- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Conditional Depletion of STN1 in Mouse Embryonic Fibroblasts
Published: Vol 14, Iss 8, Apr 20, 2024 DOI: 10.21769/BioProtoc.4977 Views: 4321
Reviewed by: Hélène LégerAYŞE NUR PEKTAŞ

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
CRISPR/Cas9-Based Protocol for Precise Genome Editing in Induced Pluripotent Stem Cells
Avinash Singh [...] Shauna H. Yuan
Dec 20, 2024 2932 Views
Efficient Fluorescent Labeling of Human Trophoblast Stem Cells via a CRISPR/Cas9-Mediated Knock-In Approach in a Safe Harbor Locus
Hengshan Zhang [...] Danny J. Schust
Jan 5, 2026 285 Views
Abstract
The CTC1-STN1-TEN1 (CST) complex is a single-strand DNA-binding protein complex that plays an important role in genome maintenance in various model eukaryotes. Dysfunction of CST is the underlying cause of the rare genetic disorder known as Coats plus disease. In addition, down regulation of STN1 promotes colorectal cancer development in mice. While prior studies have utilized RNAi to knock down CST components in mammalian cells, this approach is associated with off-target effects. Attempts to employ CRISPR/Cas9-based knockout of CST components in somatic cell lines have been unsuccessful due to CST's indispensable role in DNA replication and cell proliferation. To address these challenges, we outline a novel approach utilizing a Cre-loxP-based conditional knockout in mouse embryonic fibroblasts (MEFs). This method offers an alternative means to investigate the function and characteristics of the CST complex in mammalian systems, potentially shedding new light on its roles in genome maintenance.
Key features
• Conditional depletion of mammalian STN1 using mouse embryonic fibroblast (MEFs).
• Analysis of oxidative damage sensitivity using STN1-depleted MEFs.
• This protocol requires Stn1flox/flox mice.
Keywords: STN1Graphical overview
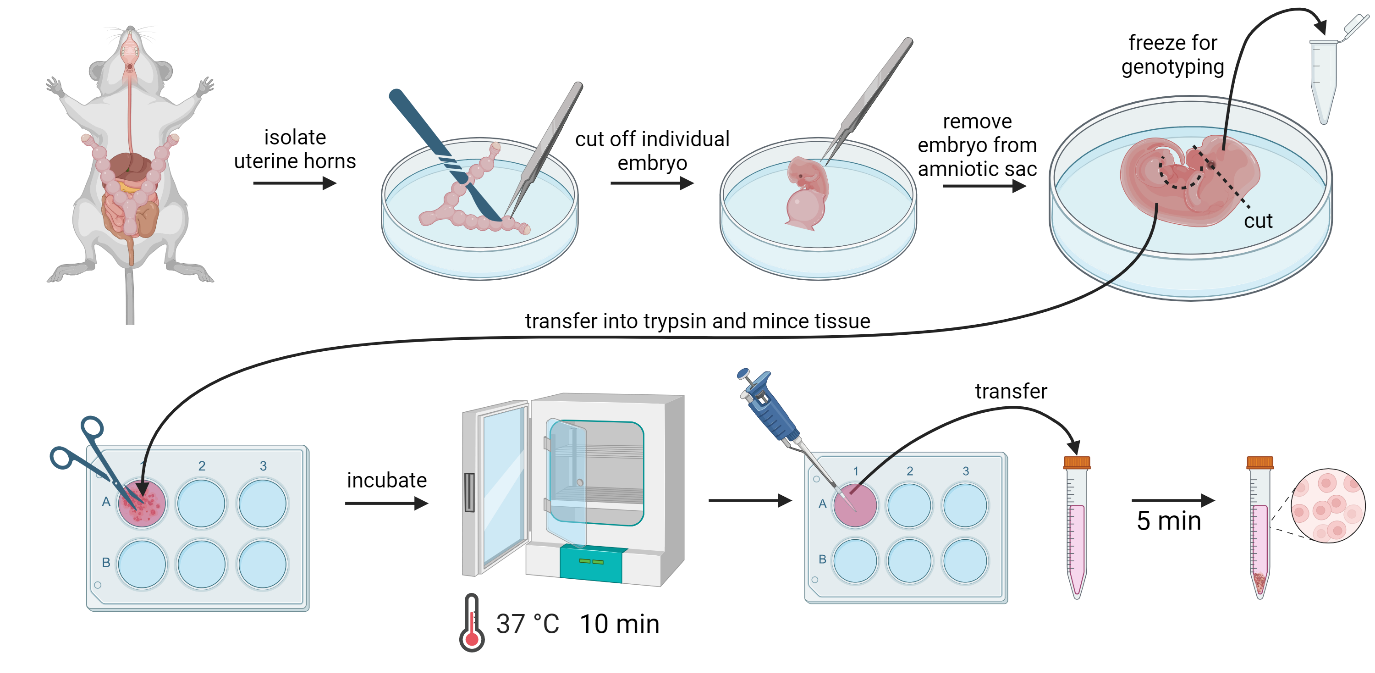
Mouse embryonic fibroblast (MEF) isolation procedure. (Images created using BioRender)
Background
The CTC1-STN1-TEN1 (CST) complex is a single-stranded DNA-binding protein complex essential in several genome maintenance pathways [1–10]. Mutations in CST components, specifically CTC1 (Conserved Telomere Maintenance Component 1) and STN1 (homolog of yeast Stn1, also known as oligonucleotide/oligosaccharide binding fold containing 1 or OBFC1, and alpha accessory factor 44 or AAF44), have been implicated in the pathogenesis of Coats plus syndrome and Dyskeratosis congenita. Coats plus syndrome is a multisystem disorder with manifestations such as retinopathy, stunted growth, and early-onset aging signs [11–13], whereas Dyskeratosis congenita presents with bone marrow failure and notable skin changes [13,14].
Initially identified as co-factors for DNA polymerase α (POLα), CST's interaction with replication machinery, including the MCM helicase complex, signifies its broader regulatory role in DNA replication [15]. Beyond POLα regulation, CST plays a critical role in telomere maintenance by binding to telomeric ssDNA and facilitating C-strand fill-in, thereby preventing telomere over-extension by telomerase and maintaining telomere stability [1,3,5,16–19]. The importance of CST is further highlighted under replication stress conditions, where it protects nascent DNA at stalled replication forks from MRE11 (meiotic recombination 11 homolog A)-mediated degradation [20]. This protection is partly due to CST interaction with RAD51 (DNA repair protein RAD51 homolog 1). CST promotes RAD51 recruitment to replication forks to protect fork stability under stress [8,9,20,21]. Beyond DNA replication, CST is recruited to double-strand breaks by the Shieldin complex, favoring the non-homologous end joining pathway, especially in BRCA1-deficient cells. CST loss in such contexts leads to increased end resection and resistance to PARP inhibitors in BRCA1-deficient cells. Thus, it appears that CST participates in cellular response to PARP inhibitors, especially in BRCA1-deficient tumors [22–25].
Genome-wide association studies have identified that STN1 variants are linked to increased risk of multiple types of cancer [26–31]. Analyses from the TCGA Pan-Cancer project show that decreased CST expression is associated with adverse clinical outcomes [32,33]. In colorectal cancer, CTC1/STN1 expression is markedly reduced, correlating with higher tumor mutation burdens and poorer survival, suggesting their potential as prognostic biomarkers [34].
The crucial role of CST in the development of genetic diseases and cancer underscores the urgency of understanding its functions in genome maintenance pathways. Traditionally, most studies investigating the role of the CST complex in mammalian cells have utilized RNAi to reduce the expression of CST components. However, the effectiveness of RNAi methods may be limited due to unintended off-target effects. Here, we describe an alternative approach using a conditional knockout strategy in mouse embryonic fibroblasts (MEFs) to examine the function of Stn1. This method involves the utilization of tamoxifen-inducible Cre recombinase to delete the promoter region and the ATG start codon of the murine Stn1 gene. The tamoxifen-inducible nature of the Cre recombinase enables researchers to grow cells before administering tamoxifen and control the timing of Stn1 deletion. By employing this method, it becomes possible to specifically deplete the mammalian Stn1 protein, leading to the destabilization of the CST complex. Additionally, the study demonstrates that conditional knockout of Stn1 renders cells more susceptible to damage caused by hydrogen peroxide exposure, emphasizing the significance of the CST complex in countering DNA damage during replication stress and oxidative stress.
Materials and reagents
Biological materials
CreERT2 C57BL/6 mice harboring the conditional CreERT2 transgene (Jackson Laboratory, stock 004682)
Homozygous Stn1flox/flox (Stn1F/F) C57BL/6 mice (generated as described in Nguyen et al. [34])
CreERT2; Stn1F/F and control Stn1F/F C57BL/6 mice: generated by crossing with Stn1F/F with CreERT2 C57BL/6 mice to generate progeny heterozygous for Stn1Flox allele and hemizygous for the CreERT2 genotype. These mice were then bred to homozygous Stn1F/F mice to derive Stn1 conditional knockout CreERT2; Stn1F/F and control Stn1F/F genotypes as described previously [34].
DMEM/F-12 (Gibco, catalog number: 11320033)
Fetal bovine serum (FBS) (Biowest, catalog number: S1620)
Antibiotic/antimycotic solution (HyClone, catalog number: SV30079.01)
Trypsin-EDTA 0.05% and 0.025% (Gibco, catalog number: 2530062)
DMSO (Sigma, catalog number: D-5879)
4-Hydroxytamoxifen (4-OHT) (VWR, catalog number: 89152-604)
KOD Hot Start DNA Polymerase kit (Millipore Sigma, catalog number: 710863, solutions provided in the kit)
Oligos:
oIMR5984: 5'-GCTAACCATGTTCATGCCTTC-3' (Cre transgene forward)
oIMR8744: 5'-CAAATGTTGCTTGTCTGGTG-3' (Cre transgene reverse)
oIMR8745: 5'-GTCAGTCGAGTGCATAGTTT-3' (Cre internal positive control forward)
oIMR9074: 5'-AGGCAAATTTTGGTGTACGG-3' (Cre internal positive control reverse)
Stn1loxPShift-F: 5'-TGTAATCCCAGCGCTCAGGAG-3'
Stn1loxPShift-R: 5'-GATCTGACAGAGATCTCCTGGCT-3'
Obfc1 5'wtF2: 5'-GACTCCTTAGCCCCAGATCTCCGTCAT-3'
Obfc1 5'mtR: 5'-CTTCGTATAGCATACATTATACGAAGTTATGGATCCACCGACT-3'
Obfc1 5'mtF: 5'-CCACATAGTCGGTGGATCCATAACTTCGT-3'
Obfc1 5'wtR: 5'-GGGCTAGGGTGTAGCTCAATGGTAGA-3'
Obfc1 3'wtF: 5'-CTCTGGGGAGACTGTCTCAGATGTTCA-3'
Obfc1 3'mtR: 5'-GAGGCTCAGGATCCATAACTTCGT-3'
Obfc1 3'mtF: 5'-GCCAGCCTGGTCTACGTGACAATAACT-3'
Obfc1 3'wtR: 5'-GGTCATATGGCCTCACCCTCTAAGA-3'
Glycerol (Fisher Scientific, catalog number: BP-229-1)
Agarose (Sigma-Aldrich, catalog number: A9539-500G)
DTT (Sigma-Aldrich, catalog number: D0632-5G)
Anti-STN1 antibody (Santa Cruz, catalog number: sc-376450), 1:500 dilution
Anti-β-actin (Sigma, catalog number: A2228) 1:60,000 dilution
Secondary horseradish peroxidase antibody: TrueBlot anti-mouse IgG HRP conjugated (Rockland, catalog number: 18-8817-30) 1:2,000 dilution
SuperSignal West Femto (Thermo Fisher, catalog number: 34095)
Hydrogen peroxide (30%, ACS grade) (Ward’s Science, catalog number: 470301)
Crystal Violet (Sigma, catalog number: C3886)
CHAPS (Fisher Scientific, catalog number: BP571-5)
Ethanol (Sigma-Aldrich, catalog number: 459844-4L)
Sodium dodecyl sulfate (SDS) (Fisher Scientific, catalog number: BP166-500)
Bromophenol Blue (Sigma, catalog number: B-8026)
NaOH (Fisher Scientific, catalog number: S318-500)
NaCl (Fisher Scientific, catalog number: S271-1)
Tris-HCl (Sigma-Aldrich, catalog number: T3253-500G)
Na2HPO4 (VWR, catalog number: 0404-500G)
KH2PO4 (J.T. Baker, catalog number: 3246-01)
KCl (EMD, catalog number: PX1405-1)
Tween-20 (VWR, catalog number: M147-1L)
Non-fat dry milk (Nestle, catalog number: 12428935)
2 mM 4-OHT stock solution (see Recipes)
DNA isolation lysis buffer (see Recipes)
DNA isolation neutralization buffer (see Recipes)
Phosphate-buffered saline (PBS), pH 7.4 (see Recipes)
CHAPS lysis buffer (see Recipes)
Loading buffer (see Recipes)
Crystal violet staining solution (see Recipes)
1% SDS in 70% Ethanol
PBS-Tween (PBST)
DMEM/F-12 culture medium (see Recipes)
Recipes
All solutions are prepared with either autoclaved Milli-Q water or DMSO.
2 mM 4-OHT stock solution
Reagent Final concentration Quantity or Volume 4-OHT 2 mM 7.75 mg DMSO n/a 10 mL Total n/a 10 mL Store at -20 °C. Protect from light. For long-term storage, store the solution at -80 °C.
DNA isolation lysis buffer
Reagent Final concentration Quantity or Volume NaOH 25 mM 20 mg EDTA 0.2 mM 1.17 mg H2O n/a 20 mL Total n/a 20 mL Store at room temperature (RT).
DNA isolation neutralization buffer (pH 5.5)
Reagent Final concentration Quantity or Volume Tris-HCl 40 mM 69.9 mg H2O n/a 20 mL Total n/a 20 mL Store at RT.
PBS
Reagent Final concentration Quantity or Volume NaCl 137 mM 8 g KCl 2.7 mM 0.2 g Na2HPO4 10 mM 1.44 g KH2PO4 1.8 mM 0.24 g H2O n/a 1,000 mL Total n/a 1,000 mL Store at RT.
CHAPS lysis buffer
Reagent Final concentration Quantity or Volume CHAPS 10% 1% 10 mL Tris-HCl pH 7.4 30 mM 363.4 mg NaCl 150 mM 876.6 mg DTT 200 mM 3 g H2O n/a 90 mL Total n/a 100 mL Store at -20 °C.
Loading buffer
Reagent Final concentration Quantity or Volume SDS 20% 4% 20 mL Tris-HCl pH 6.8 100 mM 1.21 g Bromophenol Blue 0.2% 0.2 g Glycerol 20% 20 mL H2O n/a 60 mL Total n/a 100 mL Store at RT.
Crystal violet staining solution
Reagent Final concentration Quantity or Volume Crystal Violet 0.5% 2.5 g Ethanol n/a 500 mL Total n/a 500 mL Store at RT.
1% SDS in 70% Ethanol
Reagent Final concentration Quantity or Volume SDS 20% 1% 50 mL Ethanol 70% 700 mL H2O n/a 250 mL Total n/a 1,000 mL Store at RT.
PBST
Reagent Final concentration Quantity or Volume NaCl 137 mM 8 g KCl 2.7 mM 0.2 g Na2HPO4 10 mM 1.44 g KH2PO4 1.8 mM 0.24 g Tween-20 50% 0.05% 1 mL H2O n/a 999 mL Total n/a 1,000 mL Store at RT.
DMEM/F-12 culture medium
Reagent Final concentration Quantity or Volume DMEM/F-12 n/a 500 mL FBS
100× antibiotic/antimycotic
13%
1×
75 mL
5 mL
Total n/a 580 mL
Laboratory supplies
96-well plates (Genesee Scientific, catalog number: 25-109)
6-well plates (VWR, catalog number: 10062-892)
10 cm tissue culture dishes (Nest Biotechnology, catalog number: 704201)
PVDF membrane (MilliporeSigma, catalog number: ISEQ00010)
Forceps (Fisher Scientific, catalog number: 08-880)
Dissecting scissors (Fisher Scientific, catalog number: 08-940)
Scalpel (Fisher Scientific, catalog number: 12-000-164 and 12-000-161)
Equipment
Inverted microscope (Fisherbrand, model: 03000013)
SDS-PAGE and western blot transfer apparatus (Bio-Rad, model: Mini-PROTEAN Tetra System)
Thermocycler (Bio-Rad, model: T100 Thermal Cycler)
iBright imaging system (Thermo Fisher, model: CL1000)
Plate reader (BioTek, model: Synergy H1)
Software and datasets
GraphPad Prism v9.3 (GraphPad, 11/15/2021)
ENSEMBL Mouse genome browser 110 (https://useast.ensembl.org/Mus_musculus/Info/Index)
Procedure
Design of CRISPR/Cas9 to insert loxP sites
The mouse Stn1 locus contains 10 exons, with the ATG start codon residing in exon 2 (Figure 1A). When loxP insertion sites were initially designed, the most convenient approach to delete murine Stn1 appeared to be deleting exons 5 and 6, which would create a frameshift and generate a truncated STN1 protein. However, a long non-coding RNA (lncRNA) with unknown function is present in this region. To avoid affecting this lncRNA, we targeted the upstream regions of the Stn1 gene by inserting two loxP sites upstream of the promoter region and downstream of the ATG start codon in exon 2 (Figure 1A). Sequence information and the exact loxP insertion sites are provided in Supplementary File 1. Cre expression resulted in the deletion of the promoter sequence and the translation start site, thereby inhibiting both transcription and translation of the gene (Figure 1A). Deleting the promoter region would also eliminate the possible complications caused by a truncated protein that could be synthesized from a downstream ATG codon. However, we have found that the inducible Stn1 deletion is incomplete, likely due to inefficient recombination caused by the long distance between two loxP sites, resulting in partial knockout or depletion of STN1.
Isolation of mouse embryonic fibroblasts
Set up the breeding pairs of CreERT2; Stn1F/F and Stn1F/F C57BL/6 mice, either male CreERT2; Stn1F/F paired with female Stn1F/F, or male Stn1F/F paired with female CreERT2; Stn1F/F. We have not observed breeding difference.
Euthanize 12–14 days (est.) pregnant mouse with CO2 followed by cervical dislocation.
Critical: Before transfer under the cell culture hood, spray mouse down thoroughly with 70% ethanol.
Open the abdominal cavity with sterile scissors. Be careful not to disturb uterine horns.
Note: Lifting the skin with a sterile forceps before incision and doing the same with the abdominal wall ensures no accidental damage.
Critical: It is important to keep any hair from being introduced into subsequent steps. Change instruments if necessary.
Extract embryos still within the uterine horns by gently lifting them with forceps and using scissors to cut them free close to the pelvis. Then, place them in a 10 cm dish with ~10 mL of PBS prewarmed to 37 °C.
Note: If they do not lift out freely, carefully check for locations to cut.
Critical: It is important not to tear or cut any part of the bowels to avoid bacterial contamination.
Cut open uterus and extract embryos still within amniotic sac; separate embryos by cutting the area between each and remove each embryo while keeping amniotic sac intact.
Open the amniotic sac with forceps and sterile scalpel, remove the embryo, and sever the head above the eye line. Place the severed head in a separate tube on ice for later DNA extraction.
Remove red organs with scalpel and place the remaining tissue in 2 mL of 0.05% trypsin in a 6-well dish.
Gently but thoroughly disrupt tissue with scissors until no piece larger than 1 × 1 × 1 mm remains.
Repeat steps B5–7 for each embryo and place dishes into incubator at 37 °C with 5% CO2 for 10 min.
Note: Start incubation within less than 5 min of processing the first embryo, depending on the number of embryos and time required for each. Incubation for separate batches may need to be staggered.
Critical: Swift processing is recommended to minimize cell death.
Gently pipette cell suspension up and down with a P1000 to further disassociate cells mechanically before they are transferred into 2 mL of DMEM/F-12 culture medium and resuspended again.
After 5 min, transfer the supernatant to a new tube, leaving the larger tissue fragments at the bottom of the tube.
Spin down the new tube at 500× g for 2 min and discard the supernatant; resuspend the cell pellet in 2 mL of fresh DMEM/F-12 culture medium, transfer into a 6-well dish, and place into the incubator overnight.
Note: Be careful not to mix or pool cells from different embryos.
The next day, replace the medium with fresh medium.
Monitor cells until each well reaches full confluency (1–2 days later) and transfer into a 10 cm dish for expanding and subsequent freezing down of cells.
Validation of genotype
Place a small piece of the tissue from cell isolation (see B6) in 50 μL of DNA isolation lysis buffer (see Recipe 2) and incubate at 98 °C for 1 h.
After incubation, add 50 μL of DNA isolation neutralization buffer (see Recipe 3), pipette the mixture up and down thoroughly to disrupt the rest of the tissue, and transfer into a fresh tube, leaving behind any significant tissue clumps.
Pause Point: DNA can be stored at -20 °C until needed.
Use 1 μL each of the final mixture to run a PCR reaction with Cre transgene primer pairs and Stn1flox primer pairs.
To determine if animals contain the CreERT2 transgene, the PCR reactions should contain:
1 μL lysate
2.5 μL dNTPs (2.5 mM stock)
2.5 μL 10× buffer for KOD Hot Start
1.5 μL oIMR5984 (10 μM)
1.5 μL oIMR8744 (10 μM)
1.5 μL oIMR8745 (10 µM)
1.5 μL oIMR9074 (10 µM)
0.5 μL KOD
1.5 μL MgSO4 (50 mM stock)
1.25 μL DMSO
9.75 μL H2O
25 μL Total
Run the PCR under the following thermocycler conditions:
95 °C for 5 min
95 °C for 30 s
55 °C for 30 s
72 °C for 1 min
Go to 2) for 31 cycles
72 °C for 5 min
10 °C infinite hold
To determine if the animal is homozygous or heterozygous of the Stn1 flox allele, the PCR reactions should contain:
1 μL lysate
2.5 μL dNTPs (2.5 mM stock)
2.5 μL 10× buffer for KOD Hot Start
1.5 μL STN1loxPShift-F (10 μM)
1.5 μL STN1loxPShift-R (10 μM)
0.5 μL KOD
1.5 μL MgSO4 (50 mM stock)
1.25 μL DMSO
13 μL H2O
Run the PCR under the following thermocycler conditions:
95 °C for 5 min
95 °C for 20 s
60 °C for 30 s
72 °C for 40 s
Go to 2) for 34 cycles
72 °C for 2 min
10 °C infinite hold
(Optional) To validate if the animal contains the loxP insertions in the correct Stn1 loci, four additional PCR reactions can be performed:
1 μL lysate
2.5 μL ldNTPs (2.5 mM stock)
2.5 μL 10× buffer for KOD Hot Start
1.5 μL Primer a, c, e, or g (10 μM) (see primer notes below)
1.5 μL Primer b, d, f, or h (10 μM) (see primer notes below)
0.5 μL KOD
1.5 μL MgSO4 (50 mM stock)
1.25 μL DMSO
13 μL H2O
Run the PCR under the following thermocycler conditions:
95 °C for 5 min
95 °C for 20 s
60 °C for 30 s
72 °C for 40 s
Go to 2) for 34 cycles
72 °C for 2 min
10 °C infinite hold
Notes:
Primer pair #1: produces a 547 bp product if 5' loxP is present.
Primer a: Obfc1 5'wtF2, anneals upstream of the 5' loxP site (Figure 1A)
Primer b: Obfc1 5'mtR, anneals at the 5' loxP site (Figure 1A )
Primer pair #2: produces a 226 bp product if 5' loxP is present.
Primer c: Obfc1 5'mtF, anneals at the 5' loxP site (Figure 1A )
Primer d: Obfc1 5'wtR, anneals downstream of the 5' loxP site (Figure 1A)
Primer pair #3: produces a 335 bp product if 5' loxP is present.
Primer e: Obfc1 3'wtF, anneals upstream of the 3' loxP site (Figure 1A)
Primer f: Obfc1 3'mtR, anneals at the 3' loxP site (Figure 1A )
Primer pair #4: produces a 550 bp product if 5' loxP is present.
Primer g: Obfc1 3'mtF, anneals at the 3' loxP site (Figure 1A )
Primer h: Obfc1 3'wtR, anneals downstream of the 3' loxP site (Figure 1A)
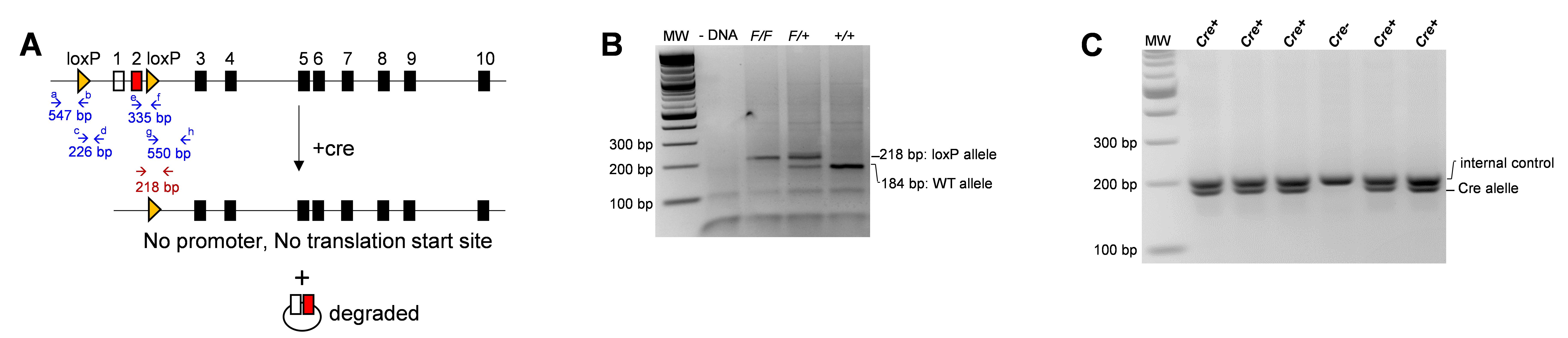
Figure 1. Genotyping of the Stn1F/F allele and the CreERT2 transgene. A. Targeting construct encoding the murine Stn1 gene, which contains 10 exons. Yellow arrowheads designate loxP sites. Arrows designate PCR primer sites. Open box: non-coding exon. The Cre protein is a site-specific DNA recombinase that recombines a pair of loxP sequences. The Cre enzyme binds to the loxP sites and brings the two sites together, catalyzing a recombination event between them, resulting in the excision of the DNA sequence between the two loxP sites. The excised DNA is usually degraded, and the ends of the remaining DNA are ligated together. B. Example of PCR products for detecting the presence of the Stn1F/F allele. C. Example of PCR products for detecting the CreERT2 transgene.Add 5 μL of 6× loading buffer (see Recipe 6) to each PCR reaction and load 12 μL of the mixture onto a 2.5% agarose electrophoresis gel. Run at 120 V for 1 h.
Pause Point: PCR reactions can be stored at -20 °C for several days before electrophoresis.
Expected results and interpretation: The WT allele will produce a 184 bp PCR product, and the recombined flox allele will produce a 218 bp product. If the PCR reaction yields a single 218 bp band, it indicates that the animal is Stn1F/F; a single 184 bp PCR product means that the animal is Stn1 +/+; and a double band of 184 bp and 218 bp means the animal is Stn1F/+ (Figure 1B). The presence of the CreERT2 transgene generates a double band (Figure 1C).
Cultivation of MEFs and induced knockout of Stn1
Grow MEF cells in DMEM/F-12 culture medium at 37 °C with 5% CO2.
Passage cells when they reach near or full confluency at a 1:4 split ratio.
Note: When cell confluency is too low after passaging, cells take a long time to grow.
To induce Cre expression to deplete Stn1, add 4-OHT into media to a final concentration of 2 μM for 48–96 h.
Immortalization of MEFs
Primary cells are spontaneously immortalized by continuous passaging in DMEM/F-12 medium at 37 °C with 5% CO2 for several months (20+ passages). Surviving cells that continue to grow and do not senesce can be considered immortalized. Immortalized cells exhibit slightly different morphology (Figure 2).
To induce Cre expression to knockout Stn1, add 4-OHT into media at 2 μM for 48–96 h.
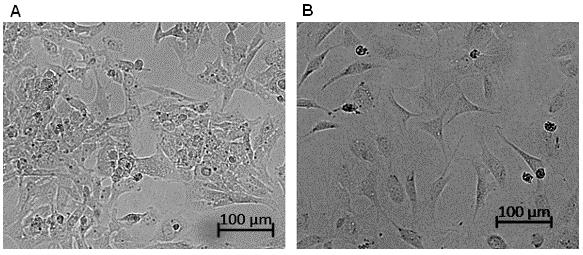
Figure 2. Cell morphology of primary and immortalized mouse embryonic fibroblasts (MEFs). A. Primary MEF cells tend to grow in aggregates. B. Immortalized MEFs grow more evenly distributed and spread out, cells are contracted and round, and spindles are narrower compared to primary MEFs. Images were taken with Zeiss inverted microscope Observer with a 10× phase contrast lens.
Analysis of protein levels after induction
Extract proteins from 20,000 cells by resuspending cells in 15 μL of CHAPS lysis buffer (see Recipe 5) and incubating on ice for 30 min. Then, spin down at 12,000× g for 15 min at 4 °C and mix the supernatant 1:1 with 2× loading buffer.
Load samples onto 10% SDS-PAGE gel and run at 160 V for 60 min in SDS gel running buffer.
Note: Exact run time may differ; refer to the location of the loading buffer band and run until it is near the bottom edge of the gel.
Transfer proteins from the gel onto a PVDF membrane in a Bio-Rad transfer chamber at ~100 V and 400 mA for 70 min.
Block membrane in PBST buffer + 5% non-fat milk for 1 h at RT.
Incubate the membrane in primary antibody (anti-STN1 and anti-β-actin) in PBST at 4 °C overnight.
Wash membrane five times in PBST for 10 min each.
Incubate the membrane in secondary HRP antibody in 5% non-fat milk PBST buffer at RT for 1 h.
Wash the membrane five times in PBST for 10 min each.
Apply SuperSignal West Femto mixture diluted 1:5 in ddH2O to the wet membrane.
Capture western blot signal with iBright CL1000 imaging system. Examples of western blot signals are shown in Figure 3A and 3B.
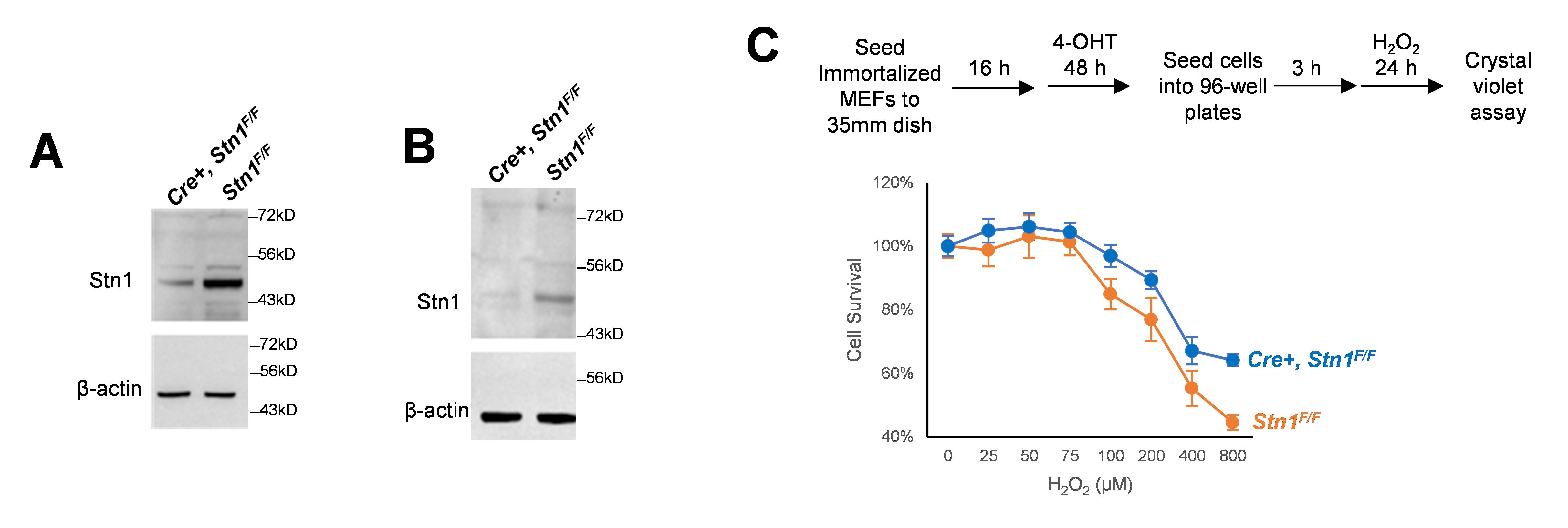
Figure 3. Induction of STN1 depletion leads to cell sensitivity to oxidative stress. A. Primary Stn1F/F and CreERT2, Stn1F/F mouse embryonic fibroblast (MEF) clones were treated with 4-OHT (2 μM) for 48 h. STN1 protein expression was analyzed by western blotting. B. Spontaneously immortalized Stn1F/F and Cre, Stn1F/F MEF clones were treated with 4-OHT (2 μM) for 48 h. STN1 protein expression was analyzed by western blotting. C. Immortalized MEF clones were seeded to 35 mm dishes and then treated with 4-OHT for 48 h. Cells were then trypsinized and seeded into 96-well plates with 5,000 cells per well. Once cells were attached, hydrogen peroxide was added at the indicated concentration for 24 h. Cell viability was determined by Crystal Violet colorimetric assay. Two independent experiments were performed. Representative results from one experiment are shown. Error bars: S.D.
Testing cell sensitivity to hydrogen peroxide
Grow MEFs in culture, trypsinized, and seed 5,000 cells into each well of a 96-well plate, following induction according to step D3. Maintain 4-OHT in the media. Seed cells in six wells for each sample and each treatment condition.
After cells have settled and have been allowed to attach to dishes for approximately 3 h, add hydrogen peroxide and incubate the plate at 37 °C and 5% CO2 overnight.
Remove media with a multichannel pipette, wash gently with 200 μL of PBS per well, and remove PBS.
Note: To avoid accidentally detaching cells, PBS should be added slowly along the side of the wells and in the same manner across the whole plate. Removal of the media and PBS should be done slowly with the plate slightly tilted and the pipette tip in one corner of the wells.
Add 100 μL of Crystal Violet solution (see Recipe 7) to each well and incubate for 15 min at room temperature.
Pour out the Crystal Violet solution and blot firmly against paper towels.
Add ddH2O with the multichannel pipette, pour out, and firmly blot against paper towels.
Immerse the plate in ddH2O for 15 min at RT, pour out, and blot against paper towels.
Air dry the plate until completely dry.
Pause Point: Dried plates can be stored in a dry and dark place at RT overnight if necessary.
Add 200 μL of 1% SDS in 70% ethanol (see Recipe 8) per well and carefully resuspend and mix each well with a multichannel pipette without introducing bubbles.
Measure absorbance at 570 nm with a plate reader. Results are shown in Figure 3C .
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Nguyen et al. [34]. Deficiency in mammalian STN1 promotes colon cancer development via inhibiting DNA repair. Science Advances (Figure 4, Figure 6, panels D and E, Figure 7, panel A).
General notes and troubleshooting
Troubleshooting
Problem 1: MEF cells grow slowly.
Possible cause: After a number of passages, primary MEFs senesce.
Solution: Use early passaged MEF cells. It is recommended to freeze down several aliquots of early-passaged primary MEFs for later use.
Problem 2: High background in anti-Stn1 western blot.
Possible cause: Use of regular HRP-conjugated secondary anti-mouse IgG antibody.
Solution: Use the TrueBlot HRP-conjugated anti-mouse secondary antibody.
Problem 3: Immortalized MEF cells lose Stn1 depletion.
Possible cause: We initially used SV40 large T-antigen to immortalize MEFs. However, we have found that MEFs immortalized with this method do not respond to 4-OHT treatment and appear to have lost STN1 depletion. Although the exact cause of this loss is unclear, we suspect that SV40 large T-antigen alters the repair of double-strand breaks, which could influence Cre-loxP recombination efficiency and precision. In addition, large T antigen expression leads to higher mutations and chromosome aberrations, which could impact the sites where Cre recombination acts.
Solution: Use spontaneously immortalized MEFs. Do not use SV40 large T-antigen to immortalize MEFs.
Problem 4: Inefficient STN1 depletion after Cre induction.
Possible cause: Tamoxifen treatment time is not long enough, or tamoxifen concentration is incorrect.
Solution: Cells need to be treated with 4-OHT for at least 48 h to achieve efficient Stn1 depletion. Check the 4-OHT concentration to make sure that it is correct.
Acknowledgments
We thank Duc Dinh Nguyen, Eugene Kim, Thanh Nguyen, and Olga Shiva for technical assistance. This protocol was described and validated in the original research paper: DOI: 10.1126/sciadv.add8023.
Competing interests
Authors declare no financial or non-financial competing interests.
Ethical considerations
Animals were housed and studied in specific pathogen-free animal facilities at Loyola University Chicago Health Sciences campuses. All studies were approved by Institutional Animal Care and Use Committee (IACUC).
References
- Chen, L. Y., Redon, S. and Lingner, J. (2012). The human CST complex is a terminator of telomerase activity. Nature 488(7412): 540–544. https://doi.org/10.1038/nature11269
- Noordermeer, S. M., Adam, S., Setiaputra, D., Barazas, M., Pettitt, S. J., Ling, A. K., Olivieri, M., Alvarez-Quilon, A., Moatti, N., Zimmermann, M., et al. (2018). The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560(7716): 117–121. https://doi.org/10.1038/s41586-018-0340-7
- Surovtseva, Y. V., Churikov, D., Boltz, K. A., Song, X., Lamb, J. C., Warrington, R., Leehy, K., Heacock, M., Price, C. M. and Shippen, D. E. (2009). Conserved Telomere Maintenance Component 1 Interacts with STN1 and Maintains Chromosome Ends in Higher Eukaryotes. Mol. Cell 36(2): 207-218. https://doi.org/10.1016/j.molcel.2009.09.017
- Stewart, J. A., Wang, F., Chaiken, M. F., Kasbek, C., Chastain, P. D., Wright, W. E. and Price, C. M. (2012). Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31(17): 3537–3549. https://doi.org/10.1038/emboj.2012.215
- Gu, P., Min, J. N., Wang, Y., Huang, C., Peng, T., Chai, W. and Chang, S. (2012). CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31(10): 2309–2321. https://doi.org/10.1038/emboj.2012.96
- Miyake, Y., Nakamura, M., Nabetani, A., Shimamura, S., Tamura, M., Yonehara, S., Saito, M. and Ishikawa, F. (2009). RPA-like Mammalian Ctc1-Stn1-Ten1 Complex Binds to Single-Stranded DNA and Protects Telomeres Independently of the Pot1 Pathway. Mol. Cell 36(2): 193–206. https://doi.org/ 10.1016/j.molcel.2009.08.009
- Nguyen, D. D., Kim, E. Y., Sang, P. B. and Chai, W. (2020). Roles of OB-Fold Proteins in Replication Stress. Front. Cell Dev. Biol. 8: 574466. https://doi.org/10.3389/fcell.2020.574466
- Wang, Y. and Chai, W. (2018). Pathogenic CTC1 mutations cause global genome instabilities under replication stress. Nucleic Acids. Res. 46(8): 3981–3992. https://doi.org/10.1093/nar/gky114
- Lei, K. H., Yang, H. L., Chang, H. Y., Yeh, H. Y., Nguyen, D. D., Lee, T. Y., Lyu, X., Chastain, M., Chai, W., Li, H. W., et al. (2021). Crosstalk between CST and RPA regulates RAD51 activity during replication stress. Nat. Commun. 12(1): 6412. https://doi.org/10.1038/s41467-021-26624-x
- Lyu, X., Sang, P. B. and Chai, W. (2021). CST in maintaining genome stability: Beyond telomeres. DNA Repair (Amst) 102: 103104. https://doi.org/10.1016/j.dnarep.2021.103104
- Anderson, B. H., Kasher, P. R., Mayer, J., Szynkiewicz, M., Jenkinson, E. M., Bhaskar, S. S., Urquhart, J. E., Daly, S. B., Dickerson, J. E., O'Sullivan, J., et al. (2012). Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44(3): 338–342. https://doi.org/10.1038/ng.1084
- Simon, A. J., Lev, A., Zhang, Y., Weiss, B., Rylova, A., Eyal, E., Kol, N., Barel, O., Cesarkas, K., Soudack, M., et al. (2016). Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects. J. Exp. Med. 213(8): 1429–1440. https://doi.org/10.1084/jem.20151618
- Han, E., Patel, N. A., Yannuzzi, N. A., Laura, D. M., Fan, K. C., Negron, C. I., Prakhunhungsit, S., Thorson, W. L. and Berrocal, A. M. (2020). A unique case of coats plus syndrome and dyskeratosis congenita in a patient with CTC1 mutations. Ophthalmic Genet. 41(4): 363–367. https://doi.org/10.1080/13816810.2020.1772315
- Keller, R. B., Gagne, K. E., Usmani, G. N., Asdourian, G. K., Williams, D. A., Hofmann, I. and Agarwal, S. (2012). CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer 59(2): 311–314. https://doi.org/10.1002/pbc.24193
- Wang, Y., Brady, K. S., Caiello, B. P., Ackerson, S. M. and Stewart, J. A. (2019). Human CST suppresses origin licensing and promotes AND-1/Ctf4 chromatin association. Life Sci. Alliance 2(2). https://doi.org/10.26508/lsa.201800270.
- Huang, C., Dai, X. and Chai, W. (2012). Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22(12): 1681–1695. https://doi.org/10.1038/cr.2012.132
- Lue, N. F., Chan, J., Wright, W. E. and Hurwitz, J. (2014). The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat. Commun. 5: 5762. https://doi.org/10.1038/ncomms6762
- Feng, X., Hsu, S. J., Bhattacharjee, A., Wang, Y., Diao, J. and Price, C. M. (2018). CTC1-STN1 terminates telomerase while STN1-TEN1 enables C-strand synthesis during telomere replication in colon cancer cells. Nat. Commun. 9(1): 2827. https://doi.org/10.1038/s41467-018-05154-z
- Dai, X., Huang, C., Bhusari, A., Sampathi, S., Schubert, K. and Chai, W. (2010). Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 29(16): 2788–2801. https://doi.org/10.1038/emboj.2010.156
- Lyu, X., Lei, K. H., Biak Sang, P., Shiva, O., Chastain, M., Chi, P. and Chai, W. (2020). Human CST complex protects stalled replication forks by directly blocking MRE11 degradation of nascent-strand DNA. EMBO J 40(2): e103654. https://doi.org/10.15252/embj.2019103654
- Chastain, M., Zhou, Q., Shiva, O., Fadri-Moskwik, M., Whitmore, L., Jia, P., Dai, X., Huang, C., Ye, P. and Chai, W. (2016). Human CST Facilitates Genome-wide RAD51 Recruitment to GC-Rich Repetitive Sequences in Response to Replication Stress. Cell Rep. 16(5): 1300–1314. https://doi.org/10.1016/j.celrep.2016.06.077
- Barazas, M., Annunziato, S., Pettitt, S. J., de Krijger, I., Ghezraoui, H., Roobol, S. J., Lutz, C., Frankum, J., Song, F. F., Brough, R., et al. (2018). The CST Complex Mediates End Protection at Double-Strand Breaks and Promotes PARP Inhibitor Sensitivity in BRCA1-Deficient Cells. Cell Rep. 23(7): 2107–2118. https://doi.org/10.1016/j.celrep.2018.04.046
- Ray Chaudhuri, A., Callen, E., Ding, X., Gogola, E., Duarte, A. A., Lee, J. E., Wong, N., Lafarga, V., Calvo, J. A., Panzarino, N. J., et al. (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535(7612): 382–387. https://doi.org/10.1038/nature18325
- Mirman, Z., Sasi, N. K., King, A., Chapman, J. R. and de Lange, T. (2022). 53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polalpha-primase fill-in synthesis. Nat. Cell Biol. 24(1): 51–61. https://doi.org/10.1038/s41556-021-00812-9
- Mirman, Z., Lottersberger, F., Takai, H., Kibe, T., Gong, Y., Takai, K., Bianchi, A., Zimmermann, M., Durocher, D. and de Lange, T. (2018). 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature 560(7716): 112–116. https://doi.org/10.1038/s41586-018-0324-7
- Phelan, C. M., Kuchenbaecker, K. B., Tyrer, J. P., Kar, S. P., Lawrenson, K., Winham, S. J., Dennis, J., Pirie, A., Riggan, M. J., Chornokur, G., et al. (2017). Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 49(5): 680–691. https://doi.org/10.1038/ng.3826
- Li, C., Zhao, Z., Zhou, J., Liu, Y., Wang, H. and Zhao, X. (2017). Relationship between the TERT, TNIP1 and OBFC1 genetic polymorphisms and susceptibility to colorectal cancer in Chinese Han population. Oncotarget 8(34): 56932–56941. https://doi.org/10.18632/oncotarget.18378
- Ojha, J., Codd, V., Nelson, C. P., Samani, N. J., Smirnov, I. V., Madsen, N. R., Hansen, H. M., de Smith, A. J., Bracci, P. M., Wiencke, J. K., et al. (2016). Genetic Variation Associated with Longer Telomere Length Increases Risk of Chronic Lymphocytic Leukemia. Cancer Epidemiol. Biomarkers Prev. 25(7): 1043–1049. https://doi.org/10.1158/1055-9965.EPI-15-1329
- Gudmundsson, J., Thorleifsson, G., Sigurdsson, J. K., Stefansdottir, L., Jonasson, J. G., Gudjonsson, S. A., Gudbjartsson, D. F., Masson, G., Johannsdottir, H., Halldorsson, G. H., et al. (2017). A genome-wide association study yields five novel thyroid cancer risk loci. Nat. Commun. 8: 14517. https://doi.org/10.1038/ncomms14517
- Valimaki, N., Kuisma, H., Pasanen, A., Heikinheimo, O., Sjoberg, J., Butzow, R., Sarvilinna, N., Heinonen, H. R., Tolvanen, J., Bramante, S., et al. (2018). Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability. Elife 7. https://doi.org/10.7554/eLife.37110
- Duffy, D. L., Zhu, G., Li, X., Sanna, M., Iles, M. M., Jacobs, L. C., Evans, D. M., Yazar, S., Beesley, J., Law, M. H., et al. (2018). Novel pleiotropic risk loci for melanoma and nevus density implicate multiple biological pathways. Nat. Commun. 9(1): 4774. https://doi.org/10.1038/s41467-018-06649-5
- Dos Santos, G. A., Viana, N. I., Pimenta, R., de Camargo, J. A., Guimaraes, V. R., Romão, P., Candido, P., Ghazarian, V., Reis, S. T., Leite, K. R. M., et al. (2022). Pan-cancer analysis reveals that CTC1-STN1-TEN1 (CST) complex may have a key position in oncology. Cancer Genet. 262–263: 80–90. https://doi.org/10.1016/j.cancergen.2022.01.006
- Wang, L., Ma, T., Liu, W., Li, H., Luo, Z. and Feng, X. (2022). Pan-Cancer Analyses Identify the CTC1-STN1-TEN1 Complex as a Protective Factor and Predictive Biomarker for Immune Checkpoint Blockade in Cancer. Front. Genet. 13: 859617. https://doi.org/10.3389/fgene.2022.859617
- Nguyen, D. D., Kim, E., Le, N. T., Ding, X., Jaiswal, R. K., Kostlan, R. J., Nguyen, T. N. T., Shiva, O., Le, M. T. and Chai, W. (2023). Deficiency in mammalian STN1 promotes colon cancer development via inhibiting DNA repair.Sci. Adv. 9(19): eadd8023. https://doi.org/doi:10.1126/sciadv.add8023
Supplementary information
The following supporting information can be downloaded here:
- Sequence of murine Stn1 (aka OBFC1) locus including the upstream region. Location of the loxP insertion sites and inserted loxP sequence are highlighted. Long red arrows indicate the location of primers used in genotyping.
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Knowles, S. and Chai, W. (2024). Conditional Depletion of STN1 in Mouse Embryonic Fibroblasts. Bio-protocol 14(8): e4977. DOI: 10.21769/BioProtoc.4977.
- Nguyen, D. D., Kim, E., Le, N. T., Ding, X., Jaiswal, R. K., Kostlan, R. J., Nguyen, T. N. T., Shiva, O., Le, M. T. and Chai, W. (2023). Deficiency in mammalian STN1 promotes colon cancer development via inhibiting DNA repair.Sci. Adv. 9(19): eadd8023. https://doi.org/doi:10.1126/sciadv.add8023
Category
Cancer Biology > Genome instability & mutation > Genetics
Cancer Biology > Genome instability & mutation > Animal models
Biological Sciences > Biological techniques > CRISPR/Cas9
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link