- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mobilization of Plasmids from Bacteria into Diatoms by Conjugation Technique
Published: Vol 14, Iss 5, Mar 5, 2024 DOI: 10.21769/BioProtoc.4945 Views: 1915
Reviewed by: Noelia ForesiEmilia KrypotouIsmail Tahmaz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Biosynthesis and Genetic Encoding of Non-hydrolyzable Phosphoserine into Recombinant Proteins in Escherichia coli
Philip Zhu [...] Richard B. Cooley
Nov 5, 2023 2507 Views
A Step-by-step Protocol for Crossing and Marker-Assisted Breeding of Asian and African Rice Varieties
Yugander Arra [...] Wolf B. Frommer
Sep 20, 2024 2325 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1561 Views
Abstract
Diatoms serve as a source for a variety of compounds with particularbiotechnological interest. Therefore, redirecting the flow to a specific pathwayrequires the elucidation of the gene’s specific function. The mostcommonly used method in diatoms is biolistic transformation, which is a veryexpensive and time-consuming method. The use of episomes that are maintained asclosed circles at a copy number equivalent to native chromosomes has become auseful genetic system for protein expression that avoids multiple insertions,position-specific effects on expression, and potential knockout of non-targetedgenes. These episomes can be introduced from bacteria into diatoms viaconjugation. Here, we describe a detailed protocol for gene expression thatincludes 1) the gateway cloning strategy and 2) the conjugation protocol for themobilization of plasmids from bacteria to diatoms.
Keywords: ConjugationBackground
Diatoms are unicellular, predominantly photosynthetic, free-living microorganisms that play a central role in trophic webs as primary producers [1]. Diatoms represent the most abundant group within the phytoplankton community, contributing to 40% of global CO2 fixation in the oceans. These organisms are found in freshwater bodies, making them one of the most ecologically successful microalgae worldwide [2,3]. One physiological aspect that explains its ecological success is the energetic-metabolic coupling between mitochondria and chloroplasts [4,5].
From a biotechnological point of view, these organisms produce a variety of compounds of interest such as silica frustule, used as filtering and abrasive materials, and terpenes like fucoxanthin, lupeol, and betulin with antitumoral and antioxidant properties [6]. Furthermore, these organisms are a source of traditional biofuels including methane, through the anaerobic digestion of algae biomass, and biodiesel derived from oil. These organisms show an interesting lipid profile, rich in very long– chain polyunsaturated fatty acids (ω-3 and ω-6), which are of great interest to the industry, especially because they are not usual in other organisms like chlorophytes and flowering plants [7].
The advancement of genetic tools and increasing knowledge on metabolic pathways have facilitated the development of strategies to enhance the productivity of diatoms byredirecting the metabolic flow towards desired products [8]. Thus, diatom genetic manipulation is essential for the elucidation of specific gene functions. Biolistic transformation methods are standard for many diatom species; however, they are time-consuming and require high-yield plasmid DNA preparations and access to expensive specific equipment and reagents (e.g., gene gun). On the other hand, episodes provide a reliable, consistent, and predictable platform for protein expression by avoiding the complications of random chromosomal integration including multiple insertions, position-specific effects on expression, and potential knockout of non-targeted genes. They can be efficiently transferred from bacteria into the diatom via the conjugation method developed by Karas et al. [9] and improved by Diner et al. [10]. Here, we thoroughly describe the procedure previously published [11] and frequently employed in our lab for cloning genes of interest and the subsequent mobilization of vector constructions from bacteria into diatoms by conjugation.
Materials and reagents
Biological materials
Pipette tips: 1–2 µL, 2–200 μL, and 100–1,000 μL (Deltalab, catalog number: 200070 and 301-09)
90 mm diameter Petri dishes (Deltalab, catalog number: 200209)
1.5 mL microcentrifuge tubes (Deltalab, catalog number: 200400P)
50 mL conical centrifuge tubes (Deltalab, catalog number: 42993)
0.22 μm nylon membrane filters (e.g., GVS, catalog number: FJ13BNPNY002AD01)
Reagents
Phaeodactylum tricornutum liquid cultures [e.g., Culture collection of algae, University of Texas, Austin (Utex), catalog number: 646]
PCR kit
dNTPs (Promega, catalog number: U123A)
Platinum Pfx (Invitrogen, catalog number: 11708-013)
10× Pfx amplification buffer (Invitrogen, catalog number: 52806)
MgCl2 25 mM (Promega, catalog number: A351H)
Taq Pegasus (Productos Bio-Logicos, catalog number: EA01M)
GoTaq green buffer 5× (Promega, catalog number: M791A)
Destination plasmid pFcpB (Addgene, catalog number: 90098)
pTA-Mob (Addgene, catalog number: 149662)
Escherichia coli DH10B (Thermo Fisher, catalog number: Eco 113)
E. coli pTA-Mob liquid cultures (homemade)
pENTR/D-TOPO Vector kits (Thermo Fisher Scientific, catalog number: K240020SP)
LR Clonase Enzyme mix (Thermo Fisher Scientific, catalog number: 11791019)
Agar medium 2% (Britannia, catalog number: B0101406)
Luria Broth (LB) medium (homemade)
Gentamicin (Gn) stock solution 25 mg/mL (1:1,000) (Merck, catalog number: G3632)
Kanamycin (Kn) stock solution 50 mg/mL (1:1,000) (Merck, catalog number: BP861)
Ampicillin (Amp) stock solution 100 mg/mL (1:1,000) (Merck, catalog number: A9518)
Zeocin (Zeo) stock commercial solution (Invitrogen, catalog number: R25001); use a concentration of 7.5 µL commercial solution stock in 10 mL of BG11 medium
Amphotericin (Amph) stock solution B 2.5 mg/ mL (1:1,000) (Richet S.A laboratory, https://www.richet.com.ar/en/list?t=n&i=13); use a concentration of 2.5 µg/ mL
BG11 medium (homemade)
Selection plate (0.5× BG11, 1% agar + antibiotics (Amp + Kn + Amph + Gn + Zeo)
Conjugation plate (0.5× BG11 + 0.9% agar + 5% LB + antibiotics (Amp + Amph + Gn)
Tryptone (BD, catalog number: 211705)
Yeast extract (Oxoid, catalog number: LP0021)
NaCl (J.T. Baker, catalog number: 3624-19)
Na2Mg EDTA (Sigma-Aldrich, catalog number: 14402-88-1)
Ferric citrate (Merck, catalog number: 3522-50-7)
CaCl2·2H2O (Merck, catalog number: 10035-04-8)
MgSO4·7H2O (Cicarelli, catalog number: 1054214)
K2HPO4 (Cicarelli, catalog number: 1015214)
H3BO3 (Cicarelli, catalog number: 771214)
MnCl2·4H2O (Merck, catalog number: 13446-34-9)
ZnSO4·7H2O (Merck, catalog number: 7446-20-0)
CuSO4·5H2O (Merck, catalog number: 1.02790.1000.1026)
CoCl2·6H2O (Merck, catalog number, 7791-13-1)
Na2MoO4·2H2O (Merck, catalog number: 10102-40-6)
NaCO3 (Merck, catalog number: 497-19-8)
NaNO3 (Merck, catalog number: 7631-99-4)
Solutions
LB medium (1 L)
BG11 medium (1 L):
Stock I (1 L)
Stock II (1 L)
Stock III (1 L)
Stock V - microelements (1 L)
Carbonate supplement (50 mL)
Nitrate supplement (50 mL)
Recipes
LB medium (1 L)
10 g of tryptone
5 g of yeast extract
10 g of NaCl
1 L of dH2O
BG11 medium (1 L)
Stock I (1 L):
0.1 g of Na2Mg EDTA
0.553 g of ferric citrate
3.6 g of CaCl2·2H2O
Filter sterilize into a sterile bottle or autoclave.
Stock II (1 L):
7.5 g of MgSO4·7H2O
Filter sterilize into a sterile bottle or autoclave.
Stock III (1 L):
3.05 g of K2HPO4
Filter sterilize into a sterile bottle or autoclave.
Stock V - microelements (1 L):
2.86 g of H3BO3
1.81 g of MnCl2·4H2O
0.222 g of ZnSO4·7H2O
0.074 g of CuSO4·5H2O
0.05 g of CoCl2·6H2O
0.4451 g of Na2MoO4·2H2O
Filter sterilize into a sterile bottle or autoclave.
Carbonate supplement (50 mL)
1 g of NaCO3
Filter sterilize into a sterile bottle.
Nitrate supplement (50 mL)
15 g of NaNO3
Filter sterilize into a sterile bottle.
For basic BG11 (1 L), combine the following stock solutions:
Add 10 mL of stock I, II, and III.
Add 1 mL of stock V and carbonate supplement.
Add 5 mL of nitrate supplement.
Add sterilized dH2O to complete to 1 L.
Equipment
Pipettes
Neubauer counting chamber
Laminar air flow equipment
Centrifuge with 50 mL tube capacity
Erlenmeyer
Room or chamber at 37 ºC
Room or chamber at 18 ºC with light 100-200 PAR
Shaker (Vicking, model: Shaker Pro)
Spectrophotometer (Gene Quant, model: 1300)
Binocular light microscope
Applied Biosystems Veriti Thermocycler
Procedure
Preparation of a donor bacterial strain (E. coli pTA-Mob)
The donor bacterial strain should contain a plasmid that allows the formation of the conjugative pili between the bacterium and the diatom. This plasmid, called pTA-Mob (Gnr) [12], is a large plasmid of 52.7 kb. Transform competent E. coli cells (e.g., DH10B) with pTA-Mob plasmid using the appropriate form (chemical/electro competents). Mix DH10B competent cells with ~50 ng of pTA-Mob plasmid and gently flick the tube several times. Incubate the cells on ice for 2 min. Transfer the tube from ice to a 42 °C water bath and heat shock for exactly 45 s. After treatment, add 700 µL of LB medium and incubate at 37 °C for 1 h. Plate on LB solid medium (LB medium + 10 g/L agar) containing gentamicin and incubate at 37 ºC overnight. Pick a resistant colony and check the presence of plasmid by colony PCR using a specific primer, e.g., Gentamycin_fw: TTAGGTGGCGGTACTTGGGT and Promoter_Rv: GTTGACATAAGCCTGTTCGGT (expected PCR product of 752 bp), and/or by digestion with restriction enzymes, e.g., EcoRI (expected digested fragments: 29 kb, 11,7 kb, 8,8 kb, 6,84 kb). Once corroborated, prepare ultracompetent cells from DH10B pTA-Mob strain (Gn r) following the Inoue method for preparation of competent cells (Figure 1A) [13].
Preparation of constructs to mobilize from bacteria into diatoms
Amplify the sequence of interest with a high-fidelity polymerase (Pfx) using a forward primer containing a CACC sequence in the 5′ end by PCR and clone it into a pENTRTM TOPO® entry vector. Once the construct is obtained, check it by PCR using a primer from your gene of interest and Universal M13 primers (M13_fw: GTAAAACGACGGCCAG, M13_rv: CAGGAAACAGCTATGAC), and digestion by restriction enzymes. Finally, corroborate the construct by sequencing.
The pENTR vector has a recombination site (attL1/attL2) that allows recombination with other plasmids containing attR1/attR2 sequences (destination plasmids). The recombination reaction is facilitated by the use of Gateway LR Clonase Enzyme mix. The pFcpB vector is used as a destination plasmid to express proteins in diatoms. Note that this vector contains a light-induced promoter. Check the destination plasmid construct by PCR (using Taq polymerase), using a primer from your gene of interest and PfcpB primer (pFcpB_fw: TTCACGGTTGCCAGAAGTCAAGTCG, pFcpB_rv: TCGAGGTAGCTCAGAATTCACCAC), and digestion by restriction enzymes (Figure 1B).
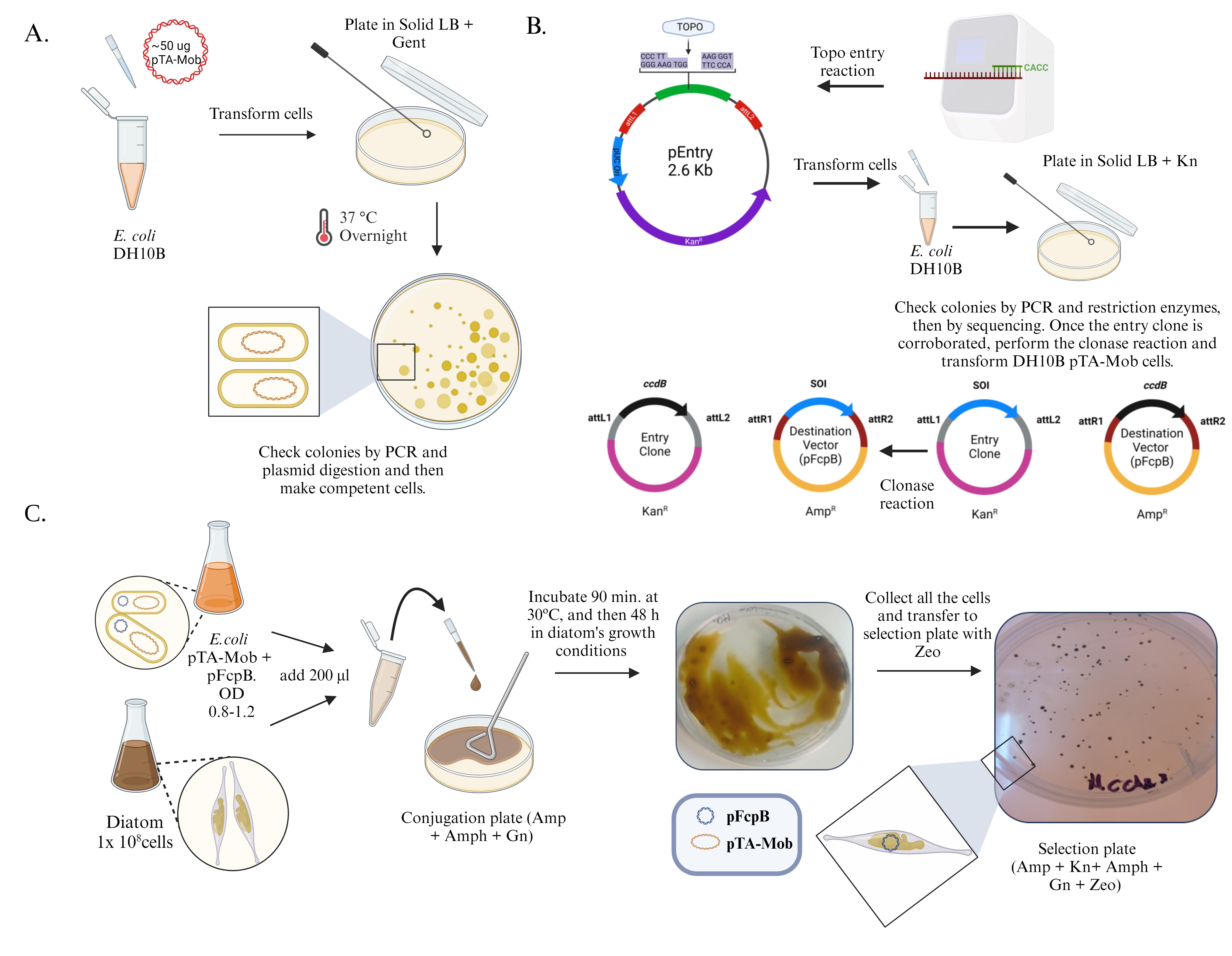
Figure 1. Simplified diagram of the conjugation protocol. A. Transform DH10B competent cells with ~50 ng of pTA-Mob plasmid, plate on LB solid medium containing gentamicin, and incubate at 37 ºC overnight. Pick a resistant colony and check the presence of plasmid by colony PCR or/and digestion with restriction enzymes. B. Amplify the sequence of interest (SOI) using a forward primer containing a CACC to clone it into a pENTR™ TOPO® entry vector. Transform competent cells and check the entry clone by PCR and digestion with restriction enzymes. With the entry clone corroborated, make a Gateway LR Clonase Enzyme Mix reaction to obtain a destination plasmid. Once the destination plasmid is corroborated by sequencing, transform the pTA-Mob competent cells. C. Centrifuge fresh diatoms (1 × 108 cells) and E. coli (OD = 0.8–1.2) cultures and resuspend in 500 µL of the corresponding culture medium. Then, combine and plate in conjugation plates. After 20 days, exconjugant diatom colonies should appear.Transformation of DH10B pTA-Mob (Gnr) strain with a destination plasmid
Transform the E. coli pTA-Mob strain with ~50 ng of destination plasmid and plate on LB solid medium with both antibiotics (Gn for pTA-Mob plasmid and Amp for pFcpB) to select both plasmids (three days before the conjugation step).
Incubate overnight at 37 °C.
Pick a colony and inoculate 5 mL of fresh LB medium with antibiotics (Gn + Amp).
Grow overnight at 37 °C with shaking.
In the afternoon of the day before the conjugation, inoculate 50 mL of fresh LB medium (with antibiotics Amp + Gn) using a 1/100 dilution of the overnight culture.
Let the culture grow overnight at 20 °C and 190 rpm instead of 3 h at 37 °C as described in Karas et al. (2015) [9]. This modification results in significantly higher conjugation efficiency, presumably because the lower temperature favors the expression of recombinant proteins. In both cases, the E. coli pTA-Mob strain with the destination plasmid at the time of harvest must have an OD between 0.8 and 1.2.
Growth of diatom (Phaeodactylum tricornutum) cells
The general growth conditions for diatoms used in this protocol include long-day photoperiod (16:8 h light/dark), 100 μmol photons m-2·s-1 (100 μE m-2·s-1) light intensity, 18 °C constant temperature, and 120 rpm constant agitation.
Pick a diatom colony grown on solid BG11 and inoculate 50 mL of liquid BG11 containing ampicillin and amphotericin.
After approximately seven days, this culture will be in exponential state (1 × 106 cells/mL). Determine the exact cell number with the Neubauer counting chamber using a binocular light microscope and take a volume of culture to obtain a final concentration of 1 × 104 cells/mL in 50 mL of liquid BG11 in an Erlenmeyer flask. Then, follow the culture until it reaches the stationary phase of the growth curve (approximately 1 × 106 cells/mL). The final cell yield in each culture may vary depending on the medium and growth conditions. In our conditions, it takes between one and two weeks. A higher cell number results in a higher number of exconjugant colonies.
Cell harvest and control plates
Take the entire volume of the Erlenmeyer flask for both bacteria and diatoms and centrifuge them at room temperature (always work under the laminar flow). Centrifuge at low speed (2,000 × g) for 5 min. Do not use a brake in order to ensure cell viability.
Resuspend the pellets in 500 µL of the corresponding culture medium: BG11 for diatoms and LB for E. coli.
Take 50 µL of diatoms and plate:
Plate without Zeo to check growth (positive control).
Plate with Zeo to corroborate wild-type diatom antibiotic susceptibility (negative control).
Incubate these plates at 18 °C under 100 μmol photons m-2·s-1 light intensity.
Mix diatoms and DH10BpTA-Mob bacteria (conjugation)
In a 1.5 mL Eppendorf tube, combine 200 µL of diatoms with 200 µL of E. coli DH10B pTA-Mob bacteria strain containing the construct of interest. Spread the mixture on conjugation plates (Figure 1C).
Incubate for 90 min in darkness at 30 °C.
Incubate at 18 °C under 100 μmol photons m-2·s -1 light intensity for 48 h. During this time, the conjugation is carried out.
Collect all the cells grown on the plate by adding 200 µL of BG11 medium and then take the entire volume and plate it on a selection plate containing Zeo.
Incubate at 18 °C under 100 μmol photons m-2·s -1 light intensity. After 20 days, exconjugant colonies resistant to Zeo should appear.
Check the presence of the plasmid in the exconjugant diatom cells by colony PCR.
To perform colony PCR, proceed as a standard colony PCR. Take a colony with a toothpick and introduce it into a PCR tube with 20 μL of PCR solution mix containing primers (the best combination will be one of the particular contrast and a second PfcpB primer, as suggested in section B).
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Cainzos, M., Marchetti, F., Popovich, D., Leonardi, P, Pagnussat, G and Zabaleta E. (2021) Gamma Carbonic Anhydrases are subunits of the Mitochondrial Complex I of diatoms. Mol. Microbiology 116(1), pp. 109–125
General notes and troubleshooting
The efficiency of conjugation strongly depends on the length of the construct, which is in turn determined by the length of the sequence of interest cloned into the destination vector. The highest number of exconjugants is obtained with small plasmids.
Acknowledgments
This work was financially supported by ANPCyT grant PICT 18 0652.
Competing interests
The authors declare no conflicts of interest.
References
- Sommer, U., Stibor, H., Katechakis, A., Sommer, F. and Hansen, T. (2002). Pelagic food web configurations at different levels of nutrient richness and their implications for the ratio fish production:primary production. Sustainable Increase of Marine Harvesting: Fundamental Mechanisms and New Concepts : 11–20.
- Falciatore, A. and Mock, T. (Eds.). (2022). The Molecular Life of Diatoms. Springer International Publishing.
- Wilhelm, C., Büchel, C., Fisahn, J., Goss, R., Jakob, T., LaRoche, J., Lavaud, J., Lohr, M., Riebesell, U., Stehfest, K., et al. (2006). The Regulation of Carbon and Nutrient Assimilation in Diatoms is Significantly Different from Green Algae. Protist 157(2): 91–124.
- Allen, A. E., LaRoche, J., Maheswari, U., Lommer, M., Schauer, N., Lopez, P. J., Finazzi, G., Fernie, A. R. and Bowler, C. (2008). Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U.S.A. 105(30): 10438–10443.
- Murik, O., Tirichine, L., Prihoda, J., Thomas, Y., Araújo, W. L., Allen, A. E., Fernie, A. R. and Bowler, C. (2018). Downregulation of mitochondrial alternative oxidase affects chloroplast function, redox status and stress response in a marine diatom. New Phytol. 221(3): 1303–1316.
- Butler, T., Kapoore, R. V. and Vaidyanathan, S. (2020). Phaeodactylum tricornutum: A Diatom Cell Factory. Trends Biotechnol. 38(6): 606–622.
- Zulu, N. N., Zienkiewicz, K., Vollheyde, K. and Feussner, I. (2018). Current trends to comprehend lipid metabolism in diatoms. Prog. Lipid Res. 70: 1–16.
- Rodolfi, L., Chini Zittelli, G., Bassi, N., Padovani, G., Biondi, N., Bonini, G. and Tredici, M. R. (2008). Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low‐cost photobioreactor. Biotechnol. Bioeng. 102(1): 100–112.
- Karas, B. J., Diner, R. E., Lefebvre, S. C., McQuaid, J., Phillips, A. P., Noddings, C. M., Brunson, J. K., Valas, R. E., Deerinck, T. J., Jablanovic, J., et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6(1): e1038/ncomms7925.
- Diner, R. E., Bielinski, V. A., Dupont, C. L., Allen, A. E. and Weyman, P. D. (2016). Refinement of the Diatom Episome Maintenance Sequence and Improvement of Conjugation-Based DNA Delivery Methods. Front. Bioeng. Biotechnol. 4: e00065.
- Cainzos, M., Marchetti, F., Popovich, C., Leonardi, P., Pagnussat, G. and Zabaleta, E. (2021). Gamma carbonic anhydrases are subunits of the mitochondrial complex I of diatoms. Mol. Microbiol. 116(1): 109–125.
- Strand, T. A., Lale, R., Degnes, K. F., Lando, M. and Valla, S. (2014). A New and Improved Host-Independent Plasmid System for RK2-Based Conjugal Transfer. PLoS One 9(3): e90372.
- Sambrook, J. and Russell, D. W. (2006). The Inoue Method for Preparation and Transformation of CompetentE. Coli: “Ultra-Competent” Cells. Cold Spring Harb. Protoc. 2006(1): 10–1101.
Article Information
Copyright
© 2024 CONICET: Consejo Nacional de Investigaciones Científicas y Técnicas; This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Berdun, F., Valiñas, M., Pagnussat, G. C. and Zabaleta, E. J. (2024). Mobilization of Plasmids from Bacteria into Diatoms by Conjugation Technique. Bio-protocol 14(5): e4945. DOI: 10.21769/BioProtoc.4945.
Category
Microbiology > Heterologous expression system
Biological Engineering > Synthetic biology
Molecular Biology > DNA > Conjugation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









