- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Simplified Protocol to Demonstrate Gene Expression in Nicotiana benthamiana Using an Agrobacterium-Mediated Transient Assay
Published: Vol 14, Iss 10, May 20, 2024 DOI: 10.21769/BioProtoc.4987 Views: 2787
Reviewed by: Samik BhattacharyaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2272 Views
Production of Homogeneous, Functional Zinc-Finger Arrays in High Yield With Two Chromatographic Steps
Jingchang Liang [...] Pierre-Jean Matteï
Aug 20, 2025 1430 Views
Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1125 Views
Abstract
Agrobacterium-mediated transient gene expression in Nicotiana benthamiana is widely used to study gene function in plants. One dramatic phenotype that is frequently screened for is cell death. Here, we present a simplified protocol for Agrobacterium-mediated transient gene expression by infiltration. Compared with current methods, the novel protocol can be done without a centrifuge or spectrometer, thereby suitable for K-12 outreach programs as well as rapidly identifying genes that induce cell death.
Key features
• The protocol simplifies the widely used Agrobacterium-mediated transient gene expression assay [1] and can be completed within one week when plants are available.
• Rice XB3 gene can induce a dramatic and easily identifiable cell death phenotype in Nicotiana benthamiana.
• Allows identification of cell death–inducing genes and is suitable for teaching.
• Compared to the currently used methods, our protocol omits the use of agroinfiltration buffer, pH meter, temperature-controlled growth chamber, centrifuge, and spectrophotometer.
Keywords: AgrobacteriumGraphical overview

Agrobacterium infiltration (agroinfiltration) of Nicotiana benthamiana. The photo demonstrates the method of agroinfiltration into the abaxial side of leaves using a needleless syringe.
Background
Agrobacterium-mediated transient transformation of the plant Nicotiana benthamiana is a powerful tool widely used in molecular biology studies [2,3]. This assay is ideal for rapidly demonstrating gene expression, especially when used for the visible cell death phenotype induced by specific plant genes from various species. While holding summer professional development workshops for pre-college educators, we found that the current protocols for Agrobacterium-mediated transient gene expression are difficult to use in pre-college classrooms due to lack of the necessary equipment and reagents.
The rice (Oryza sativa) cell surface innate immune receptor XA21 specifies resistance to bacterial strains of Xanthomonas oryzae pv. oryzae [4]. XA21 interacts with a number of proteins in vitro and in vivo. One protein that XA21 interacts with is called XB3 for XA21 binding protein 3 [5]. XB3 induces highly visible cell death in N. benthamiana when over-expressed [1]. To support public education and quickly identify cell death–inducing genes, we developed a novel protocol to simplify the current Agrobacterium-mediated transient gene expression assay.
Materials and reagents
N. benthamiana seeds (available from numerous plant science research labs)
pC1300S-XB3: Agrobacterium tumefaciens strain EHA105 (Intact Genomics, catalog number: 1084-06) harboring the construct pC1300S-XB3 [1]
EV: A. tumefaciens strain EHA105 harboring pC1300S (empty vector)
P19: A. tumefaciens carrying the p19 construct. The P19 protein from tomato bushy stunt virus is an efficient suppressor of posttranscriptional gene silencing, which enables higher expression of the gene of interest [6]
Luria-Bertani broth (LB Broth) (Fisher Scientific, catalog number: 10855001)
Kanamycin sulfate (Fisher Scientific, catalog number: BP906-5)
Rifampicin (PhytoTechnology Lab, catalog number: R501)
100 mM acetosyringone (PhytoTechnology Lab, catalog number: A1104)
Filtered water
Soilless potting mix (Growing Mix, PRO-LINE HFC/B HydraFiber, from Jolly Gardener)
Solutions
Kanamycin sulfate stock solution (50 mg/mL) (see Recipes)
Rifampicin stock solution (50 mg/mL) (see Recipes)
Infiltration buffer [1] (see Recipes)
Recipes
Kanamycin sulfate stock solution (50 mg/mL)
Dissolve 0.5 g of kanamycin sulfate (powder) in 10 mL of distilled water at room temperature. Filter through a 0.2 μm filter. Aliquot in 1 mL fractions and store in a -20 °C freezer.
Rifampicin stock solution (50 mg/mL)
Dissolve 0.5 g of rifampicin (powder) in 10 mL of 100% methanol at room temperature. Aliquot in 1 mL fractions and store in a -20 °C freezer.
Infiltration buffer [1]
Dissolve 1.95 g of MES [2-(N-Morpholino)ethanesulfonic acid] and 1.96 g of MgCl2 in 1,000 mL of sterile water at room temperature. Adjust pH to 5.6. Aliquot in 1 mL fractions and store in a -20 °C freezer.
Laboratory supplies
9-cm square disposable pots with drainage holes (any manufacturer)
15 mL culture tubes (Carolina Biological Supply, catalog number: 215090)
50 mL conical screw cap tubes (Carolina Biological Supply, catalog number: 215095)
Syringes (1 cc) (Carolina Biological Supply, catalog number: 697765)
10 mm cork borer (Carolina Biological Supply, catalog number: 712202)
Equipment
Forceps for transplanting seedlings (Carolina Biological Supply, catalog number: 624790)
Incubator shaker (New Brunswick Scientific, model: C24KC)
Conductivity meter with electrode (Eutech, model: Cond 6+ ECCON603PLUS)
Deep freezer with -20 °C (any manufacturer)
Bunsen burner (Carolina Biological Supply, catalog number: 706706)
Laminar flow hood (any manufacturer)
Procedure
Growth conditions for N. benthamiana plants
Fill a 9 cm disposable square pot with moist potting soil and sprinkle 20–50 N. benthamiana seeds on the surface of the soil. Cover the pot with plastic wrap or a plastic dome to create a humid environment. Place the pot for two weeks in a warm location with a day length of 16 h (24 °C) and a period of darkness of 8 h (21 °C) each day. Once the seeds have germinated and the seedlings are 1–1.5 cm tall, remove the plastic wrap. Water the plants twice a week (~20 mL water/pot).
After two weeks, transplant the germinated seedlings to new pots containing moist potting soil (one seedling per pot) and grow them in the above-mentioned conditions.
Water the plants regularly (~20 mL water/pot, every 48 h), keeping the soil consistently moist but not waterlogged and growing the plants under the same conditions as described above for another five weeks until they reach the 4–6 true-leaf stage.
Note: Keep an eye on the plants for pests and diseases. Fertilize plants with a balanced liquid fertilizer every 2–3 weeks during the growing season, if required. The optimal stage for Agrobacterium infiltration (agroinfiltration) for the experiment occurs when the plants reach a young vegetative stage, characterized by actively dividing and expanding leaf area.
Preparation of A. tumefaciens culture
Inoculate individual A. tumefaciens EHA105 strains carrying p19, pC1300S-XB3, or pC1300S (EV) (Figure 1) into three separate 15 mL culture tubes each containing 3 mL of LB broth supplemented with kanamycin (100 μg/mL) and rifampicin (30 μg/mL) in a laminar flow hood. Place the culture tubes in a shaker set at 28 °C and a rotation rate of 220 rpm and incubate for 36–40 h in the dark.
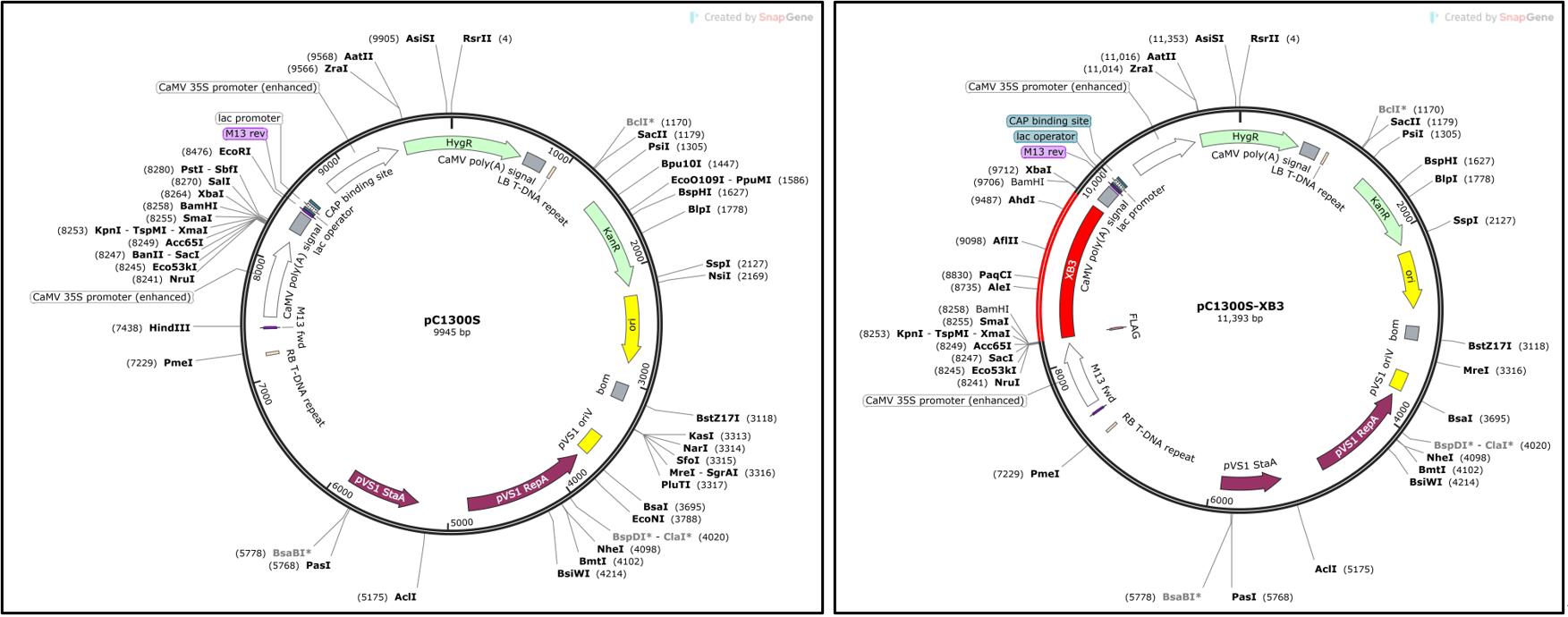
Figure 1. Maps of the pC1300S (EV, kindly provided by Dr. Yinong Yang) and pC1300S-XB3 [1] constructs used in this study. P19 is described in Voinnet et al. [6].Establish secondary cultures by taking 25 μL from the primary culture and inoculating into 50 mL conical screw cap tubes filled with 15 mL of LB broth supplemented with kanamycin (100 μg/mL) and rifampicin (30 μg/mL). Grow the new culture at 28 °C overnight in the dark reaching the logarithmic growth phase of the bacteria.
Dilute the overnight bacterial culture into LB broth supplemented with kanamycin (100 μg/mL) and rifampicin (30 μg/mL) based on the visual guidance provided in Figure 2, which covers a range of dilutions of the control LB broth supplemented with kanamycin (100 μg/mL) and rifampicin (30 μg/mL) to translucent and turbid cultures containing bacterial cells. Add 30 μL of 100 mM acetosyringone into 15 mL of bacterial culture.

Figure 2. Reference for bacterial dilution for agroinfiltration experiments. An overnight Agrobacterium culture containing pC1300S-XB3 in LB broth supplemented with kanamycin (100 μg/mL) and rifampicin (30 μg/mL) is diluted to different bacterial densities (within the range of OD600 0.1 to 1). The visual representation of Agrobacterium at varying densities offers a visual target for dilution without a spectrometer.Incubate the diluted bacterial suspensions for an additional 3 h at room temperature to activate the virulence machinery and prime the cells for successful genetic transformation.
Note: If a laminar flow hood is not available, bacterial inoculation can be performed using a Bunsen burner. A temperature range of 28–30 °C is optimal for the growth and replication of A. tumefaciens. Overnight growth of Agrobacterium culture often leads to an OD600 value within the range of 0.8–1.0.
Agroinfiltration using pC1300S-XB3 and controls
Two Agrobacterium cultures will be combined: either pC1300 (EV) and p19 or pC1300S-XB3 and p19 in a proportion of 3:2 to prepare samples for infiltration.
Inject the abaxial side (lower surface) of N. benthamiana leaves with the two prepared Agrobacterium mixtures using needleless syringes under controlled pressure. Separate the injection sites of each test mixture.
Incubate the infiltrated plants at room temperature with humidity for a defined period of 12–72 h. This incubation is essential for the plants to acclimatize to the introduced genetic material and to enable the expression of the transferred genes.
Notes:
The volume of bacterial cultures used for infiltration can be scaled accordingly. Always include the empty vector control (pC1300S) as one of the infiltration spots in your experiments to validate the success of the infiltration and to identify any potential issues with the transformation process. We have noticed that healthy plant conditions are crucial for the success of the infiltration experiments. Sometimes, this is not easy to visualize from plant appearance. The empty vector control serves as the “baseline” to phenotype the effect of XB3.
As shown in Figure 3, similar results were obtained when LB broth was used to replace infiltration buffer for diluting bacterial cultures.
Monitor cell death response
The infiltrated leaves are tested for the expression of the XB3 gene by monitoring cell death detected as visible grey patches 12–48 h after agroinfiltration (Figures 3A and 3B).
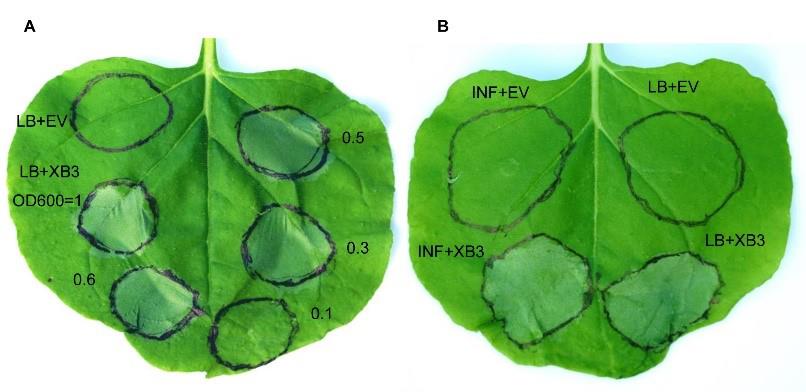
Figure 3. Leaf collapse induced by expression of XB3. (A) Cell death phenotypes induced by various densities of the agrobacterial culture harboring pC1300S-XB3. Photo taken 48 h after infiltration. EV, empty control. Visible cell death phenotypes were observed when the leaves were infiltrated with the Agrobacterium at densities ranging from OD600 = 0.3 to 1. (B) Cell death phenotypes induced by the Agrobacterium harboring pC1300S-XB3 in LB medium and infiltration buffer at OD600 of 0.5. The figure shows an N. benthamiana leaf 48 h after infiltration by four different solutions. Top-left quadrant of the leaf was infiltrated by Agrobacterium containing the empty vector diluted in infiltration buffer and the bottom-left quadrant was infiltrated with Agrobacterium containing XB3 expression plasmid (pC1300S-XB3) diluted in infiltration (INF) buffer at OD600 of 0.5. Similar cell death was observed when LB was used to dilute the Agrobacterium cultures for infiltration (Right).Ion leakage assay
Electrolyte leakage of infiltrated leaves can be quantified to assess cellular mortality. Punch out three leaf discs, each 10 mm in diameter, from each region infiltrated by Agrobacterium using a 10-mm cork borer.
Submerge these leaf discs in 10 mL of water. Incubate for 2 h at room temperature with gentle agitation of 160 rpm.
Measure the conductivity of the resulting solution using the COND 6+ conductivity meter according to the manufacturer’s instructions (Figure 4). In order to calibrate the conductivity meter, rinse the electrode with sterile water and calibrate it with water, as in our experiment, water served as standard.
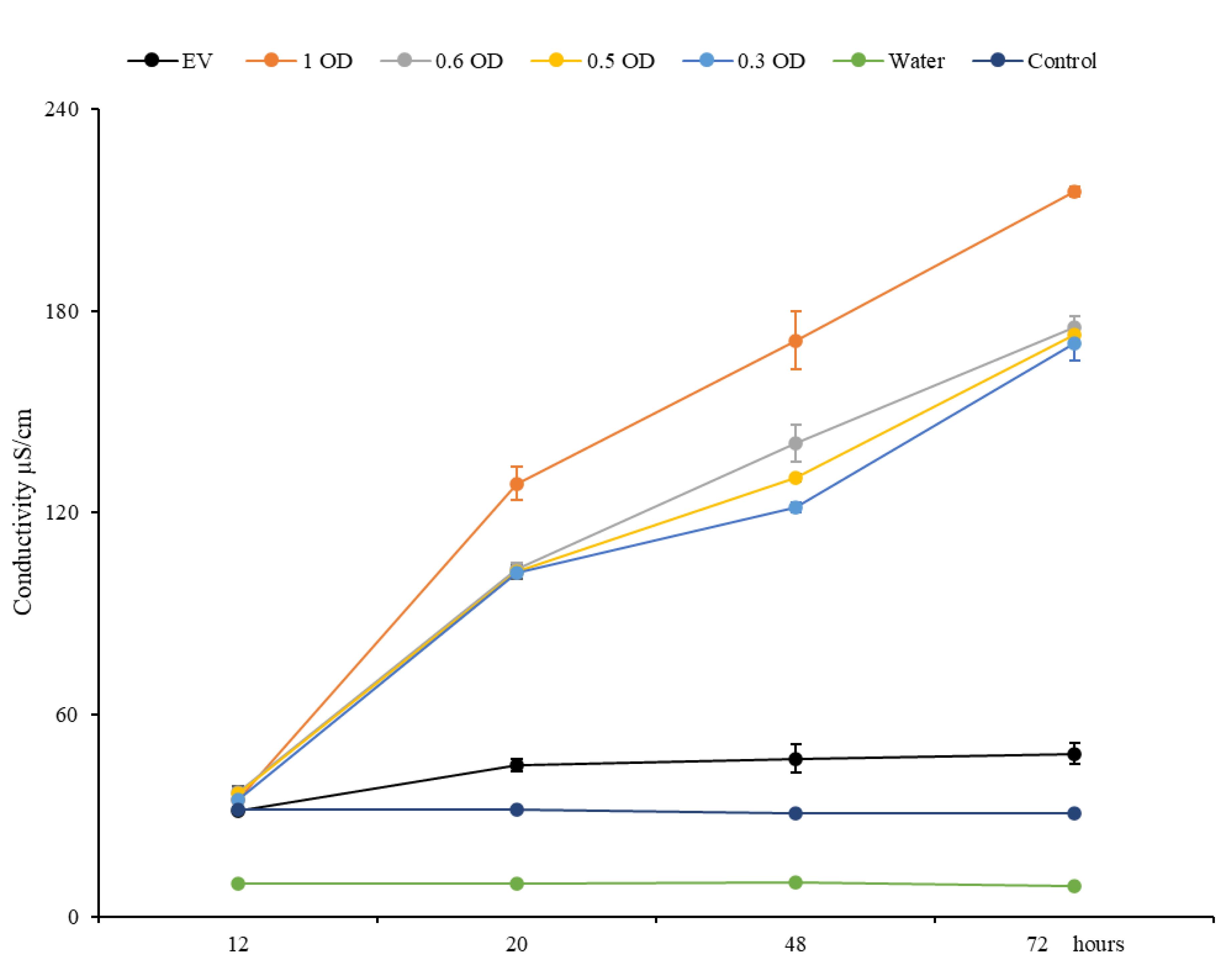
Figure 4. Ion leakage induced by the expression of XB3. The graph represents the ion leakage from N. benthamiana leaf tissues isolated at different time points (12, 20, 48, and 72 h) after infiltration with Agrobacterium cultures containing pC1300-XB3 at OD600 values of 1.0, 0.6, 0.5, and 0.3 along with Agrobacterium culture containing the empty vector (EV) pC1300S. Water, conductivity of water. Control, ion leakage from healthy, non-infiltrated leaves.
Validation of protocol
This protocol was modified from Huang et al. [1] and reproducible (Figures 3 and 4).
General notes and troubleshooting
General notes
Our protocol presents advantages over conventional agroinfiltration methods [1,7–9]. We eliminate the centrifugation step, as it is cost-prohibitive for many school programs, and bypass the use of a spectrophotometer to measure OD at a specific wavelength. Additionally, we replace the agroinfiltration buffer with LB to reduce chemical costs. Target bacterial density is instead assessed using Figure 2. The figures demonstrate the efficiency of LB-based agroinfiltration, and the cell death response induced by XB3 expression.
EHA105 possesses a wide host range and is suitable for delivering genetic material to various plant species. Alternatively, researchers may opt for other Agrobacterium strains with similar capabilities, ensuring compatibility with the specific plant species under investigation and the intended genetic modifications.
A wide range of the concentrations of agrobacterial cultures (OD600 from 0.3 to 1) harboring the XB3 gene can induce cell death response. If an incubator shaker is not available, a flask with a magnetic stir bar can be used to culture agrobacterial cells on a magnetic stir plate.
For the ion leakage assay, if a COND 6+ conductivity meter is not available, a less expensive TDS sensor might be used [10].
Troubleshooting
Reduced transformation efficiency. Overgrown Agrobacterium cultures may lead to a reduction in plant transformation efficiency. Avoid plant leaves with visible signs of disease, stress, or damage. We have noticed that certain types of soil affect the growth of N. benthamiana. If plants grow poorly, we recommend using Growing Mix, PRO-LINE HFC/B HydraFiber.
Acknowledgments
This research was supported by the National Science Foundation (MCB-2114833). This protocol was modified from Huang et al. [1]. We extend our sincere gratitude to Beatriz de Toledo Franceschi, Maggie Paxson, Vincent Newman, Paul Gleason, Lilybeth Moreno, William Bartenslager, Heather Sinclair, and Michael Cartamil for their valuable discussion. We also thank Anita K. Snyder for her critical reading of the manuscript and invaluable comments on the work.
Competing interests
The authors declare no competing interests.
References
- Huang, X., Liu, X., Chen, X., Snyder, A. and Song, W. Y. (2013). Members of the XB3 Family from Diverse Plant Species Induce Programmed Cell Death in Nicotiana benthamiana. PLoS One 8(5): e63868.
- Jacob, P., Kim, N. H., Wu, F., El-Kasmi, F., Chi, Y., Walton, W. G., Furzer, O. J., Lietzan, A. D., Sunil, S., Kempthorn, K., et al. (2021). Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373(6553): 420–425.
- Qi, T., Seong, K., Thomazella, D. P. T., Kim, J. R., Pham, J., Seo, E., Cho, M. J., Schultink, A. and Staskawicz, B. J. (2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 115(46): e1814856115.
- Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., Gardner, J., Wang, B., Zhai, W. X., Zhu, L. H., et al. (1995). A Receptor Kinase-Like Protein Encoded by the Rice Disease Resistance Gene, Xa21. Science 270(5243): 1804–1806.
- Wang, Y. S., Pi, L. Y., Chen, X., Chakrabarty, P. K., Jiang, J., De Leon, A. L., Liu, G. Z., Li, L., Benny, U., Oard, J., et al. (2006). Rice XA21 Binding Protein 3 Is a Ubiquitin Ligase Required for Full Xa21-Mediated Disease Resistance. Plant Cell 18(12): 3635–3646.
- Voinnet, O., Pinto, Y. M. and Baulcombe, D. C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U.S.A. 96(24): 14147–14152.
- Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., Xiao, W., Liu, Y., Chai, J., Takken, F. L. W., et al. (2012). Structure-Function Analysis of Barley NLR Immune Receptor MLA10 Reveals Its Cell Compartment Specific Activity in Cell Death and Disease Resistance. PLoS Pathog. 8(6): e1002752.
- del Pozo, O., Pedley, K. F. and Martin, G. B. (2004). MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23(15): 3072–3082.
- Melech-Bonfil, S. and Sessa, G. (2010). Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 64(3): 379–391.
- Jain, N., Khurana, P. and Khurana, J. P. (2022). Overexpression of a rice Tubby–like protein-encoding gene, OsFBT4, confers tolerance to abiotic stresses. Protoplasma 260(4): 1063–1079.
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Vergish, S., Wolf, R. and Song, W. Y. (2024). Simplified Protocol to Demonstrate Gene Expression in Nicotiana benthamiana Using an Agrobacterium-Mediated Transient Assay. Bio-protocol 14(10): e4987. DOI: 10.21769/BioProtoc.4987.
Category
Plant Science > Plant biochemistry
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








