- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Simple Rescue of Opaque Tissue Previously Cleared by iDISCO
(*contributed equally to this work) Published: Vol 14, Iss 5, Mar 5, 2024 DOI: 10.21769/BioProtoc.4948 Views: 2210
Reviewed by: Hong LianOneil Girish BhalalaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
A Versatile Pipeline for High-fidelity Imaging and Analysis of Vascular Networks Across the Body
Stephen Vidman [...] Andrea Tedeschi
Feb 20, 2024 2996 Views
Visualization and Analysis of Neuromuscular Junctions Using Immunofluorescence
You-Tian Hsieh and Show-Li Chen
Oct 5, 2024 2599 Views

Identification of Neurons Containing Calcium-Permeable AMPA and Kainate Receptors Using Ca2+ Imaging
Sergei G. Gaidin [...] Sultan T. Tuleukhanov
Feb 5, 2025 1676 Views
Abstract
Recent advancements in tissue-clearing techniques and volumetric imaging have greatly facilitated visualization and quantification of biomolecules, organelles, and cells in intact organs or even entire organisms. Generally, there are two types of clearing methods: hydrophobic and hydrophilic (i.e., clearing with organic or aqueous solvents, respectively). The popular iDISCO approach and its modifications are hydrophobic methods that involve dehydration, delipidation, decolorization (optional), decalcification (optional), and refractive-index (RI) matching steps. Cleared samples are often stored for a relatively long period of time and imaged repeatedly. However, cleared tissues can become opaque over time, which prevents accurate reimaging. We reasoned that the resurgent haziness is likely due to rehydration, residual lipids, and uneven RI deep inside those tissue samples. For rescue, we have developed a simple procedure based on iDISCO. Beginning with a methanol dehydration, samples are delipidated using dichloromethane, followed by RI matching with dibenzyl ether (DBE). This simple method effectively re-clears mouse brains that have turned opaque during months of storage, allowing the user to effectively image immunolabeled samples over longer periods of time.
Key features
• This simple protocol rescues previously cleared tissue that has turned opaque.
• The method does not cause detectable loss of immunofluorescence from previously stained samples.
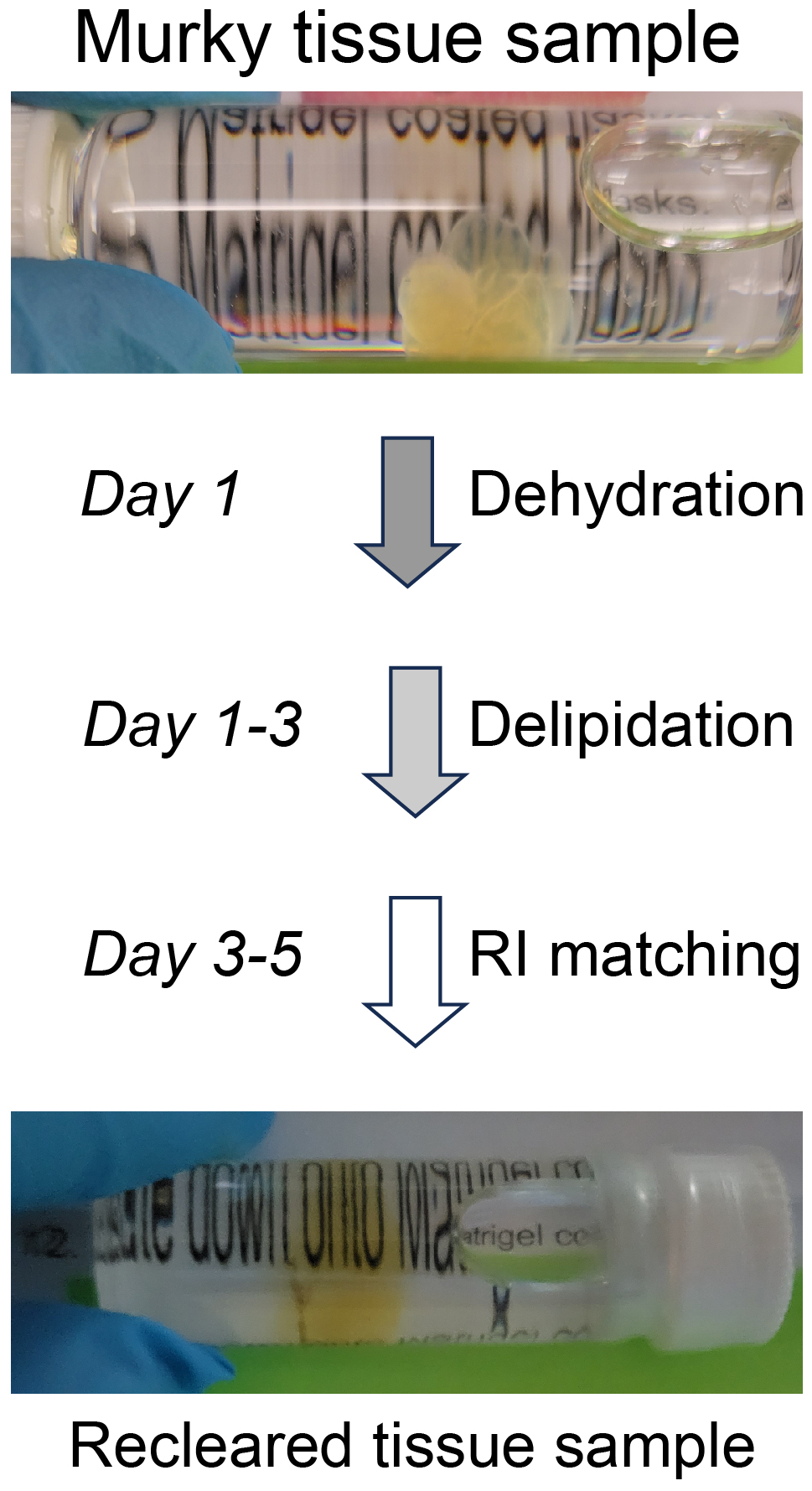
Graphical overview
Background
Tissue clearing paired with light-sheet microscopy rejuvenated the century-old histochemistry field [1]. Clearing can render organs or even whole animals transparent [2], while the light-sheet microscopy can image fluorescent labels deep inside tissues or bodies [3]. Available clearing techniques generally fall into two categories based on the solvents used, specifically whether using hydrophilic or hydrophobic (including hydrogel-based methods) solutions [4]. Despite technical differences, the underlying principles between the two are the same, i.e., substituting molecules that diffract lights (e.g., lipids, pigments, and calcium phosphate) with other molecules that match the refractive index (RI) of the imaging media [2]. The transparency of cleared tissues allows fluorescence imaging of biomolecules deep inside the tissue (i.e., intrinsically expressed fluorescent proteins or after labeling with fluorescently tagged antibodies), which is further facilitated by light-sheet microscopy that illuminates the sample using an adjustable thin sheet of UV light [3]. Notably, fluorophore-tagged antibodies are often used to boost the detection of endogenous fluorescent proteins. iDISCO, a hydrophobic clearing method integrated with immunohistochemistry [5], has become very popular and given rise to multiple derivatives including uDISCO [6] and vDISCO [7]. Conventionally, cleared samples are stored in RI-matching media, often for months, for repeated imaging (please see https://idisco.info for more information). However, during storage, iDISCO-cleared specimens can turn cloudy and some even become opaque, which makes them unsuitable for imaging accurately by light-sheet microscopy. We suspected that such changes in tissue opacity with storage arose from residual lipids, a loss of RI matching deep inside the tissue, or the rehydration of samples by moisture in the air or dissolved in the storage solvent. To counteract those changes, we developed a simple rescue protocol based on immunofluorescence-compatible iDISCO. The approach is comprised by three steps—dehydration, delipidation, and RI rematching. The protocol we describe can successfully re-clear entire, murky mouse brains that had been previously cleared by iDISCO. We imaged the re-cleared brains by light-sheet microscopy, observing little or no loss of immunofluorescence as compared to imaging prior to storage, allowing the samples to be stored and studied far beyond the time of initial scanning.
Materials and reagents
Biological materials
Murky tissues previously cleared by iDISCO and possibly other hydrophobic clearing methods
Methanol (Sigma-Aldrich, catalog number: 179337)
Dichloromethane (Sigma-Aldrich, catalog number: 270997)
Dibenzyl ether (Sigma-Aldrich, catalog number: 33630)
Dichloromethane and methanol mix (see Recipes)
Recipes
Dichloromethane and methanol mix (per sample)
Reagent Final concentration (vol%) Volume Dichloromethane 66% 3.3 mL Methanol 34% 1.7 mL Total n/a 5.0 mL The mix should be made freshly and kept at room temperature before use.
Laboratory supplies
Organ holding forceps (e.g., Fine Science Tools, catalog number: 11031-15)
Screw-cap glass vials (e.g., Millipore Sigma, catalog number: 27151)
Nitrile gloves (any brand)
Kimwipes and paper towels (any brand)
Equipment
Fume hood (e.g., Hamilton Lab, model: SafeAire II)
Orbital shaker (e.g., Stovall Life Science, Belly Dancer, model: BDRAA115S)
Light-sheet microscope (e.g., Miltenyi Biotec, model: UltraMicroscope II Blaze)
Computer workstation equipped with, at least, Intel Xeon CPU, 128 G RAM, GPU with 16 GB memory, 2 TB storage, and USB3.0 port.
Software and datasets
Cloud-based lab management software (e.g., Labguru.com) is recommended to track and record the rescue procedure
Microscope system control and image acquisition software (e.g., ImSpector 7.5.3, free)
Image processing software (e.g., ImageJ1.53, free)
Atlas-matching and immunofluorescence quantification software (e.g., NeuroInfo, licensed by MBF Bioscience)
Procedure
Dehydration
Note: Please see note 2 and 3 about waste disposal and light avoidance.
Transfer the murky tissue sample (Figure 1) to a 5 mL screw-cap glass vial filled with 100% methanol and seal tightly.
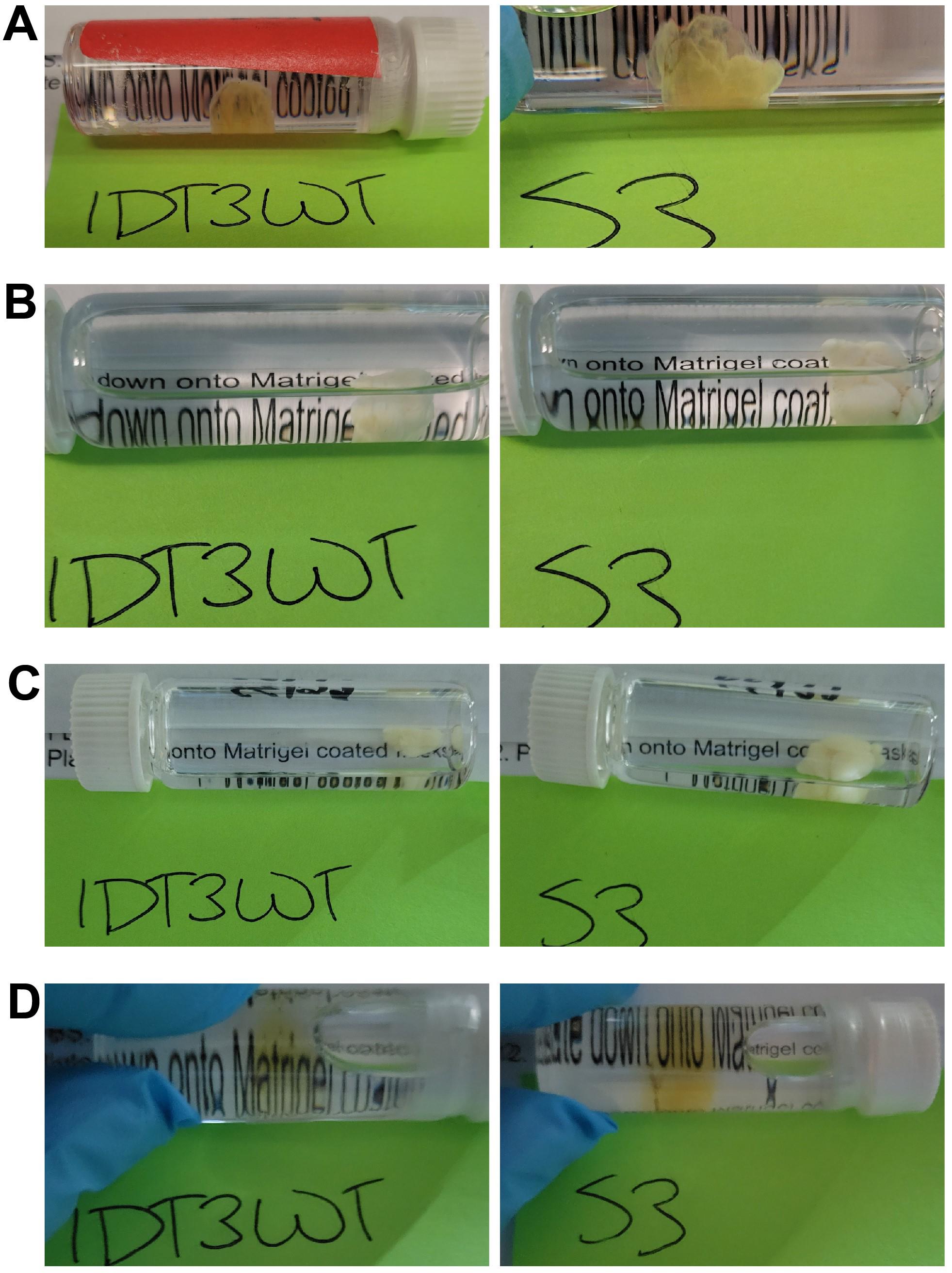
Figure 1. Two previously cleared but murky brain samples over the course of rescue. Brain hemisphere (IDT3WT) and whole brain (S3) previously cleared by iDISCO but that turned murky during months of storage. Samples in A) and B) become totally opaque after dehydration; in C) they remain totally opaque after delipidation, with transparency restored with a light-yellow tinge, D) after refractive index (RI) matching.Place the vial on an orbital shaker (e.g., Belly Dancer) and shake at 70 rpm at room temperature for 1 h.
Discard methanol and repeat steps A1 and A2 three more times.
Delipidation
Transfer tissue sample (Figure 1B) to a 5 mL screw-cap glass vial filled with 66% dichloromethane + 34% methanol and seal tightly.
CAUTION: Wear double nitrile gloves and work in a ventilation hood when handling dichloromethane. Wipe the exterior of the vials with paper towels.
Place the vial on an orbital shaker and shake at 70 rpm at room temperature for 48 h.
Transfer tissue sample to a 5 mL screw-cap glass vial filled with 100% dichloromethane and seal tightly.
CAUTION: One can always keep the sample in the same vial and change the solution instead (see note 4).
Place the vial on an orbital shaker and shake at 70 rpm at room temperature for 1 h.
RI rematching
Transfer tissue sample (Figure 1C) to a 5 mL screw-cap glass vial filled with 100% dibenzyl ether and seal tightly.
CAUTION: Wear double nitrile gloves and work in a ventilation hood when handling dibenzyl ether. Wipe the exterior of the vials with paper towels.
Place the vial on an orbital shaker and shake at 70 rpm at room temperature for 48 h.
Storage
Transfer tissue sample (Figure 1D) to a 5 mL screw-cap glass vial filled with fresh 100% dibenzyl ether and seal tightly.
CAUTION: Ensure no air bubbles are visible in the storage vials, which can be done by slight overfill of the glass vial and closing the cap very slowly.
Store the sample at room temperature and refresh dibenzyl ether once a month.
Data analysis
Tissue samples are imaged by light-sheet microscopy (e.g., Blaze) to localize immunofluorescence. Compiling of Z-stack and stitching of tiles is carried out using image processing software (e.g., Imaris). Atlas-matching and immunofluorescence measuring are achieved by image analysis programs (e.g., NeuroInfo). Figure 2A shows fluorescence light-sheet images of mouse brain before and after rescue. Figure 2B summarizes the fluorescence changes in different brain regions before and after rescue.
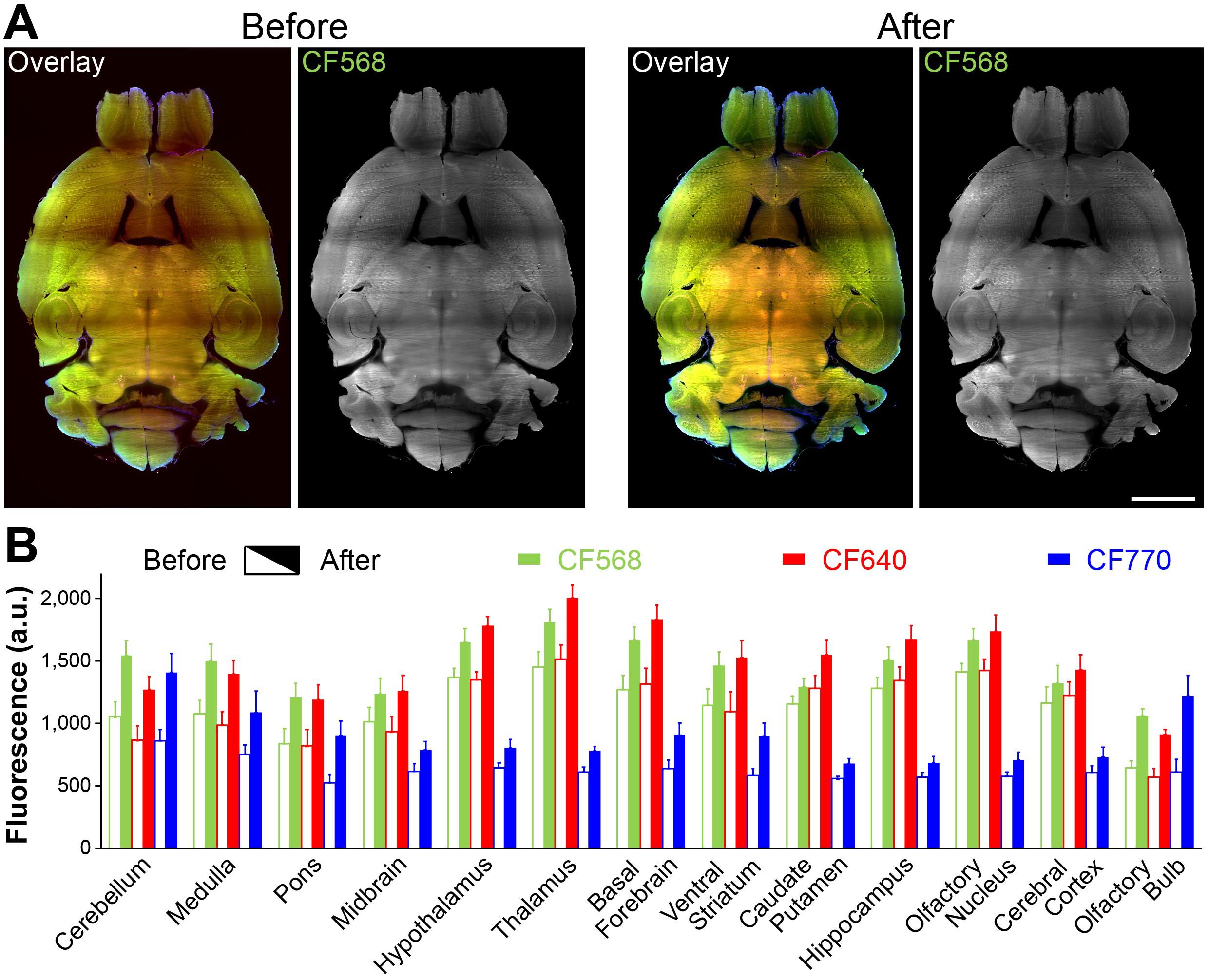
Figure 2. Rescued mouse brain. A. Sample images before and after the rescue. Overlay images are the composite of CF568, 640, and 770 signals. CF568 images show an apparent haze in midbrain and thalamus (before), which was cleared after rescue. Scale bar, 2 mm. B. Average fluorescence for all three immunofluorescent labels (arbitrary unit, a.u.; calculated from 15–30 randomly selected regions of interest within those brain regions). The increases are likely due to significantly lowered background (i.e., much less diffraction) after the rescue.
Validation of protocol
We have applied this rescue protocol to three different batches of murky brain tissues previously cleared by iDISCO and two more batches of murky brain tissues similarly cleared by a different researcher from another lab. In total, we tested this protocol using eight murky brain samples and successfully re-cleared all of them while preserving immunofluorescent signal.
General notes and troubleshooting
Unless specified, all steps are carried out at room temperature.
Waste disposal. As with initial iDISCO clearing, users should follow institutional requirements for waste disposal as established by local Environmental Health and Safety officers. All reagents used in this protocol should be disposed of in appropriate liquid chemical waste containers.
Light avoidance. If the tissue samples to be rescued have immunofluorescence labels, caution is needed to reduce loss of fluorescence. Options include always using aluminum foil to cover the glass vials containing tissue samples and operating under reduced light.
Solution change. Although the protocol above describes solution change as involving a transfer of samples to new vials containing the needed solutions, one can also discard the solution with retention of the tissue in the original vials, followed by adding new solutions to the original vial. It is important that containers (i.e., glass vials) holding samples are completely filled with solutions.
This rescue protocol is tested for the rescue of iDISCO-cleared samples. In rescued mouse brains, we used the following primary antibodies: one is the combination of guinea pig anti-GFAP antibody, chicken anti-Tuj-1 antibody, and rabbit anti-SREBP2 antibody; the other is rabbit anti-cFos alone. Accordingly, the secondary antibodies are highly cross-absorbed CF770-conjugated goat anti-guinea-pig IgG, CF640-conjugated goat anti-chicken IgG, and CF-568-conjugated goat anti-rabbit IgG (all from Biotium).
The timeline of this protocol is adjustable depending on the size of the samples.
Like iDISCO, the survival of different epitopes as well as corresponding antibodies during the rescue needs to be evaluated on a case-by-case basis. Since the rescue procedure and solutions used are largely the same to those of iDISCO, it is likely that antibodies suitable for iDISCO should be compatible with this rescue procedure.
Because this protocol is based on the hydrophobic iDISCO method, it may also work for samples prepared by other hydrophobic clearing methods but not for samples cleared by hydrophilic methods.
Acknowledgments
H.M., J.M., and Q.Z. were supported by NIH award GM147912, a Florida Department of Health award 21A04, and a pilot award from the Stiles-Nicholson Brain Institute. P.G.-K. and R.B. were supported by NIH Award MH094527.
Competing interests
The authors declare no competing interests.
Ethical considerations
Animal use and related procedures were approved by the Florida Atlantic University Institutional Animal Care and Use Committee (IACUC Protocol #A21-06). Mice were housed in pairs. A total of eight male and female C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) between the age of 6 and 30 months were used for collecting brain tissues for this study.
References
- Magaki, S., Hojat, S. A., Wei, B., So, A., and Yong, W. H. (2019). An introduction to the performance of immunohistochemistry. In: Yong, W. (Eds) Biobanking. Methods in Molecular Biology, vol 1897. Humana Press, New York, NY.https://doi.org/10.1007/978-1-4939-8935-5_25
- Ueda, H. R., Ertürk, A., Chung, K., Gradinaru, V., Chédotal, A., Tomancak, P. and Keller, P. J. (2020). Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21(2): 61–79. https://doi.org/10.1038/s41583-019-0250-1
- Stelzer, E. H. K., Strobl, F., Chang, B. J., Preusser, F., Preibisch, S., McDole, K. and Fiolka, R. (2021). Light sheet fluorescence microscopy. Light sheet fluorescence microscopy. Nat. Rev. Methods Primers 1(1): e1038/s43586–021–00077–4. https://doi.org/10.1038/s43586-021-00077-4
- Tainaka, K., Kuno, A., Kubota, S. I., Murakami, T. and Ueda, H. R. (2016). Chemical Principles in Tissue Clearing and Staining Protocols for Whole-Body Cell Profiling. Annu. Rev. Cell Dev. Biol. 32(1): 713–741. https://doi.org/10.1146/annurev-cellbio-111315-125001
- Renier, N., Wu, Z., Simon, D. J., Yang, J., Ariel, P. and Tessier-Lavigne, M. (2014). iDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging. Cell 159(4): 896–910. https://doi.org/10.1016/j.cell.2014.10.010
- Pan, C., Cai, R., Quacquarelli, F. P., Ghasemigharagoz, A., Lourbopoulos, A., Matryba, P., Plesnila, N., Dichgans, M., Hellal, F., Ertürk, A., et al. (2016). Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13(10): 859–867. https://doi.org/10.1038/nmeth.3964
- Cai, R., Pan, C., Ghasemigharagoz, A., Todorov, M. I., Förstera, B., Zhao, S., Bhatia, H. S., Parra-Damas, A., Mrowka, L., Theodorou, D., et al. (2018). Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat. Neurosci. 22(2): 317–327. https://doi.org/10.1038/s41593-018-0301-3
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Mesa, H., Meade, J., Gajewski-Kurdziel, P., Blakely, R. D. and Zhang, Q. (2024). Simple Rescue of Opaque Tissue Previously Cleared by iDISCO. Bio-protocol 14(5): e4948. DOI: 10.21769/BioProtoc.4948.
Category
Neuroscience > Neuroanatomy and circuitry > Fluorescence imaging
Cell Biology > Cell imaging > Fixed-tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








