- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Multicellular 3D Liver Organoids From Induced Pluripotent Stem Cells as a Tool for Modelling Liver Diseases
Published: Vol 14, Iss 15, Aug 5, 2024 DOI: 10.21769/BioProtoc.5042 Views: 2801
Reviewed by: Komuraiah MyakalaHsih-Yin TanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
In situ Hybridization of miRNAs in Human Embryonic Kidney and Human Pluripotent Stem Cell-derived Kidney Organoids
Filipa M. Lopes [...] Ioannis Bantounas
Sep 5, 2021 3337 Views
Multiplex Genome Editing of Human Pluripotent Stem Cells Using Cpf1
Haiting Ma
Nov 20, 2024 2644 Views
A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1249 Views
Abstract
The liver is an essential organ that is involved in the metabolism, synthesis, and secretion of serum proteins and detoxification of xenobiotic compounds and alcohol. Studies on liver diseases have largely relied on cancer-derived cell lines that have proven to be inferior due to the lack of drug-metabolising enzymes. Primary human hepatocytes are considered the gold-standard for evaluating drug metabolism. However, several factors such as lack of donors, high cost of cells, and loss of polarity of the cells have limited their widescale adoption and utility. Stem cells have emerged as an alternative source for liver cells that could be utilised for studying liver diseases, developmental biology, toxicology testing, and regenerative medicine. In this article, we describe in detail an optimised protocol for the generation of multicellular 3D liver organoids composed of hepatocytes, stellate cells, and Kupffer cells as a tractable robust model of the liver.
Key features
• Optimising a protocol for generating multicellular 3D liver organoids from induced pluripotent stem cells.
Keywords: Stem cellsGraphical overview
Background
The liver is a vital organ that is involved in the metabolism, synthesis, and production of serum proteins and bile acid and the detoxification of xenobiotics [1]. Earlier liver models relied heavily on cancer-derived cell lines, which have been proven to be inferior due to the suboptimal activity of drug-metabolising enzymes [2,3]. Isolated primary human hepatocytes (PHH) are considered to be the “gold standard” for drug metabolism and toxicity screening [4,5]. However, these cells display a rapid decline in the phenotypic function when cultured in traditional two-dimensional monolayer cell cultures; also, there is a scarcity of donors [5]. To overcome these limitations of using PHHs, researchers have developed various approaches including genetic modification of the cells and three-dimensional cultures combined with tissue engineering and media compositions [1,6,7]. Additionally, advances in cell culture systems offer a great opportunity to generate patient-specific hepatocytes using 2D cell-based [2] and 3D organoid platforms [1,8].
The liver cells can be obtained by direct differentiation of iPSCs [7,9] and tissue-derived stem cells [1,6,10] by exploiting inductive and repressive signals essential for liver ontogenic development. The 3D cultured cells display better physiologic and metabolic features of the native liver tissue in comparison with 2D cell-based cultures [1,9]. However, the vast majority of the reported methods predominantly differentiate cells into hepatic epithelial lineage only, thus lacking essential supportive cellular lineages such as profibrotic hepatic stellate cells (hematopoietic stem cells; HSCs) and inflammatory cells (Kupffer cells; KCs). Hence, they lack the capacity to model inflammatory diseases [1,11]. To circumvent these challenges, other researchers developed co-culture cell models based on mixing both the epithelial cells and supportive lineages from iPSCs [12,13]. However, these methods suffer from artefactual inflammation and fibrosis resulting from the difficulty in choosing the appropriate culture medium and extracellular matrix in which multiple cell lineages can be co-maintained [12,13].
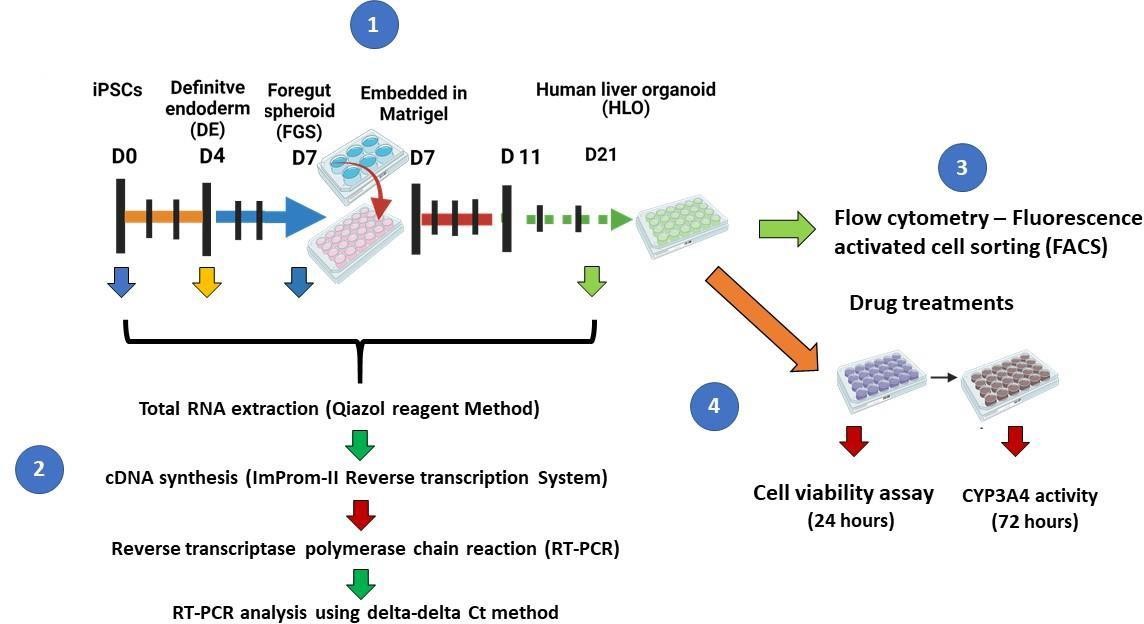
A recent study by Ouchi and colleagues described a protocol for the generation of a multicellular human liver organoid (HLO) composed of hepatocyte-like cells, hepatic stellate–like cells, and Kupffer-like cells from iPSCs [9]. The cells aggregated to form a 3D liver organoid that was used to model steatohepatitis after the addition of free fatty acids to the culture media [9]. In this paper, we describe a method for generating multicellular 3D liver organoids (Figure 1) adapted from the protocols published by Ouchi and colleagues [9] and Mun and colleagues [8]. We further describe in detail how to generate these organoids and demonstrate their utility as a tool to study downstream assays for liver disease models.
Materials and reagents
Biological materials
Schistosome eggs antigen (SEA) (Schistosome Biological Supply Center, Theodor Bilharz Research Institute)
Reagents
A83-01 (R&D Systems, catalog number: R2939)
Activin A (R&D Systems, catalog number: 338-AL)
Accutase (Sigma-Aldrich, catalog number: A6964)
CellTiter-GLO® cell viability assay (Promega, catalog number: PRG9681)
CHIR99021 (Sigma-Aldrich, catalog number: SML1046)
Advanced DMEM (Gibco, catalog number: 12634-010)
B27 supplement (Gibco, catalog number: A1486701)
Bone morphogenetic protein 4 (BMP-4) (R&D Systems, catalog number: 314-13P)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A8531)
Dexamethasone (Sigma-Aldrich, catalog number: D4902)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650)
Dulbecco’s phosphate buffered saline (DPBS) (Gibco, catalog number: 14190-094)
Essential 8 basal medium (Gibco, catalog number: A1517001)
Epidermal growth factor (R&D Systems, catalog number: 236-EG)
Ethylenediaminetetraacetic acid (EDTA) (Invitrogen, catalog number: 15575-020)
Fibroblast growth factor 4 (R&D Systems, catalog number: 325-4F)
Fibroblast growth factor 10 (R&D Systems, catalog number: F8924)
Geltrex (Life Technologies, catalog number: A1413302)
HCl (Merck, catalog number: 100317)
Hepatocyte culture medium (Lonza, catalog number: CC-3198)
Hepatocyte growth factor (Sigma-Aldrich, catalog number: H9661)
HEPES (Gibco, catalog number: 15630080)
ImProm-II reverse transcriptase (Promega, catalog number: PRA3800)
Knockout serum replacer (KSR) (Gibco, catalog number: 10828028)
N2 supplement (Gibco, catalog number: A1370701)
Oncostatin M (R&D Systems, catalog number: 295-OM)
P450-Glo CYP3A4 assay with Luciferin-IPA (Promega, catalog number: PRV9002)
Penicillin-streptomycin (Gibco, catalog number: 15140-12221)
PowerUPTM SYBRTM Green Master Mix (Applied Biosystems, catalog number: A25741)
Retinoic acid (RA) (Sigma-Aldrich, catalog number: R2625)
Rho kinase (ROCK) inhibitor (Y-27632) (Sigma-Aldrich, catalog number: Y0503)
RPMI-1640, GlutaMAX supplement (Gibco, catalog number: 61870-036)
TRIzolTM reagent (Thermo Fisher Scientific, catalog number: 15596026)
TryLETM Express enzyme (Thermo Fisher Scientific, catalog number: 12604013)
Vascular endothelial growth factor (R&D Systems, catalog number: 293-VE)
Vitronectin (Gibco, catalog number: A27940)
Antibody list for organoid phenotyping (Table 1)
Table 1. List of antibodies used to phenotype human liver organoids
Phenotype Antibody Concentration Fluorophore Host Vendor Dilution Epithelial cells EpCam 0.2 mg/mL BV421 Mouse BioLegend 1:80 Stellate cells CD166 5 µL (0.06 µg) PE-ALCAM Mouse eBioscience 1:160 Kupffer cells CD68 5 µL (0.05 µg) PE-Cy7 Mouse BioLegend 1:80 Antiretroviral and anti-tuberculosis drugs (Table 2)
Table 2. List of anti-retroviral and anti-tuberculosis drugs used to study liver injury
Drug Manufacturer Catalogue number Final concentration Efavirenz (EFV) Sigma-Aldrich 025M478 7.39 mM Tenofovir (TDF) Sigma-Aldrich SML1795 34.2 mM Lamivudine (3TC) Sigma-Aldrich L1295 27.91mM Rifampicin (RIF) Sigma-Aldrich R3501 28.42 mM Isoniazid (INH) Sigma-Aldrich MKCF2223 20.04 mM Primer sequences for RT-qPCR (Table 3)
Table 3. Primer sequences for stage-specific markers for liver organoids
Human stem cell markers Gene name Forward primer sequence (5'-3') Reverse primer sequence (5'-3') Oct-4 CAGGAGATATGCAAAGCAGAAAC GGCACTGCAGGAACAAATT Nanog AGCCTAATCAGCGAGGTTTC CAGAGCAAGACTCCGTTTCA Sox2 GCTACAGCATGATGCAGGACCA TCTGCGAGCTGGTCATGGAGTT Definitive endoderm (DE) markers Sox17 CGCACGGAATTTGAACAGTA GGATCAGGGACCTGTCACAC GSC GAGGAGAAAGTGGAGGTCTGGTT CTCTGATGAGGACCGCTTCTG Immature human liver organoids markers CYP3A7 GAAACACAGATCCCCCTGAA TCAGGCTCCACTTACGGTCT Mature human liver organoids markers HNF4α CATGGCCAAGATTGACAACCT TTCCCATATGTTCCTGCATCAG ALB TTG GCA CAA TGA AGT GGG TA AAA GGC AAT CAA CAC CAA GG A1AT CCACCGCCATCTTCTTCCTGCCTGA GAGCTTCAGGGGTGCCTCCTCTGTG CYP3A4 TGTGCCTGAGAACACCAGAG GTGGTGGAAATAGTCCCGTG GADPDH CCATCTTCCAGGAGCGAG GCAGGAGGCATTGCTGAT
Solutions
Vitronectin (see Recipes)
0.5 mM ultrapure EDTA (see Recipes)
Activin A (see recipe)
Bone morphogenetic protein 4 (see Recipes)
Fibroblast growth factor 4 (see Recipes)
0.1% (m/v) bovine serum albumin (see Recipes)
CHIR99021 (see Recipes)
Retinoic acid (see Recipes)
Hepatocyte growth factor (see Recipes)
Dexamethasone (see Recipes)
Oncostatin M (see Recipes)
Fibroblast growth factor 10 (see Recipes)
Epidermal growth factor (see Recipes)
A83-01 (see Recipes)
Rho kinase protein (ROCK) inhibitor (Y-27632) (see Recipes)
Cell culture media
hiPSCs maintenance media: complete essential 8 medium (see Recipes)
Definitive endoderm induction (days 1–3) media: Complete RPMI medium (see Recipes)
Foregut spheroid (FGS) induction (days 4–6) media: Complete Advanced DMEM/F2 (see Recipes)
Human liver organoids (HLO) formation (days 7–10) media 1: Advanced DMEM/F12 + retinoic acid (see Recipes)
Hepatocyte maturation (days 11–25) media: Hepatocyte culture medium (HCM) (see Recipes)
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
First strand CDNA synthesis cocktail 1 (see Recipes)
cDNA synthesis master mix 2 (see Recipes)
qRT-PCR reagents mix (see Recipes)
Recipes
Vitronectin solution
Reagents Final concentration Amount Vitronectin 9 µg/mL 0.9 mg/mL DPBS 1× 990 mL Total n/a 1,000 mL 0.5 mM ultrapure EDTA
Reagents Final concentration Amount Ethylenediaminetetraacetic acid 0.5 mM 10 mL DPBS 1× 9,990 mL Total n/a 10,000 mL Store at room temperature (RT) and use on the day of preparation.
Activin A solution
Reagents Final concentration Amount Activin A 100 µg/mL 10 µg HCl 4 mM 100 mL Total n/a 100 mL Prepare 1,000 ng/mL working solution in RPMI-1640 + GlutaMAX and store at -20 °C until needed for the experiment.
Bone morphogenetic protein 4 (BMP-4) solution
Reagents Final concentration Amount BMP-4 100 µg/mL 10 µg 0.1% BSA in 4 mM HCl n/a 100 mL Total n/a 100 mL Prepare 1,000 ng/mL working solution in RPMI-1640 + GlutaMAX and store at -20 °C until needed for experiment.
Fibroblast growth factor 4 (FGF-4) solution
Reagents Final concentration Amount FGF-4 100 µg/mL 25 µg 0.1% BSA in PBS (Recipe 6) n/a 100 mL Total n/a 100 mL Prepare 1,000 ng/mL working solution in Advanced DMEM/F12 and store at -20 °C until needed for the experiment.
0.1% (m/v) bovine serum albumin (BSA) in PBS
Reagents Final concentration Amount Bovine serum albumin (BSA) 0.1% (m/v) 0.1 g H2O n/a 100 mL Total n/a 100 mL CHIR99021 solution
Reagents Final concentration Amount CHIR99021 5 mg/mL 5 mg DMSO n/a 1,000 mL Total n/a 1,000 mL Prepare 1 mM working solution in advanced DMEM/F12 and store at -20 °C until needed for experiments.
Retinoic acid (RA) solution
Reagents Final concentration Amount Retinoic acid 50 mg/mL 50 mg DMSO n/a 1,000 mL Total n/a 1,000 mL Prepare 1 mM working solution in advanced DMEM/F12 and store at -20 °C until needed for experiments.
Hepatocyte growth factor solution
Reagents Final concentration Amount Hepatocyte growth factor (HGF) 100 mg/mL 5 mg 0.1% BSA in PBS (Recipe 6) n/a 50 mL Total n/a 50 mL Prepare 1,000 ng/mL working solution in hepatocyte culture medium (HCM) and store at -20 °C until needed for the experiment.
Dexamethasone (Dex) solution
Reagents Final concentration Amount Dexamethasone (Dex) 10 mM 5 mg DMSO n/a 1,000 mL Total n/a 1,000 mL Prepare 1,000 µM working solution in HCM and store at -20 °C until needed for the experiment.
Oncostatin M (OSM) solution
Reagents Final concentration Amount Oncostatin M (OSM) 100 mg/mL 10 µg 0.1% BSA in PBS (Recipe 6) n/a 100 mL Total n/a 100 mL Prepare 1,000 ng/mL working solution in HCM and store at -20 °C until needed for the experiment.
Fibroblast growth factor 10 (FGF-10) solution
Reagents Final concentration Amount Fibroblast growth factor 10 (FGF-10) 250 mg/mL 25 µg 0.1% BSA in PBS (Recipe 6) n/a 100 mL Total n/a 100 mL Prepare 1,000 ng/mL working solution in advanced DMEM/F12 and store at -20 °C until needed for experiments.
Epidermal growth factor (EGF) solution
Reagents Final concentration Amount Epidermal growth factor 500 mg/mL 200 µg/mL 0.1% (m/v) BSA in PBS n/a 400 mL Total n/a 400 mL Prepare 1,000 ng/mL working solution in advanced DMEM/F12 and store at -20 °C until needed for experiments.
A 83-01 solution
Reagents Final concentration Amounts A 83-01 10 mg/mL 10 mg DMSO n/a 1,000 mL Total n/a 1,000 mL Prepare 1 mM working solution in advanced DMEM/F12 and store at -20 °C until needed for experiments.
Rho kinase protein (ROCK) inhibitor (Y-27632) solution
Reagents Final concentration Amount Y-27632 10 mM 5 mg DMSO n/a 1,000 mL Total n/a 1,000 mL Store at -20 °C for up to six months.
Cell culture media
hiPSCs maintenance media: complete essential 8 medium
Reagents Final concentration Amount E8M 1× 485 mL E8 supplement 1× 10 mL P/S 1× 5 mL Total n/a 500 mL Alternatives: other hiPSC culture media can be used such as mTeSR1, StemFlex, and TeSR-E8. Swirl the bottle to mix contents properly, label with name and date prepared, filter, and store at 4 °C for up to 14 days. Alternatively, 50 mL aliquots can be made and stored at -20 °C.
Definitive endoderm induction (days 1–3) media: Complete RPMI medium
Regents Final concentration Amount RPMI-1640 1× 482 mL P/S 1× 5 mL HEPES 25 mM 12.5 mL Activin A 100 ng/mL - BMP-4 50 ng/mL - KSR 1× - Total n/a 500 mL Amount of Activin A, BMP4, and KSR are not shown on the Table as we prepare them fresh every day and they are dependent on the number of wells and volumes you are working with.
Swirl the bottle to mix contents thoroughly, label with name and date prepared, filter, and store at 4 °C for up to 14 days. Alternatively, 50 mL aliquots can be made and stored at -20 °C. Day 1: RPMI media supplemented with 100 ng/mL Activin A and 50 ng/mL BMP-4. Day 2: RPMI media supplemented with 100 ng/mL Activin A and 0.2% KSR. Day 3: RPMI media supplemented with 100 ng/mL Activin A and 2% KSR without BMP4 (see Notes 1–3).
Foregut spheroid (FGS) induction (days 4–6) media: Complete Advanced DMEM/F2
Reagents Final concentration Amount Advanced DMEM/F12 1× 475 mL B27 supplement 1× 5 mL N2 supplement 1× 10 mL GlutaMAX 1× 5 mL P/S 1× 5 mL FGF-4 - - CHIR99021 - - Total n/a 500 mL Amounts of FGF-4 and CHIR99021 are not shown on the Table as we prepare them fresh every day and they are dependent on the number of wells and volumes you are working with. Swirl the bottle to mix contents thoroughly, label with name and date prepared, filter, and store at 4 °C for up to 14 days. Alternatively, 50 mL aliquots can be made and stored at -20 °C. Days 4–6 media: advanced DMEM/F12 supplemented with 3 µM CHIR99021 and 500 ng/mL FGF-4 (see Notes 1–3).
Human liver organoids (HLO) formation (days 7–10) media 1: Advanced DMEM/F12 + retinoic acid
Reagents Final concentration Amount Advanced DMEM/F12 1× 475 mL B27 supplement 1× 5 mL N2 supplement 1× 10 mL GlutaMAX 1× 5 mL P/S 1× 5 mL Retinoic acid 2 µM 1 mL Total n/a 500 mL Place the media in the dark or cover with foil.
Hepatocyte maturation (days 11–25) media: Hepatocyte culture medium (HCM)
Reagent Final concentration Amount HCM basal medium 1× 500 mL HGF - - OSM - - Dex - - Total n/a 500 mL Amounts for HGF, OSM, and Dex are not shown on the Table as we prepare them fresh every day in small quantities depending on the number of wells and volumes you are working with. Swirl the bottle to mix contents thoroughly, label with name and date prepared, filter, and store at 4 °C for up to 14 days. Alternatively, 50 mL aliquots can be made and stored at -20 °C. Days 11–25 media: supplement HCM medium with 10 ng/mL HGF, 0.1 µM Dex, and 20 ng/mL OSM.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
First-strand CDNA synthesis cocktail 1
Reagents Final concentration Amount mRNA 2 µg - Oligo dT primer n/a 1 µL ddH2O n/a - Total n/a 9 µL Note: The amount of mRNA will depend on the concentration of RNA per sample; the volume of water will be calculated based on the concentration value of RNA.
cDNA synthesis master mix 2
Reagents Amount 5 first synthesis strand buffer 5 µL dNTPs mix 1 µL RNase inhibitor 1 µL MgCl2 2 µL ImPromp II reverse transcriptase 1 µL ddH2O 6 µL Total 16 µL qRT-PCR reagents mix
Reagents Final concentration Amount SYBR Green PCR Master Mix n/a 6.25 µL Forward primer (10 µM) 10 µM 0.5 µL Reverse primer (10 µM) 10 µM 0.5 µL ddH2O n/a 4.25 µL cDNA n/a 1 µL Total n/a 12.5 µL
Laboratory supplies
Nunclon Sphera multi-well plates (Thermo Fisher Scientific, catalog number: 174930)
Sterile 15 mL Falcon tubes (Nest Biotechnology, catalog number: 602072)
Sterile 50 mL Falcon tubes (Nest Biotechnology, catalog number: 601001)
Equipment
Flow Cytometer (BD Biosciences, model: BD LSR Fortessa)
Biosafety cabinet suitable for cell culture (Nuaire)
Centrifuge (United Scientific, model: Orto Alresa)
Microplate Luminometer (Promega, model: GLOMAX 96)
Heat block (Acorn Scientific, model: Stuart)
Spectrophotometer (Thermo Fisher Scientific, model: NanoDropTM 2000)
Phase contrast/inverted microscope (Leica Microsystems, model: Leica DM IL LED)
Thermal Cycler (Thermo Fisher Scientific, model: QuantStudio 3 RT-PCR Systems)
Shellab CO2 Water Jacket HEPA Filter Incubator (United Scientific, model: SC05W-2)
Vortex machine (Labnet International)
Software and datasets
GraphPad Prism (GraphPad Software Inc.; version 5)
FlowJo software (Treestar; version 10)
Procedure
Coating plates with matrix
Vitronectin (VTN) solution
Thaw the stock vial of vitronectin (VTN) solution at RT or overnight at 4 °C.
Prepare 60 µL aliquots of 1 mL VTN and store at -80 °C until needed for the experiment.
For coating 6-well tissue culture plates or 35 mm dishes, dilute VTN (1:100 dilution, e.g., 60 µL in 6,000 µL in DBPS. Gently swirl the bottle to allow the solution to mix thoroughly.
Immediately dispense 1 mL of the VTN solution to as many wells of the 6-well plate as required, rock the plate to allow even distribution of the VTN solution across the surface of the plate, and incubate for 1 h at RT (see Note 1).
Geltrex
Thaw a 5 mL stock bottle of Geltrex overnight (16–20 h) on ice.
Working on ice and using chilled pipettes tips, mix well the contents without creating bubbles and aliquot into 1.5 mL cryotubes.
Label with name and date of preparation and store at -20 °C until needed for an experiment.
hiPSCs culture and organoid generation
Resuscitation of hiPSCs
Remove iPSCs from liquid nitrogen storage and thaw quickly in a 37 °C water bath.
Carefully add 1 mL of E8 + ROCK solution dropwise to the cryovial and transfer cells to a sterile 15 mL tube containing 8 mL of warm E8 + ROCK solution.
Pellet the cells by centrifugation at 398× g for 3 min at 4 °C.
Aspirate the supernatant and gently resuspend the pellet in 1 mL of warm E8 + ROCK solution (mix thoroughly by pipetting up and down). Transfer the cell suspension to one well of the pre-coated plate.
Agitate the plate gently within the tissue culture hood to ensure an even distribution of cells throughout the well and incubate cells at 37 °C and 5% CO2 overnight (see Note 5).
Check cell attachment under phase contrast microscope (see Note 6).
Change media daily by removing 95% of old medium using aspirator pipette. Cells require passaging upon 70%–80% confluency (see Note 7).
Passaging
Aspirate the old media and rinse the cells with 2 mL of DBPS.
Add 1 mL of 0.5 mM EDTA solution to the well to be passaged and rock the plate to cover the entire surface of the well.
Incubate at RT for 4–8 min and observe cells under a phase-contrast microscope.
Aspirate the 0.5 mM EDTA solution by tilting the plate forward slightly to collect the EDTA solution at the bottom edge of the well.
Immediately add 2 mL of complete E8 medium to the well and pipette the media to dislodge cell clusters (see Note 8).
Use the 2 mL of medium to gently wash the cells from the plate by pipetting the medium around the well approximately three times using a 5/10 mL serological pipette (see Note 6).
Dilute the cell suspension with complete E8 medium in a 15/50 mL tube at an appropriate cell density (in accordance with your desired split ratio, see Note 9).
Maintenance
Aspirate complete E8M from the hiPSCs and add 1 mL of Accutase solution per well to a 6-well plate. Place the plate in the incubator for 3–5 min, checking if cells detach. The cells are ready when they easily detach with gentle pipetting. Add 5 mL of complete E8 medium to a 15 mL Falcon tube.
Remove the plate from the incubator, add 1 mL of complete E8 medium, and lift the cells by gentle pipetting. Add this cell suspension in Accutase solution to the 5 mL complete E8 medium in the 15 mL Falcon tube. Gently pipette up and down twice to rinse off Accutase on the cells with complete E8 medium.
Centrifuge the cells at 398× g for 3 min at 4 °C.
Measure viable cells using Trypan blue exclusion method and calculate the number of viable cells/mL in the cell suspension.
For each cell line, optimal seeding cell density will need to be determined to achieve 90% confluency on day one.
For our hiPSCs cell line (N-N), 1.5 × 106 cells/well in a 6-well plate was ideal.
Aspirate the medium from the pellet and add 1 mL of complete E8M supplemented with 10 µM ROCK inhibitor solution to the VTN-coated 6-well plate. The ROCK inhibitor increases cell viability of the single cells in suspension. Add 1 mL of the cell suspension and the complete E8M supplemented with ROCK inhibitor solution and rock the plate back and forth and sideways to ensure even distribution of cells. Place the plate back in the incubator for 24 h.
At Day 0, warm complete E8M at RT. Confirm that the plated cells are healthy and growing at a uniformly high density. Aspirate the old medium and replenish the cells with warm complete E8M without ROCK inhibitor solution.
Place the plate back in the incubator for 24 h.
Step-by-step liver organoid method
Definitive endoderm (DE) induction (days 1–3)
To ensure a successful DE induction of hiPSCs, it is critical to start with high-quality hiPSCs. The cells should be at 80%–90% confluency with no sign of differentiation. Before beginning, warm complete E8 medium, Accutase, and base medium at RT.
Day 1: Cells should have reached 80%–90% confluency. Too low cell density will result in poor differentiation and cell death after Activin A solution treatment. Therefore, before beginning, titrate the cell number to ensure 80%–90% confluency of cells. It is possible to wait for an extra 24 h until the cells have reached the desired density.
Warm day 1 media at 37 °C. As Activin A solution treatment will result in cell death, prewarming the media at 37 °C increases the survival of cells. Aspirate complete E8M and add 2 mL/well of a 6-well plate of day 1 media and place the plate back in the incubator for 24 h.
Day 2: Check under the microscope if there still are layers of cells attached to the plate. If satisfied, aspirate floating dead cells and day 1 medium and replace with 2 mL/well for a 6-well plate of the warm day 2 media. Place the plate back in the incubator for 24 h. However, if there is little to no layer of cells attached, do not continue with the differentiation process and restart the protocol.
Day 3: Warm day 3 media at 37 °C. Aspirate Day 2 medium and replace with 2 mL/well for a 6-well plate of day 3 medium. Place the plate back in the incubator for 24 h.
Day 4: Endoderm cells should be confluent after differentiation (see Figure 1B). At this stage of the protocol, RT-qPCR and immunofluorescence can be performed to confirm gene-specific stage markers and cell localisation markers. Cells should express SRY-box transcription factor 17 (SOX17), Goosecoid (GSC), and Forkhead box protein A2 (FOXA2), and 85%–95% of the cells should co-stain double-positive for SOX17 and FOXA2.
Warm freshly prepared days 4–6 media at 37 °C. Each day, aspirate the media and replace with 2 mL/well of a 6-well plate of the prewarmed days 4–6 media. Owing to the number of cells present in the well, the media will turn yellow. Confirm the presence of 3D structures forming from the monolayer culture, including attached and floating spheroids (see Figure 1B).
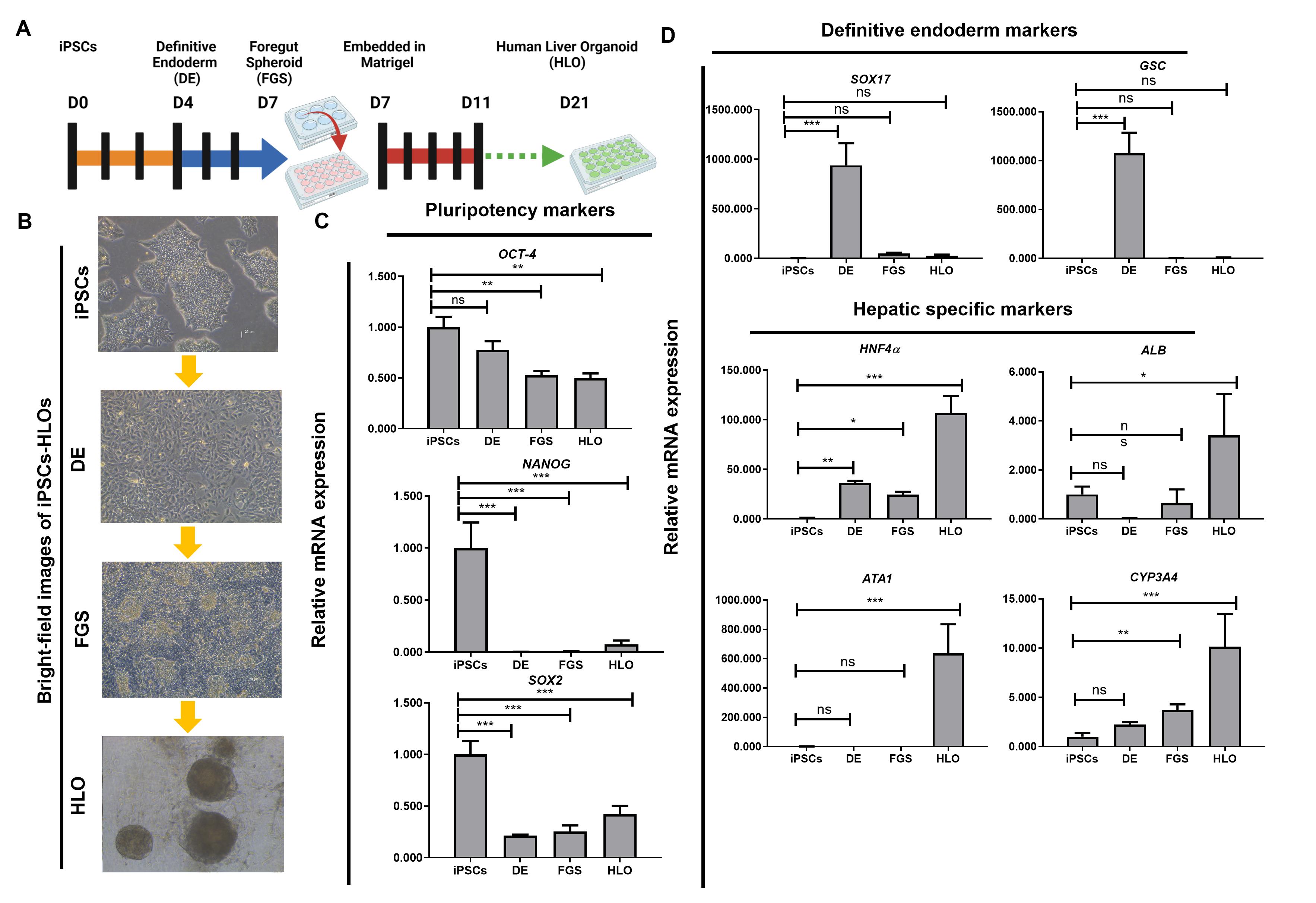
Figure 1. Schematic of liver organoid differentiation protocol. (A) Schematic overview of the differentiation method for liver organoids. (B) Phase-contrast images of iPSCs-HLO depicting change in cell morphology. Scale bar = 100 µm. (C) Reverse transcription qPCR analysis of represented genes related to pluripotency or undifferentiated state and (D) definitive endoderm (DE) state and hepatic function. Undifferentiated iPSCs (iPSCs, n = 3), DE (n = 3), posterior foregut spheroid (foregut, n = 3), and HLO (n = 3). Data are presented as mean ± SEM (n = 3) and analysed by Student t-tests and one-way analysis of variance (ANOVA) using Dunnett’s multiple comparison as a post-hoc test. p < 0.05*, p < 0.001**, and p < 0.0001***.HLO formation: enzymatic dissociation
Warm base medium, Accutase, and freshly made HLO formation medium at RT. In this step, collect the day 6 medium and floating spheroids into a 15 mL Falcon tube containing 5 mL of base medium. Add 2 mL of Accutase solution to each well and place the plate in the incubator for 3 min. Detach cells and pipette up and down four times. Aspirate and add cells–Accutase suspension to 5 mL of base medium.
Centrifuge cells at 398× g for 3 min at 4 °C. Completely aspirate the supernatant so it does not interfere with Geltrex.
Remove Geltrex from 4 °C and keep it on ice in the biosafety cabinet. Using chilled 1,000 µL pipette tips, use a P1000 pipette to mix the Geltrex thoroughly without introducing bubbles and add 1 mL of Geltrex to the pellet of cells in the 15 mL Falcon tube. Gently mix the contents by pipetting up and down several times to ensure even distribution of cells in the suspension. Keep the tube on ice.
For plating, use 24-well Nunclon Sphera multi-well plates. To each well of 24-well plate, add 40 µL (two 20 µL) drops of Geltrex–cell suspension at the centre of the plate forming a dome-shape.
Gently place the plate back in the incubator for 15–20 min to solidify Geltrex.
Add 500 µL of day 7 medium to each well of a 24-well plate and place it back into the incubator. There will be different sizes of 3D structures. Change medium every two days.
On day 9, 48 h later, warm the freshly made HLO formation medium at RT. Gently, aspirate old medium without disturbing the Geltrex drops and add 500 µL of fresh HLO formation medium per well of a 24-well plate.
Mechanical dissociation: cryopreservation of FGS
Alternative to HLO enzymatic dissociation step, we cryopreserve the FGS cells post mechanical dissociation of cells. Longer culture periods often introduce potential contaminations. This step is necessary for resting the FGS and is optional.
Use day 6 medium in each well and pipette up and down forcefully (5×) using a P1000 pipette.
Collect cell clusters and transfer into a 15 mL Falcon tube.
Centrifuge cells at 398× g for 3 min at 4 °C.
Aspirate supernatant and resuspend pellet in 1 mL of cryopreservation medium (30% basal advanced DMEM/F12, 60% FBS, and 10% DMSO).
Carry the cryopreservation tubes in a box containing cotton cloth dampened with isopropanol.
Store at -80 °C for two weeks or in liquid nitrogen for long-term storage.
FGS resuscitation & formation medium 2
FGS resuscitation & formation 2 consists of advanced DMEM/F12 + 5 factors (5 µM A83-01 solution, 20 ng/mL FGF-10 solution, 3 µM CHIR99021 solution, 20 ng/mL EGF solution, and 50 ng/mL VEGF solution).
Thaw FGS cells in organoid formation medium 2 supplemented with Y27632 solution (10 µM).
Centrifuge cells at 398× g for 3 min at 4 °C.
Completely aspirate the supernatant so it does not interfere with Geltrex.
Remove the Geltrex from 4 °C and keep it on ice in the biosafety cabinet. Using chilled 1,000 µL pipette tips, use a P1000 pipette to mix the Geltrex thoroughly without introducing bubbles and add 1 mL of Geltrex to the pellet of cells in the 15 mL Falcon tube. Gently mix the contents by pipetting up and down several times to ensure an even distribution of cells in the suspension. Keep the tube on ice.
For plating, see enzymatic dissociation section above.
On day 9, 48 h later, warm the freshly made HLO formation medium for 30 min at RT. Gently, aspirate old medium without disturbing the Geltrex drops and add 500 µL of fresh HLO formation medium per well of a 24-well plate every two for four days (see Note 11).
HLO maturation
On day 11, switch to HCM medium, which is hepatocyte-specific medium. Warm HCM to RT, aspirate HLO formation medium, and add 500 µL of HCM to each well of a 24-well plate.
Every 3 days, exchange the media by aspirating media in the well and adding 500 µL of fresh HCM.
Day 18: On this day, do not aspirate the media but pipette the media up and down to dislodge the cell drop. Add 1 mL of cold DBPS to dissociate the Geltrex until no pieces are visible. Place 250 µm cell strainer on a 15 mL tube and collect cells from each well and sieve through the cell strainer. This step ensures the removal of extra mesenchymal-like larger aggregates of cells. Also, this step is necessary where organoids will need to be individually sorted for downstream applications and sorted into 96- or 384-well plates. Warm fresh HCM at RT.
Add 4 mL of DBPS to the 15 mL Falcon tube with organoids and centrifuge at 398× g for 3 min at 4 °C.
Aspirate the supernatant, add HCM with 10% Geltrex to pelleted organoids, and pipette up and down to mix the cells. The cells will still be supported by the Geltrex matrix in suspension. Add 40 µL of Geltrex–cell suspension to each well of a new 24-well plate and place the plate back in the incubator.
Day 21: On this day, organoids can start to be used for downstream assays such as functional assays (see Figure 2A–2C) and staining. Confirm if the organoids are healthy and growing (see Figure 1B and 1D). No aspirations are needed for organoids not chosen for downstream assays; add 500 µL of fresh HCM and place the plate back in the incubator.
Perform quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) for hepatic gene markers (Figure 1C–1D), enzyme linked immunosorbent assay (ELISA) to detect cytokines (Figure 4), and flow cytometry (Figure 2) to analyse the organoids.
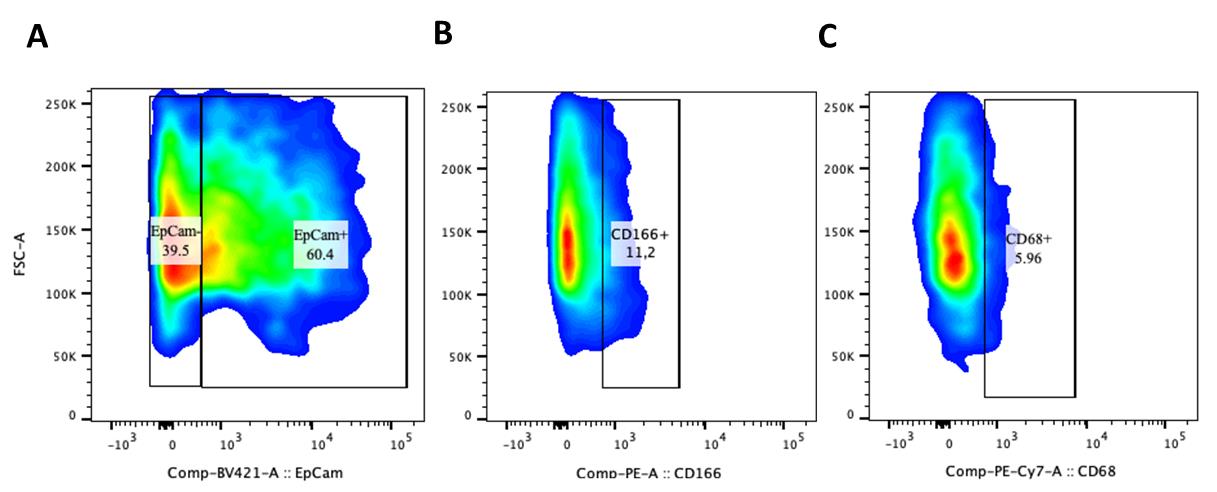
Figure 2. Plot showing the percentage of EpCAM+, CD166+, and CD68+ cells. Human liver organoids (HLO) were examined for the co-expression of EpCAM+, CD166+, and CD68+ cells. Briefly, HLO were rinsed with DBPS, followed by dissociation into single organoids using the TrypLE express solution. Subsequently, dissociated HLO were incubated with FACS buffer solution containing EpCAM (1:80), CD166 (1:160), and CD68 (1:80) antibodies. The y-axis represents forward scatter (FSC-A), where an increased signal indicates the frequency of cells in HLO. The x-axis indicates the GFP fluorescence. The black lines are included to guide the eye to distinguish the fluorescence frequencies between EpCAM- cells and EpCAM+, CD166+, and CD68+ stained cells present in our HLO. (A) Percentage of EpCAM+ stained cells. (B) Percentage of CD166+ stained cells. (C) CD68+ stained cells of the whole organoids cell population after 21 days of HLO culture. EpCAM+ cells (60.4%), CD166+ (11.2%), and CD68+ (5.96%), respectively.
Total RNA isolation and RT-qPCR
Total RNA isolation
First, isolate the organoids from Geltrex using 1 mL of TrypLETM Express Enzyme solution in the 24-well plate and collect them from each well into a 15 mL Falcon tube.
Subsequently, rinse the organoids twice with 3 mL of DBPS followed by centrifugation at 398× g for 3 min at 4 °C until a thin layer of Geltrex remains.
Add 1 mL of TRIzolTM reagent to the pelleted organoids to extract total RNA following the manufacturer’s instructions.
Quantify extracted RNA using the ND-1000 Nanodrop spectrometer and store RNA at -80 °C until needed for further experiments.
Complementary DNA (cDNA) synthesis
For first-strand cDNA synthesis, reverse transcribe the total RNA using the ImProm-II reverse transcriptase following manufacturer's instructions, as shown in Recipes 1 and 2 of the Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
Mix 2 µg of template RNA, 1 µL Oligo dT, and ddH2O to a final volume of 9 µL. Heat the mixture for 10 min at 70 °C in a thermal block cycler with a heated lid to denature any secondary structure in the RNA and to minimise evaporation of solutions. Place the mixture on ice for 5 min.
Prepare 16 µL of the second cocktail [see Recipe 2 of Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)] by adding and mixing reagents outlined in Recipe 1 of Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), followed by centrifugation.
Incubate the mixture at 25 °C for 5 min to anneal oligo DT primers to the template cDNA, followed by 2 h of incubation at 42 °C.
Inactivate reverse transcriptase by heating the incubated mixture for 10 min at 70 °C, followed by stopping reaction and placing the tubes on ice. Synthesised cDNA may be diluted and stored at -20 °C until needed for further experiments.
qRT-PCR
Perform qRT-PCR in triplicates [see Recipe 3 of Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)] using PowerUpTM SYBRTM Green Master Mix and amplify synthesised cDNA on a thermal cycler with heated lid.
Calculate relative mRNA gene expression of 2-∆∆Ct method with GAPDH as a normalisation control. All primers (Table 3) were purchased from Integrated DNA Technologies (IDT).
Flow cytometry analysis
Optional: Before starting, coat the tubes and pipette tips with 1% BSA to reduce the loss of organoids that adhere to the sides of tips and tubes.
Collect organoids from wells of the 24-well plate into 15 mL Falcon tubes. If you want to process only one well, then collect from one well only. Add 6 mL of cold DBPS and gently pipette up and down to wash the Geltrex from the organoids.
Centrifuge the Falcon tube(s) at 398× g for 3 min at 4 °C. There will be a pellet of organoids at the bottom of the tube, a small layer of Geltrex in the middle, and media layered on top. Gently aspirate the media without disturbing the pellet and Geltrex layer.
Wash for a second time with 6 mL of DBPS and centrifuge the cells at 398× g for 3 min at 4°C.
After aspirating the DBPS, add 1 mL of TrypLETM solution and incubate for 10 min at RT to dissociate the organoids into single cells. After 10 min of incubation, add 4 mL of DBPS and centrifuge at 398× g for 3 min at 4 °C.
Use DBPS to wash out TrypLETM solution and centrifuge the cells again.
Aspirate the supernatant and resuspend cells in 100 µL of antibody + FACS buffer solution.
Incubate for 30 min at RT in the dark or cover the tubes with foil.
Add 100 µL of FACS buffer and centrifuge the cells at 398× g for 3 min at 4 °C.
Aspirate the supernatant and resuspend the pellet in 200 µL of FACS buffer.
Transfer the entire 200 µL of resuspended pellet into FACS tubes.
Acquire the samples on the BD LSR Fortessa flow cytometer (Figure 2).
Modelling liver injury
Antiretroviral (ARV), anti-tuberculosis drugs-induced hepatotoxicity
Perform steps E1–E5.
Aspirate the supernatant and resuspend cells in a mix of antiretroviral drugs or a mix of anti-retroviral drugs and anti-tuberculosis drugs.
Incubate the organoids with the drugs for 24 h at 37 °C in a 5% CO2 incubator.
After 24 h, take images of the organoids using a phase-contrast microscope (Figure 3A).
Collect the supernatants and analyse cytokine expression using ELISA (Figure 4).
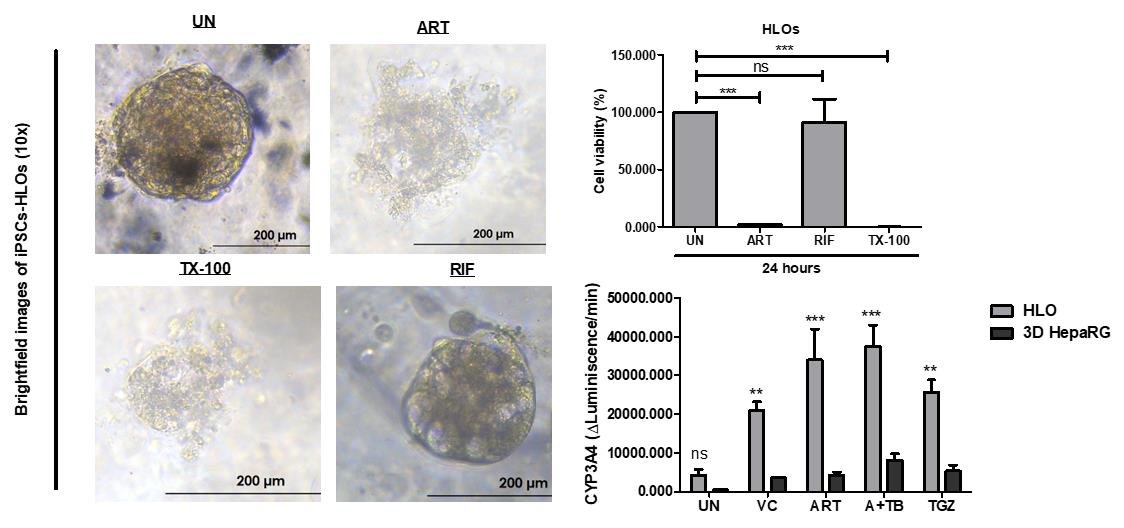
Figure 3. Drug treatment of human liver organoids (HLO), day 23. HLO were rinsed with DBPS, followed by dissociation into single organoids using the TrypLE express solution. Subsequently, dissociated HLO were embedded in Geltrex and cultivated in 96-well sphere U-bottom plates for 24 h. HLO were further incubated with media only (UN), ART, Rifampicin (50 µM), and 1% Triton X-100 (TX-100) to determine cell viability using 3D GLO cell viability assay and CYP3A4 activity using the CYP3A4 GLO assay. (A) HLO phase contrast images of different treatment groups: untreated (UN), ART, rifampicin (RIF), TX-100 (Triton X-100). (B) Cell viability percentages. (C) CYP3A4 enzyme activity in untreated HLO, ART, and ART+TB treated HLO and 3D HepaRG model post 72 h of treatment. Data are presented as mean ± SEM (n = 3) and were analysed by one-way analysis of variance (ANOVA) using Dunnett’s multiple comparison as a post-hoc test. Ns: non-significant, p < 0.05*, p < 0.001**, and p < 0.0001***.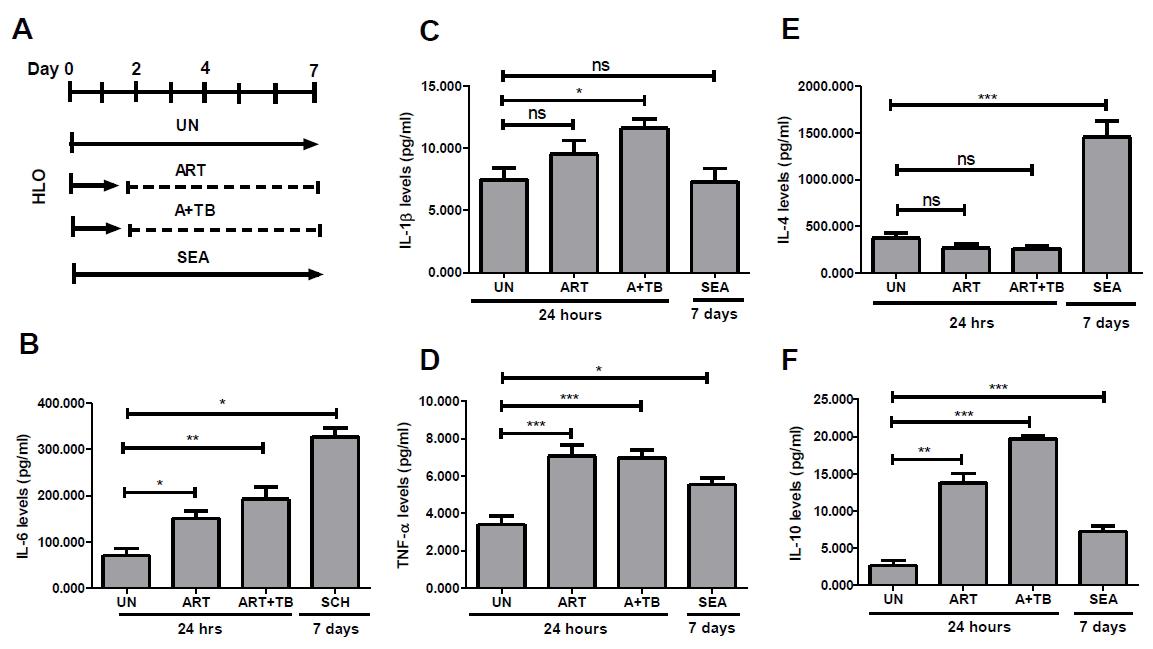
Figure 4. Inflammatory response after drug and pathogen treatment in human liver organoids (HLO). HLO were rinsed with DBPS, followed by dissociation into single organoids using the TrypLE express solution. Subsequently, dissociated HLO were embedded in Geltrex and cultivated in 96-well sphere U-bottom plates for 24 h. HLO were incubated with media only, ART & A+TB for 24 h, and SEA for 7 days. After each incubation time or treatment period, supernatants were aspirated and aliquoted onto the wells of a corresponding 96-well plate. Supernatants were used to carry out ELISA assay using BD Bioscience ELISA kit according manufacturer’s protocol. (A) Schematic diagram showing treatment duration for ART, A+TB- and SEA-induced liver injury. (B) Levels of proinflammatory mediators IL-6, (C) IL-1β, and (D) IL-4 in ART, A+TB post 24 h, and SEA post 7 days compared to UN counterpart. (D) Levels of the pathological mediator (E) TNF-α and (F) the anti-inflammatory mediator IL-10 compared to UN group. Data are presented as mean ± SEM (n = 7) and analysed by one-way analysis of variance (ANOVA) using Dunnett’s multiple comparison as a post-hoc test. p > 0.05ns, p < 0.05*, p < 0.001**, and p < 0.0001***.Induction of inflammation with Schistosome eggs antigen (SEA)
Follow steps F1a–e described in the induction of hepatotoxicity with drugs.
Stimulate the organoids with 20 µg/mL of SEA and incubate the organoids for seven days at 37 °C in a 5% CO2 incubator.
After seven days, take the images using a phase contrast microscope (Figure 3A).
Collect the supernatants and analyse cytokine expression using the enzyme linked immunosorbent assay (ELISA) (Figure 4).
Cell viability determination using CellTiter-GLO® cell viability assay
Treat organoids with 0.05% DMSO, 1% Triton X-100, and a combination of antiretroviral drugs (ARVs) or rifampicin (RIF) for 24 h in a 5% CO2 incubator.
Remove the supernatant from each well. Add 100 µL of CellTiter-GLO® reagent to each well and 100 µL of hepatocyte culture media (HCM).
Mix the contents vigorously for 5 min to induce cell lysis.
Incubate the plate for 25 min at RT to stabilise the luminescent signal.
Read the plate using the GLOMAXTM 96 Microplate Luminometer (Figure 3B).
Cytochrome P450 Assay
Treat organoids or 3D HepaRG cells (control cell line) with troglitazone (TGZ) or rifampicin (RIF) for 24, 48, and 72 h. Vehicle control is 0.05% DMSO.
Remove the supernatants from each well. Wash cells twice with ice-cold PBS.
Add 50 µL of 50 µM P450-GloTM CYP3A4 (5 mM luciferin-BE) dissolved either in advanced DMEM or full HCM to each well.
Incubate the mixture at 37 °C in a 5% CO2 incubator for 4 h.
Collect the supernatants and transfer 25 µL of the supernatant to white-walled clear-bottom plates. Add 25 µL of luciferin detection reagent (reconstitution buffer with luciferase detection reagent).
Incubate the plate at 37 °C in a 5% CO2 incubator for 30 min.
Read the plate using the GLOMAXTM 96 Microplate Luminometer (Figure 4C).
Validation of protocol
This protocol or parts of it has been used and validated in the following research articles:
Mun et al. [8]. Generation of expandable human pluripotent stem cell derived hepatocyte-like liver organoids. Journal of Hepatology. 2019; 71:970–985. See Figure 2A.
Ouchi et al. [9]. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metabolism. 2019; 30: 374–384.e6. See Figure 1A–D.
The robustness and reproducibility of our protocol can be evidenced from the above research articles.
General notes and troubleshooting
General notes
Store all media at 4 °C. Aliquot and store at -20 °C if required.
The growth factors (e.g., Activin, BMP-4, FGF-4, CHIR99021, Dex, HGF, RA, and OSM) should be added fresh every day.
Complete essential 8 medium, RPMI-1640 medium, advanced DMEM medium, and all growth factors require filtering before use. HCM does not require filtering.
Plates that have been coated with Vitronectin or Geltrex should be covered with parafilm and stored at 4 °C for up to one week. They should be clearly labelled with the date they were coated. Discard any plates not used within one week.
When plating cells, agitate the plate gently within the tissue culture hood to ensure even distribution of cells across the well, as colonies tend to settle in the centre of the plate affecting cell replating and differentiation.
After 24 h of plating iPSCs, check for cell attachment. If cell attachment is good, change medium to 2 mL of E8 media. If there are more cells floating than attached, then top up with 1 mL of freshly made E8 + ROCK solution.
Cells should be passaged when they have reached 70% confluency and are well compacted and the colonies have well-defined edges. Cells may also require passaging if levels of differentiation exceed that of iPSC or colonies start to look overgrown or unhealthy.
Split cells in the ratio of 1:4 to 1:6 (e.g., transferring all colonies from four or six wells).
When washing cells with 2 mL of E8 media after aspirating 0.5 mM EDTA solution, ensure to pipette the media up and down as this should dislodge cell clusters without dislodging any differentiated cells. Do not over pipette because it may result in single cells rather than cell clusters.
Forty-eight hours post FGS resuscitation, replace the resuscitation & organoid medium with organoid formation medium (supplemented with RA) every two days for a period of four days. Proceed with the protocol to HLO maturation.
Cryopreserved FGS-formed organoids get ready for downstream assays on day 23 instead of 21.
Acknowledgments
We would like to thank Dr Janine Scholefield from the Council for Scientific and Industrial Research (CSIR) in South Africa for generously gifting us with the induced pluripotent stem cell line. This study was funded by the South African Medical Research Council (SA-MRC) Self-Initiated Research grant and the National Research Foundation Competitive Support for Unrated Researchers (CSUR) grant (grant number 116260) awarded to Associate Professor Hlumani Ndlovu. This protocol was adapted from protocols developed by Mun et al. [8] and Ouchi et al. [9].
Competing interests
There are no competing interests to disclose.
Ethical considerations
No ethical considerations to disclose.
References
- Huch, M., Gehart, H., van Boxtel, R., Hamer, K., Blokzijl, F., Verstegen, M. M., Ellis, E., van Wenum, M., Fuchs, S. A., de Ligt, J., et al. (2015). Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell. 160: 299–312.
- Zeilinger, K., Freyer, N., Damm, G., Seehofer, D. and Knöspel, F. (2016). Cell sources for in vitro human liver cell culture models. Exp Biol Med. 241(15): 1684–1698.
- Yokoyama, Y., Sasaki, Y., Terasaki, N., Kawataki, T., Takekawa, K., Iwase, Y., Shimizu, T., Sanoh, S. and Ohta, S. (2018). Comparison of Drug Metabolism and Its Related Hepatotoxic Effects in HepaRG, Cryopreserved Human Hepatocytes, and HepG2 Cell Cultures. Biol Pharm Bull. 41(5): 722–732.
- Godoy, P., Hewitt, N. J., Albrecht, U., Andersen, M. E., Ansari, N., Bhattacharya, S., Bode, J. G., Bolleyn, J., Borner, C., Böttger, J., et al. (2013). Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 87(8): 1315–1530.
- Pfeiffer, E., Kegel, V., Zeilinger, K., Hengstler, J. G., Nüssler, A. K., Seehofer, D. and Damm, G. (2014). Featured Article: Isolation, characterization, and cultivation of human hepatocytes and non-parenchymal liver cells. Exp Biol Med. 240(5): 645–656.
- Broutier, L., Andersson-Rolf, A., Hindley, C. J., Boj, S. F., Clevers, H., Koo, B. K. and Huch, M. (2016). Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc. 11(9): 1724–1743.
- Siller, R., Greenhough, S., Naumovska, E. and Sullivan, G. J. (2015). Small-Molecule-Driven Hepatocyte Differentiation of Human Pluripotent Stem Cells. Stem Cell Rep. 4(5): 939–952.
- Mun, S. J., Ryu, J. S., Lee, M. O., Son, Y. S., Oh, S. J., Cho, H. S., Son, M. Y., Kim, D. S., Kim, S. J., Yoo, H. J., et al. (2019). Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J Hepatol. 71(5): 970–985.
- Ouchi, R., Togo, S., Kimura, M., Shinozawa, T., Koido, M., Koike, H., Thompson, W., Karns, R. A., Mayhew, C. N., McGrath, P. S., et al. (2019). Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 30(2): 374–384.e6.
- Garnier, D., Li, R., Delbos, F., Fourrier, A., Collet, C., Guguen-Guillouzo, C., Chesné, C. and Nguyen, T. H. (2018). Expansion of human primary hepatocytes in vitro through their amplification as liver progenitors in a 3D organoid system. Sci Rep. 8(1): 8222.
- Kruitwagen, H. S., Oosterhoff, L. A., Vernooij, I. G., Schrall, I. M., van Wolferen, M. E., Bannink, F., Roesch, C., van Uden, L., Molenaar, M. R., Helms, J. B., et al. (2017). Long-Term Adult Feline Liver Organoid Cultures for Disease Modeling of Hepatic Steatosis. Stem Cell Rep. 8(4): 822–830.
- Takebe, T., Sekine, K., Enomura, M., Koike, H., Kimura, M., Ogaeri, T., Zhang, R. R., Ueno, Y., Zheng, Y. W., Koike, N., et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 499(7459): 481–484.
- Nie, Y. Z., Zheng, Y. W., Ogawa, M., Miyagi, E. and Taniguchi, H. (2018). Human liver organoids generated with single donor-derived multiple cells rescue mice from acute liver failure. Stem Cell Res Ther. 9(1): 5.
Supplementary information
The following supporting information can be downloaded here:
- Figure S1. Workflow for establishment, expansion, and differentiation of iPSCs-HLOs.
- Figure S2. Phase-contrast images representative of iPSCs-HLO at different days of the differentiation protocol.
- Table S1. Drugs and antigen treatment groups.
Article Information
Publication history
Received: Dec 5, 2023
Accepted: Apr 30, 2024
Available online: Jul 18, 2024
Published: Aug 5, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Maepa, S. W., Marakalala, M. J. and Ndlovu, H. (2024). Generation of Multicellular 3D Liver Organoids From Induced Pluripotent Stem Cells as a Tool for Modelling Liver Diseases. Bio-protocol 14(15): e5042. DOI: 10.21769/BioProtoc.5042.
Category
Stem Cell > Pluripotent stem cell > Regenerative medicine
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link







