low. Here, we showed that BHK-21 cells are more sensitive than Vero cells in the detection of all DENV-1–4 plaques and foci. In addition, we developed an improved FFA protocol for the quantification of all four DENV serotypes. Using a pan-flavivirus envelope (E) antibody, we reduce the possibility of false positives by defining a focus to consist of a minimum of eight infected cells. We outlined a protocol using the Operetta® high-content imaging system to automate the digital capture of these infected cells. A pipeline was also designed using the CellProfilerTM automated image analysis software to detect these foci. We then compare the results of the improved FFA with plaque assay. Notably, the improved FFA detected clear foci of the DENV-4 strain that does not form distinct plaques. We subsequently demonstrated the potential application of the improved FFA protocol in antiviral testing, utilizing a nucleoside inhibitor of DENV, NITD008 as a control. The protocol is amenable to a diverse array of applications, including high-throughput compound screening (HTS).
Key features
• An improved focus-forming assay that has the potential to quantify the DENV-4 strain, which was previously hard to plaque.
• Improvements have been made to reduce the possibility of false positives.
• Improved workflow using automated digital imaging process and counting of foci.
• Applicable to antiviral compounds screening and is amenable to high-throughput screening.
Keywords: DENV clinical isolatesBackground
Dengue represents a significant challenge to global public health systems, with over 40% of the global population at risk of infection. It is estimated that approximately 400 million people are infected with dengue virus (DENV) each year, of whom 100 million manifest clinical symptoms of dengue hemorrhagic fever with 25,000 deaths reported annually [1,2]. The symptoms include the abrupt onset of fever, which is followed by a multitude of other critical complications, including circulatory failure, hemorrhages in the skin and gastrointestinal tract, and shock. DENV comprises four antigenically distinct serotypes, exhibiting variations in both structural and nonstructural proteins. The four antigenically distinct serotypes of dengue virus are designated DENV-1, DENV-2, DENV-3, and DENV-4 [3,4]. The necessity for efficient, accurate, and robust assays for the quantification of infectious viruses, which contribute to the discovery of antiviral drugs, has thus increased. A variety of techniques have been explored and developed over the years, with plaque and/or focus-forming assays (FFA) still representing the gold standard for such assays. A variety of cell lines have been employed in these assays, with Vero (African green monkey kidney fibroblasts) cells being the most frequently utilized in plaque assay [5]. Furthermore, a number of optimization protocols for FFA have been documented for a range of viruses, including DENV [6,7]. However, this method typically necessitates a great number of steps, including the use of multiple distinct monoclonal antibodies for foci visualization; moreover, the method was not extended to DENV-3 and DENV-4 detection. In this work, we delineate the methodology for employing the FFA for the quantification of infectious virus particles using E protein detection in all serotypes and compare it with plaque assay quantification of infectious virus production. We demonstrate that the FFA approach was more sensitive than the plaque assay in detecting DENV-4, which exhibited poor plaque formation. In addition, all four DENV serotypes form clearer plaques and foci in BHK-21 cells than in Vero cells. Moreover, improvements have been made to the existing FFA. For example, we defined a focus to comprise a minimum of eight cells, thereby reducing the possibility of false positives. We also outlined a protocol using the Operetta® high-content imaging system and a pipeline using the CellProfilerTM software to automate foci imaging and foci counting. In addition, we demonstrate the immediate application of FFA as a tool for assessing the antiviral efficacy of compounds using a known adenosine nucleoside DENV inhibitor, NITD008 [9,10], as a control. Altogether, the improved FFA is a precise and reliable assay not only for quantifying clinical isolates but also for high-throughput screening of compounds for dengue drug discovery.
Materials and reagents
Biological materials
Representative Dengue virus strains from EDEN study [8]: DENV-1 clinical isolate (EDEN1: GenBank accession EU081230), DENV-2 clinical isolate (EDEN2: GenBank accession EU081177), DENV-3 clinical isolate (EDEN3: GenBank accession EU081190), DENV-4 clinical isolate (EDEN4: GenBank accession GQ398256)
Aedes albopictus C6/36 cell line (ATCC, CRL-1660TM)
Baby hamster kidney fibroblast, BHK-21 (ATCC, CCL-10TM)
African green monkey kidney fibroblast, Vero cell line (ATCC, CCL-81TM)
Hybridoma cells; 4G2 (ATCC, HB-112TM)
Reagents
RPMI1640 medium (Thermo Fisher Scientific, Gibco®, catalog number: 11875093)
DMEM (1×), high glucose, pyruvate medium (Thermo Fisher Scientific, Gibco®, catalog number: 11995-040)
Heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific, Gibco®, catalog number: 10082147)
200 mM L-glutamine (Thermo Fisher Scientific, Gibco®, catalog number: 25030081)
Penicillin and streptomycin (pen/strep) (Thermo Fisher Scientific, Gibco®, catalog number: 15140122)
1 M HEPES (Thermo Fisher Scientific, Gibco®, catalog number: 15630-080)
0.25% Trypsin-EDTA (Thermo Fisher Scientific, Gibco®, catalog number: 25200-056)
Protein-free hybridoma medium (PFHM-II medium) (Thermo Fisher Scientific, Gibco®, catalog number: 12040-077)
1× PBS (1st BASE, catalog number: BUF-2040-10X1L)
Ethanol (EtOH) molecular biology grade (Merck, Sigma-Aldrich, catalog number: 51976)
Hydrochloric acid (Merck, Sigma-Aldrich, catalog number: 258148)
RPMI1640 powder (Thermo Fisher Scientific, Gibco®, catalog number: 31800-022)
DMEM powder, high glucose (Thermo Fisher Scientific, Gibco®, catalog number: 12100046)
Sodium Bicarbonate (Thermo Fisher Scientific, catalog number: 25080094)
Tris (1st BASE, catalog number: BIO-1400)
Glycine (Merck, Sigma-Aldrich, catalog number: G7126)
Triton X-100 (Bio-Rad, catalog number: 161-0407)
Bovine serum albumin (BSA), heat shock isolation (Bio Basic, catalog number: AD0023)
Sodium azide (Merck, Sigma-Aldrich, catalog number: S8032)
DAPI (Merck, Sigma-Aldrich, catalog number: D9542)
37% Formaldehyde solution (Merck, Sigma-Aldrich, catalog number: F1635)
Crystal violet (Merck, Sigma-Aldrich, catalog number: C3886)
Alexa FluorTM 488 F(ab’) 2 fragment of goat anti-mouse IgG (H + L) (Thermo Fischer Scientific, Invitrogen, catalog number: A11017)
Alexa FluorTM 594 F(ab’) 2 fragment of goat anti-mouse IgG (H + L) (Thermo Fischer Scientific, catalog number: A-11020)
Alexa FluorTM 488 Phalloidin (Thermo Fisher Scientific, catalog number: A12379)
Solutions
0.8% methyl-cellulose medium supplemented with 2% FBS (see Recipes)
1% crystal violet (see Recipes)
0.1 M glycine (pH 2.7) (see Recipes)
1 M Tris- HCl (pH 9.0) (see Recipes)
3.7% formaldehyde (see Recipes)
10% formaldehyde (see Recipes)
Recipes
0.8% methyl-cellulose medium supplemented with 2% FBS
Add 8 g of methyl-cellulose powder into 500 mL of water and autoclave twice to dissolve the powder.
Prepare 500 mL of 2× RPMI1640 or DMEM media by dissolving RPMI1640 powder or DMEM powder in water followed by supplementing with 4% heat-inactivated FBS, 4 mM L-glutamine, 200 U/mL pen/strep, 0.075% sodium bicarbonate solution, and 50 mM HEPES.
After filtration of 2× RPMI1640 or DMEM media through a 0.2 μm membrane filter unit, mix well with 500 mL prepared methyl-cellulose.
Store at 4 °C.
1% crystal violet
Add 5 g of crystal violet to 100 mL of 100% EtOH and mix well to dissolve powder.
Add 400 mL of water.
Store at room temperature.
0.1 M glycine (pH 2.7)
Add 3.75 g of glycine into 500 mL of water.
Adjust the pH to 2.7 using 1 N HCl.
Store the solution at 4 °C
1 M Tris-HCl (pH 9.0)
Add 30.3 g of Tris into 250 mL of water.
Adjust the pH to 9.0 using 1 N HCl.
Store the solution at room temperature.
3.7% formaldehyde
Add 500 mL of 37% formaldehyde into 4,500 mL of Milli-Q water.
10% formaldehyde
Add 200 mL of 37% formaldehyde into 540 mL of Milli-Q water.
Laboratory supplies
NunclonTM MULTIDISH 24 (Thermo Fisher Scientific, catalog number: 142475)
NunclonTM MULTIDISH 48 (Thermo Fisher Scientific, catalog number: 150687)
PerkinElmer cell carrier ultra microplates, treated, black, 96-well with lid (Genomax Technologies, catalog number: 6055302)
Greiner Bio-One MASTERBLOCKTM 96 deep-well conical-bottom 2 mL storage plate (Fisher Scientific, catalog number: 07-000-873)
Cryotubes vials for freezing viruses (Thermo Fisher Scientific, catalog number: 368632)
175 cm2 angled-neck easy flasks (Nunc, catalog number: 159920)
50 mL centrifuge tubes (BD Falcon, catalog number: 357550)
Filter unit 0.45 µm (Merck, Sigma-Aldrich, Millex®-HP, catalog number: SLHPR33RS)
Filter unit 0.2 μm (Thermo Fisher Scientific, catalog number: 567-0020)
HiTrap protein G HP-5 mL (GE Healthcare, catalog number: 170-0405-01)
Snakeskin dialysis tubing 10 kDa (Thermo Fisher Scientific, catalog number: 68100)
15 mL centrifuge tubes (Merck, Sigma-Aldrich, catalog number: CLS430791)
Aluminum foil
Equipment
Incubator without CO2 atmosphere at 28 °C (Sanyo, model: MIR-262)
Humidified incubator with 5% CO2 atmosphere at 37 °C (Nuaire, model: NU-5710E)
VACUSAFE aspiration system (Integra Biosciences, model: 158310)
ProBlotTM 25 economy rocker single platform (Bio Laboratories, model: S2025-B-230V)
Operetta® high content imaging system (PerkinElmer, model: Operetta)
Cermax® Xenon fiber-optic light source (Excelitas Technologies, model: XL3000)
DELL computer swinging rotor centrifuge (for cells) (Thermo Electron)
-80 °C freezer (Thermo Fisher Scientific)
Visi-White transilluminator (Analytik Jena US, model: TW-26)
Autoclave (TOMY, model: SX-700)
AKTA purifierTM UPC 10 (GE Healthcare, catalog number: 28406268)
pH meter (Satorius, catalog number: PY-PW-A)
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, catalog number: ND-2000)
Software and datasets
Harmony v4.8 (Operetta, 1/1/2013)
CellProfilerTM (Broad institute, 03/12/2022)
Prism v10.0 (GraphPad, 08/23/2023)
Procedure
Generation of DENV stocks
Grow C6/36 cells in a 175 cm2 flask with RPMI1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL pen/strep, and 25 mM HEPES and incubate at 28 °C under non-CO2-atmosphere-condition until ~90% confluency.
Note: Growth media supplemented with 20% FBS improves cell growth if C6/36 cells do not proliferate desirably.
Thaw virus stock and dilute with serum-free RPMI1640 medium.
Remove the medium from the culture flask and inoculate with 10 mL of virus inoculum at MOI of 0.1 into a 175 cm2 flask.
Incubate for 1 h at 28 °C under non-CO2 atmosphere conditions.
Discard the virus inoculum and add 25 mL of RPMI1640 medium supplemented with 2% FBS into the 175 cm2 flask.
Incubate for 5–7 days at 28 °C under non-CO2 atmosphere conditions.
Note: The cytopathic effect is observed depending on the virus strain (seen only in DENV-2 infection but not in other DENV strains infection). To ascertain the optimal duration for obtaining a high virus titer, it is recommended that the virus titer in the supernatant be determined at different time points. For example, the incubation period for DENV2 is five days, whereas it is seven days for DENV-1, DENV-3, and DENV-4.
Scrape the cells and transfer them into a 1.5 mL microcentrifuge tube.
Spin down the cells at 1,800× g for 10 min at 4 °C. Pass the supernatant through a 0.45 μm membrane filter and collect it in a fresh microcentrifuge tube.
Aliquot virus supernatant into cryotubes and store in a -80 °C freezer until use.
Determination of viral titer by plaque assay (Figure 1, Table 1, Figure 2)
Grow BHK-21 cells in RPMI1640 supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL pen/strep or Vero cells in DMEM supplemented with 10% FBS, 4.5 g/L D-glucose, 2 mM L-Glutamine, and 110 mg/L sodium pyruvate and 100 U/mL pen/strep in a humidified incubator with 5% CO2 atmosphere at 37 °C.
Seed cells at 2 × 105 cells per well in 500 μL of supplemented RPMI1640 or DMEM medium into a 24-well plate.
Note: For detachment of cells during the cell seeding procedure, 2–3 mL of trypsin is used. Cells may be seeded at 5 × 104 or 2 × 104 cells per well in 500 μL for 100% confluency prior to infection after 2 or 3 days of incubation, respectively. Gently move the plate using short, back-and-forth, and side-to-side motions prior to placing it into the incubator to ensure an even distribution of cells.
Incubate cells overnight at 37 °C in a 5% CO2 incubator to allow cells to attach and reach 100% confluency.
Dilute the virus in a series of 10-fold dilutions in serum-free RPMI1640 or DMEM.
Note: Dilution in microfuge or centrifuge tubes is preferred to enable vertexing and spinning down in the centrifuge after diluting for a homogenous suspension. If unable to dilute in tubes, resuspend virus suspension with a pipette for a minimum of 30 times upon diluting.
Discard culture supernatant of BHK-21 cells or Vero cells and add 200 μL of the diluted virus into each well.
Note: Virus should be added immediately after removing culture supernatant to avoid cells drying out.
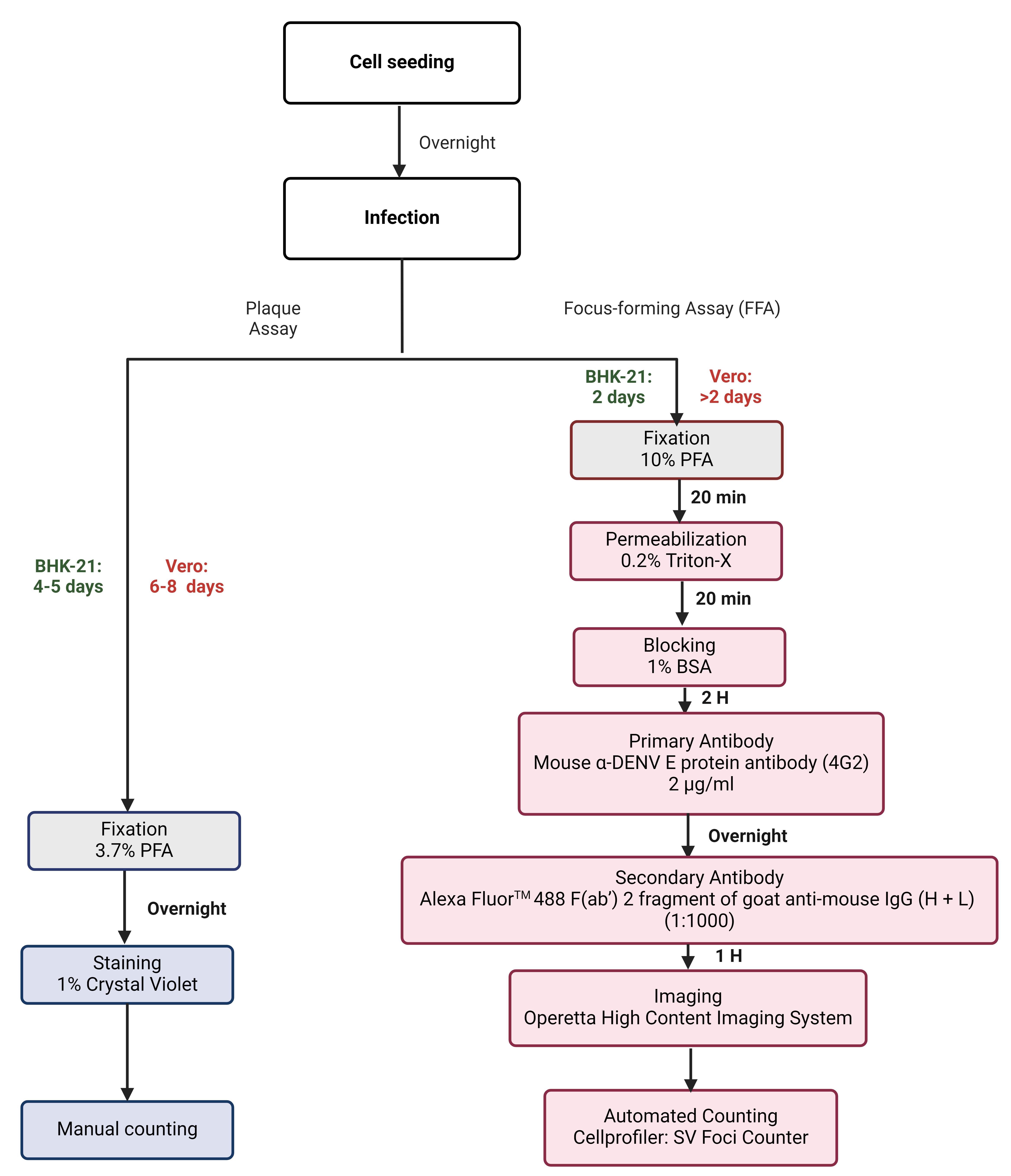
Figure 1. Flow diagram of plaque and improved focus-forming assays (FFA) to quantify dengue virus (DENV). The sequential steps were employed during plaque and FFA to quantify DENV1-4 clinical isolate titers. Following the seeding of BHK-21 or Vero cells onto 24- or 96-well plates, the plates were incubated overnight at 37 °C in 5% CO for the formation of a cell monolayer. The following day, the cells were infected with the respective virus and incubated for a period of 4–5 days (BHK-21) and 6–8 days (Vero) to allow for the development of plaques, and for a period of 2 days for both BHK-21 and Vero FFA plates, respectively. The plaque assay plates were fixed in 3.7% formaldehyde for a period of 12 h, stained with 1% crystal violet, and the number of plaques was counted manually. In contrast, FFA plates were fixed in 10% formaldehyde, permeabilized with 0.2% Triton X-100, blocked with 1% bovine serum albumin (BSA), and incubated with α-DENV E protein (4G2) (overnight) followed by the addition of Alexa FluorTM 488 antibody. Plates were then imaged using the Operetta® high-content imaging system and foci were counted using the automated System Virus (SV) Foci Counter imaging pipeline.Table 1. Comparative analysis of the plaque assay and the improved focus-forming assay. The table illustrates the differences between the two assays with regard to virus quantification from a clinical standpoint, where virus quantification is of paramount importance. In such a scenario, FFA is a highly credible method and serves as a useful tool.
Plaque assay Focus forming assay (FFA) Plate format 24-well 96-well Sample volumes 200 µL 50 µL Reagent volumes More volume of all reagents is required overall Less volume of all reagents is required overall Assay duration Two weeks 1 week Assay sensitivity for identification of viral strains Low (DENV-4 plaques are not clear) High (DENV-4 foci are clear) Assay read-out Manual Automated High throughput screening No Yes 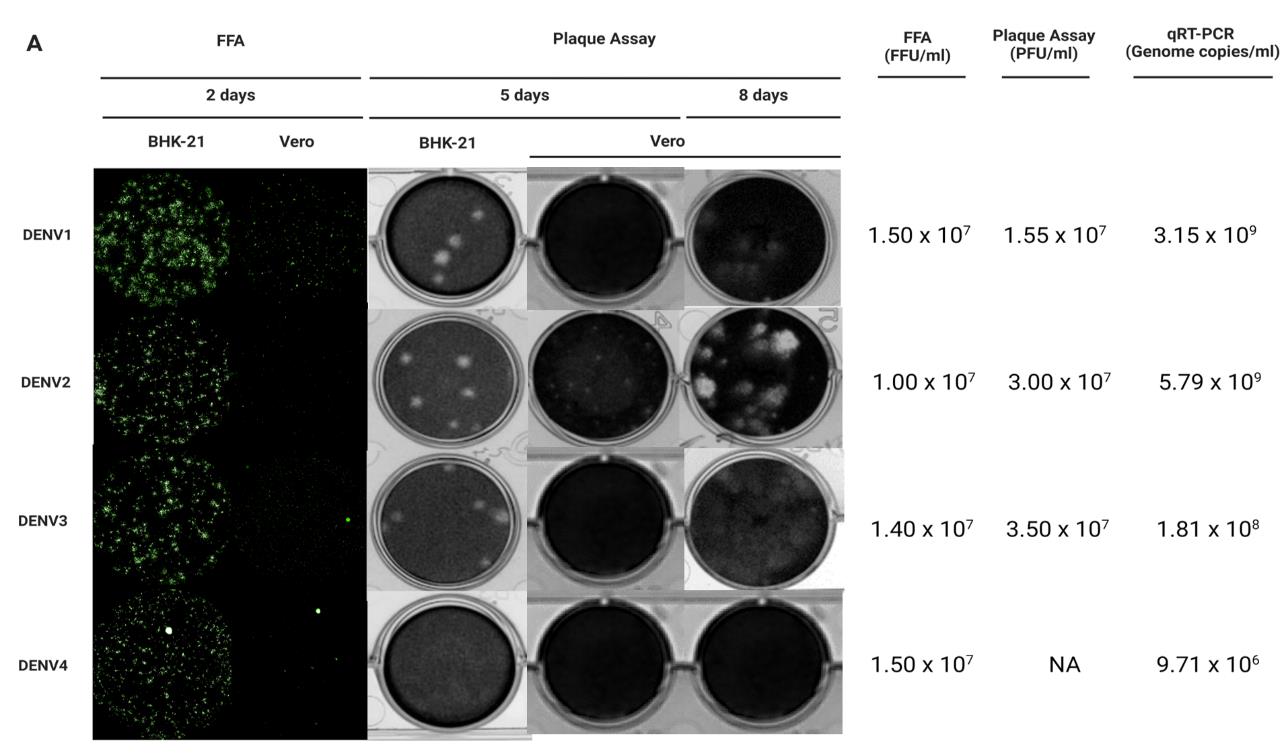
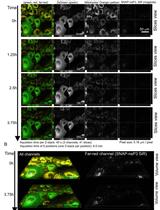
Figure 2. BHK-21 and Vero cell lines serve as in vitro cell-based infection models for DENV1-4 clinical isolates quantification. (A) BHK-21 and Vero cells were used as in vitro infection models to quantify infectious virus production by plaque assay and focus-forming assay (FFA). Viral genomic copies were detected by quantitative reverse transcription polymerase chain reaction (qRT-PCR) as a positive indication of infection. After 2 days of incubation*, DENV-1–4 foci in BHK-21 were clearer, brighter, and larger compared to the foci seen in Vero cells for all the clinical isolates. DENV-1 formed larger foci in both BHK-21 and Vero cells, while single-cell infections were seen mostly for DENV-2 and DENV-4 in Vero cells. Dashed circles indicate representative foci for each serotype in the different cell lines. After 5 days of incubation, DENV-1–3 plaques were clear and large, while DENV-4 formed unclear plaques in BHK-21. No plaques were seen for DENV-1, DENV-3, and DENV-4; however, small plaques were seen for DENV-2 in Vero after 5 days. Plaques for DENV-1–3 in Vero cells were seen after 8 days of incubation. In addition, the presence of DENV-1–4 viral RNA was detected in the respective samples. Of note, the BHK-21 infectious model is more efficient and is preferred over Vero, and FFA is a viable alternative to detect DENV-4 that does not form proper plaques. Note: All plates were incubated at 37 °C in 5% CO2. FFU: focus-forming unit; PFU: plaque-forming unit. Green: E protein.Incubate the plate for 1 h at 37 °C in a 5% CO2 incubator.
Note: Pre-equilibrate the 0.8% methyl-cellulose medium supplemented with 2% FBS to room temperature prior to use.
Discard the virus and overlay the cells with 500 μL of 0.8% methyl-cellulose medium supplemented with 2% FBS.
Note: The viscosity of methyl-cellulose medium necessitates the use of a Pasteur pipette for overlaying the medium onto the cells.
Incubate plate for 4–8 days at 37 °C in 5% CO2 incubator.
Note: The plaque size is affected by the virus replication rate. Check the plaque size visually before fixation to obtain clear plaque morphology. BHK-21-seeded plates of DENV-1 infection are normally incubated until day 4 post-infection, whereas plates of DENV-2–4 are usually incubated until day 5 post-infection. Vero seeded plates of DENV-1,3,4 infection were incubated for 8 days and DENV-2 was incubated for 5 days. However, it is recommended to incubate Vero-seeded plates of DENV-2 infection for more than 5 days as the plaques were small.
Fix cells with 500 μL of 3.7% formaldehyde (diluted in Milli-Q water) for a minimum of 3 h.
Note: Fixation overnight may be required if 3.7% formaldehyde is not freshly made.
Rinse the plate with a copious amount of running water to remove methyl-cellulose medium completely.
Add 1–2 drops of 1% crystal violet into each well and stain for 1 min.
Rinse the plate with copious amounts of running water to remove the excess crystal violet stain.
Air dry plates on a paper towel.
Visualize plaques using the white light transilluminator apparatus and count the number of plaques for determination of virus titer as follows:
Virus titer [plaque-forming units (pfu)/mL] = average number of plaques × 1,000 µL/200 µL inoculum × reciprocal of dilution factor
Preparation of α-DENV E protein antibodies (4G2) from hybridoma cells
Note: Antibodies specific to the detection of E protein of flaviviruses including DENV can be found commercially.
Culture 4G2 hybridoma cells in 50 mL of PFHM-II medium in 175 cm2 flasks in a humidified incubator with 5% CO2 atmosphere at 37 °C.
Note: It is recommended to culture the cells in RPMI1640 supplemented with 10% FBS, 2 mM L-glutamine, 100 U/mL pen/strep during the initial period of several days after thawing cells. Once the cells grow well, replace the media gradually by increasing the proportion of PFHM-II media to RPMI1640 from 10% (vol/vol) to 100% PFHM-II media.
Collect cell suspension into a centrifugal tube when the cells become confluent (the color of culture media turns to orange or yellow).
Centrifuge cells at 900× g for 5 min at room temperature.
Collect the supernatant and store it at 4 °C without filtration until a sufficient volume of supernatant is obtained.
Continue culturing the cells and repeat steps C2–C4 if a large volume of the supernatant is required.
Filter the supernatant through a 0.45 μm membrane filter unit.
Load the 4G2 supernatant onto a 5 mL Protein G column pre-equilibrated in PBS (pH 7.2).
Note: 4G2 antibody is purified using the AKTA purifier. Refer to the manufacturer’s instruction guides regarding sample loading specifications.
Wash the column with PBS using five times the column volume (i.e., 25 mL).
Prepare a Grenier 96-well master block containing 60 μL 1 M Tris-HCl pH 9.0 for collection.
Note: The standard ratio of Tris-HCl to glycine (100:6) for neutralization is subject to change depending on the concentration of buffers prepared. The volume of Tris-HCl required for neutralization (pH 7) can be adjusted by pH paper testing.
Elute antibodies using 100% 0.1 M glycine pH 2.7 and collect 1 mL fractions into the wells of the block.
Note: Check the purity of the antibody by running an SDS-PAGE.
Select fractions of high purity and collect them into a dialysis membrane; then, dialyze against PBS overnight.
Quantitate the concentration of the purified antibody using NanoDrop.
Determination of viral titer by FFA (Figure 1, Table 1, Figure 2, Figure 3)
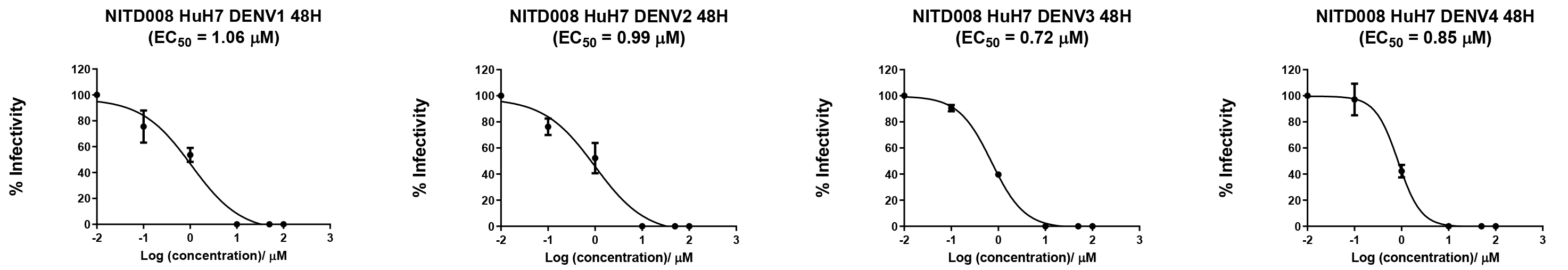
Figure 3. Application of the BHK-21 infectious model in evaluating the efficacy of antiviral compound NITD008 against DENV1-4 clinical isolates using focus-forming assay (FFA). Huh7 cells were infected with DENV-1–4 at a multiplicity of infection (MOI) of 0.3 and subsequently treated with varying concentrations of NITD008 (100, 50, 10, 1, 0.1, and 0.01 µM). Post 48 h, viral supernatants were assayed on BHK-21 cells for FFA to estimate the virus titers. The half-maximal effective concentration (EC50) of NITD008 was calculated through the plotting of a sigmoidal dose-response curve using GraphPad Prism. The known EC50 of NITD008 lies between the range of 0.16–2.61 µM [9,10], and the range of calculated EC50 from both assays was 0.62–1.18 µM, indicating the efficiency of FFA. The data was obtained from three biological replicates and verified by an independent experimenter as being reproducible. The data presented here represents a single experiment out of a total of three (n = 3, SD = 0.02–0.78).D1. Preparation of FFA samples for image acquisition
Maintain BHK-21 cells in RPMI1640 supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL pen/strep or Vero cells in DMEM supplemented with 10% FBS, 4.5 g/L D-glucose, L-Glutamine and 110mg/L Sodium Pyruvate and 100 U/mL pen/strep in a humidified incubator with 5% CO2 atmosphere at 37 °C.
Seed cells at 5 × 104 cells per well in 100 μL into the PerkinElmer cell carrier 96 ultra microplates.
Note: Cells may be seeded at 1.25 × 104 or 5 × 103 cells per well in 100 μL for 100% confluency prior to infection after 2 days or 3 days incubation, respectively. A single-cell monolayer formation is crucial to allow the capture of focused images for accurate analysis.
Incubate cells overnight at 37 °C in a 5% CO2 incubator to allow cells to adhere and reach 100% confluency.
Prepare 10-fold serial dilutions of the virus in serum-free RPMI1640 or DMEM.
Remove the culture supernatant of BHK-21 cells or Vero cells and add 50 μL of diluted virus into each well.
Note: Virus should be added immediately after removing culture supernatant to avoid cells drying out.
Incubate the plate for exactly 1 h at 37 °C in a 5% CO2 incubator.
Remove the virus and add 125 μL of 0.8% methyl-cellulose medium supplemented with 2% FBS. Incubate the plates with infected BHK-21 cells at 37°C in a 5% CO2 incubator for two days.
Note: Plates with infected Vero cells will require more than 2 days incubation, as foci are not as clear as those seen in BHK-21 with mostly single-cell infections observed (Figure 2).
Add 100 µL/well of 10% formaldehyde (diluted in Milli-Q water) and incubate for 1 h at room temperature or 20 min in the 37 °C incubator.
Wash off the 10% formaldehyde thoroughly with a copious amount of water in a container. Shake the plate vigorously to remove the methyl-cellulose medium completely.
Add 100 µL/well of 0.2% Triton X-100 in PBS and incubate on ProBlotTM 25 economy rocker with maximum speed at room temperature for 20 min.
Note: For the prevention of microbial growth, you can optionally add 0.01% sodium azide to the 0.2% Triton-X-100.
Discard 0.2% Triton X-100 in 1× PBS and wash wells three times with 1× PBS.
Note: Remove as much 1× PBS as possible when washing each time and avoid drying of cells by ensuring wells are always filled with 1× PBS.
Add 125 µL/well of 1% BSA in 1× PBS and incubate on ProBlotTM 25 economy rocker at maximum speed at room temperature for 1–2 h.
Discard 1% BSA in 1× PBS and wash wells three times with 1× PBS.
Add 50 µL/well of purified mouse anti-E 4G2 antibody (final concentration of 2 µg/mL) in 1% BSA/PBS and incubate for at least 1 h on ProBlotTM 25 economy rocker at 60–70 rpm at room temperature or overnight at 4 °C.
Collect the purified mouse anti-E 4G2 antibody from each well in a new tube after incubation and keep at -30 °C.
Note: Diluted antibody can be reused approximately 2–3 times.
Wash wells three times with 150 µL of 1× PBS.
Add 50 µL/well of Alexa FluorTM 488 goat anti-mouse antibody with a 1:1,000 dilution in 1% BSA/PBS and incubate for 1 h on ProBlotTM 25 economy rocker at 60–70 at room temperature.
Note: Protect the antibody and sample plate from light by wrapping the tube holding the diluted antibody and sample plates with aluminum foil from this step onward. Do not incubate for more than 1 h as this may result in a high background signal.
Remove the secondary antibody (Alexa FluorTM 488 goat anti-mouse antibody) and wash wells three times with 1× PBS.
Add 50 µL/well of DAPI at 1:10,000 dilution and incubate for 5 min on ProBlotTM 25 economy rocker at room temperature in the dark.
Wash wells three times with 150 µL of 1× PBS.
Maintain wells in approximately 125 µL of 1× PBS at 4 °C until imaging.
D2. Image acquisition from FFA samples using the Operetta® high content imaging systemPlace the microplate with FFA samples into the instrument.
Note: Ensure the light on the instrument turns blue before opening the lid and inserting the microplate.
Select the Setup tab and choose the following parameters:
Plate type: 96 PerkinElmer CellCarrier Ultra
Objective: 20× high NA
Note: Magnification of the objective used affects the imaging of whole wells and the time taken to acquire an image. Choose a high-magnification objective for whole well imaging; however, more time will be needed for acquisition. The automated SV Foci Counter pipeline has been optimized for the analysis of images captured with the 20× high NA objective.
Optical (Opt.) mode: Non-Confocal
Excitation: 50%
Transmission: 0%
Select channels:
Alexa 488:
Time: 20 ms
Height: 0.0 µM
Note: The term "time (ms)" is used to describe the exposure time, which plays a crucial role in determining the brightness of the image. The exposure time may require adjustment due to batch-to-batch variability in staining intensity. Height (µM) refers to the focus height above the plate bottom. The height indicated here is the default setting and may require amendment to achieve a more focused image.
DAPI:
Time: 10 ms
Height: 0.0 µM
Layout selection:
Number of wells: 96
Well:
Number of fields: 97
Overlap: 0%
Stack:
First plane at: 0.0 µM
Number of planes: 0
Distance: 0.0 µM
Last plane at: 0.0 µM
Overall height: 0.0 µM
Image control:
Coloring: highlight
Flatfield correction: None
Select the Run Experiment tab and press start.
D3. Determination of focus-forming units (FFU) by CellProfilerTM (Figure 4)
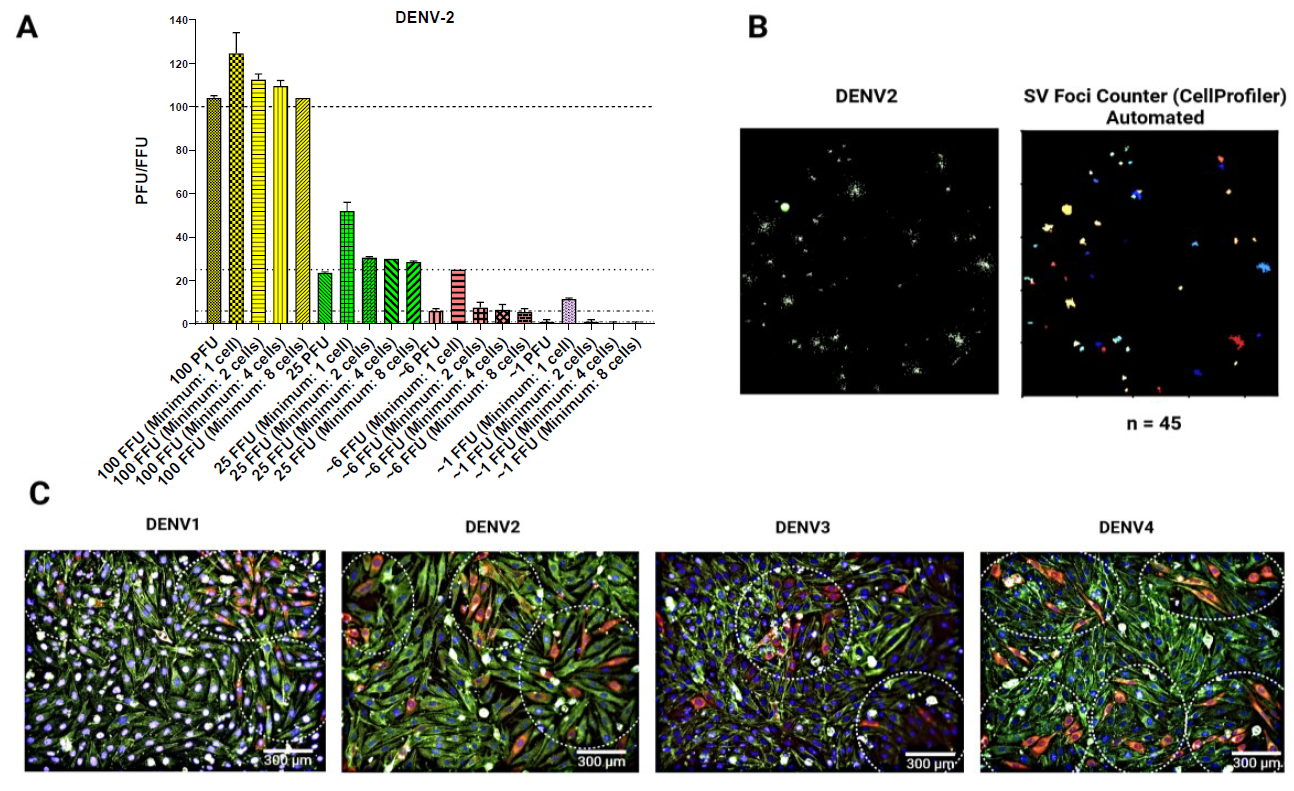
Figure 4. Characterization of foci formation in the BHK-21 infectious model. (A) The correlation between the number of plaque-forming units (PFU) and the number of foci is presented herewith. A DENV-2 inoculum of 100 PFU was added to a BHK-21 cell monolayer, and a plaque assay and focus-forming assay (FFA) were subsequently conducted. The number of plaques and foci was determined through manual counting. The number of foci was determined by employing a minimum of one, two, four, or eight infected cells per foci. As illustrated in the bar diagram (dotted lines), the foci identified with a minimum of eight cells demonstrated a stronger correlation with PFUs. In some cases, larger foci may consist of cells that are more than eight in number and may appear to be fusing together. In such instances, the foci will continue to be regarded as two discrete entities. (B) Images of foci were captured using the Operetta® high-content imaging system, and the automated foci counting process was conducted using the SV foci counter, a pipeline designed using the CellProfilerTM software. A foci plot was generated, and the number of foci was counted automatically using the SV foci counter imaging pipeline, which estimated the size of the foci to be eight cells. n = number of foci. (C) Representative images of DENV-1–4 individual foci using 20× objective. The presence of a dotted circle indicates the focus for DENV-1–4, with the number of instances quantified as follows: 3, 3, 2, and 5. Red: E Protein; green: f-actin; blue: nuclei. Scale bar: 300 µm. Data is representative of two technical replicates (n = 2).Note: See supplementary materials for an example customized image analysis pipeline termed “SV Foci Counter.”
Load images in CellProfilerTM and categorize images into DAPI- and Alexa FluorTM 488-labeled images as Nuclei and Foci under NamesAndTypes function.
Add the following modules and customize the respective settings:
CropGreen and CropBlue
Cropping shape: Rectangle
Cropping method: Coordinates
Apply which cycle’s cropping pattern: Every
Left and right rectangle positions: 0 - end – Absolute
Top and bottom rectangle positions: 0 - end – Absolute
Remove empty rows and columns: All
Identify Primary Objects:
Use advanced settings: Yes
Select the input image and name the primary objects to be identified: Foci
Typical diameter of objects in pixel units (Min, Max): 10, 1,000
Note: Diameter of foci was estimated based on 8 cells minimum per foci.
Discard objects outside the diameter range: Yes
Discard objects touching the border of the image: No
Threshold strategy: Global
Thresholding method: Manual
Manual threshold: 0.1–0.16
Note: Depending on the quality of antibody staining done, there may be a need to adjust the threshold correspondingly.
Threshold smoothing scale: 1.3488
Method to distinguish clumped objects: Intensity
Method to draw dividing lines between clumped objects: Shape
Automatically calculate the size of the smoothing filter for declumping: Yes
Automatically calculate the minimum allowed distance between local maxima: Yes
Speed up by using a lower-resolution image to find local maxima: Yes
Display accepted local maxima: No
Fill holes in identified objects: After both thresholding and declumping
Handling of objects if an excessive number of objects identified: Continue
Export to spreadsheet.
Data analysis
All experiments were conducted with a minimum of two technical replicates and biological replicates. To ensure the accuracy and reliability of the results, an independent experimenter conducted a verification process. The determination of focus-forming units from images captured with the Operetta® high-content imaging system was analyzed using the Harmony program and CellProfilerTM. Dose-response analysis for half maximal effective concentration (EC50) determination can be performed by plotting the dose-response curve using the GraphPad Prism software.
Validation of protocol
Verification of the protocol has been shown in Figures 2–4 in the procedure section. FFA usage in BHK-21 and Vero cells has been shown in Figure 2, section B. Application of the BHK-21 infectious model in evaluating the efficacy of antiviral compound NITD008 against DENV1-4 clinical isolates using FFA is shown in Figure 3, section D. Usage of the Operetta® high-content imaging system and the SV foci counter in determining foci formation is shown in Figure 4, section D3.
Acknowledgments
This research is supported by the IAF-ICP I2301E0019 administrated by Agency for Science, Technology and Research. We would like to acknowledge and give our warmest thanks to all the members of Subhash Vasudevan’s laboratory without whom the protocol established would have not been possible.
Competing interests
Authors declare no competing interests.
References
- Bhatt, S., Gething, P. W., Brady, O. J., Messina, J. P., Farlow, A. W., Moyes, C. L., Drake, J. M., Brownstein, J. S., Hoen, A. G., Sankoh, O., et al. (2013). The global distribution and burden of dengue. Nature. 496(7446): 504–507.
- Messina, J. P., Brady, O. J., Golding, N., Kraemer, M. U. G., Wint, G. R. W., Ray, S. E., Pigott, D. M., Shearer, F. M., Johnson, K., Earl, L., et al. (2019). The current and future global distribution and population at risk of dengue. Nat Microbiol. 4(9): 1508–1515.
- James, M. N. and Dubovi, E. J. (2017). Chapter 29 - Flaviviridae. (p. 525–545). In: Fenner's Veterinary Virology (Fifth Edition). Academic Press.
- Bruno, F., Abondio, P., Bruno, R., Ceraudo, L., Paparazzo, E., Citrigno, L., Luiselli, D., Bruni, A. C., Passarino, G., Colao, R., et al. (2023). Alzheimer’s disease as a viral disease: Revisiting the infectious hypothesis. Ageing Res Rev. 91: 102068.
- Baer, A. and Kehn-Hall, K. (2014). Viral Concentration Determination Through Plaque Assays: Using Traditional and Novel Overlay Systems. J Visualized Exp.: e52065.
- Bolívar-Marin, S., Bosch, I. and Narváez, C. F. (2022). Combination of the Focus-Forming Assay and Digital Automated Imaging Analysis for the Detection of Dengue and Zika Viral Loads in Cultures and Acute Disease. J Trop Med. 2022: 1–11.
- Payne, A. F., Binduga-Gajewska, I., Kauffman, E. B. and Kramer, L. D. (2006). Quantitation of flaviviruses by fluorescent focus assay. J Virol Methods. 134: 183–189.
- Low, J. G., Ooi, E. E., Tolfvenstam, T., Leo, Y. S., Hibberd, M. L., Ng, L. C., Lai, Y. L., Yap, G. S., Li, C. S., Vasudevan, S. G., et al. (2006). Early Dengue Infection and Outcome Study (EDEN) – Study Design and Preliminary Findings. Ann Acad Med Singap. 35(11): 783–789.
- Yin, Z., Chen, Y. L., Schul, W., Wang, Q. Y., Gu, F., Duraiswamy, J., Kondreddi, R. R., Niyomrattanakit, P., Lakshminarayana, S. B., Goh, A., et al. (2009). An adenosine nucleoside inhibitor of dengue virus. Proc Natl Acad Sci USA. 106(48): 20435–20439.
- Touret, F., Baronti, C., Goethals, O., Van Loock, M., de Lamballerie, X. and Querat, G. (2019). Phylogenetically based establishment of a dengue virus panel, representing all available genotypes, as a tool in dengue drug discovery. Antiviral Res. 168: 109–113.
Supplementary information
The following supporting information can be downloaded here:
- SV foci counter
Article Information
Publication history
Received: May 6, 2024
Accepted: Aug 21, 2024
Available online: Sep 29, 2024
Published: Oct 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Mahid, M. B. A., Bist, P., Sigmundsson, K., Mazlan, M. D. B. M., Watanabe, S., Choy, M. M., Vasudevan, S. G. and Chan, K. W. K. (2024). An Improved Focus-Forming Assay for Determination of the Dengue Virus Titer. Bio-protocol 14(20): e5084. DOI: 10.21769/BioProtoc.5084.
Category
Microbiology > Antimicrobial assay > Antiviral assay
Cell Biology > Cell imaging > Fluorescence
Drug Discovery > Drug Screening > Anti-infective agents
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









