- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Use of the Fluorescent Dye Thioflavin T to Track Amyloid Structures in the Pathogenic Yeast Candida albicans
(§ Technical contact: thierry.mourer@pasteur.fr) Published: Vol 14, Iss 3, Feb 5, 2024 DOI: 10.21769/BioProtoc.4932 Views: 2338
Reviewed by: Alba BlesaLucy Xie

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Substituted Cysteine Accessibility Method for Topology and Activity Studies of Membrane Enzymes Forming Thioester Acyl Intermediates in Bacteria
Sébastien Gélis-Jeanvoine and Nienke Buddelmeijer
Nov 5, 2015 8782 Views

Aggregation Prevention Assay for Chaperone Activity of Proteins Using Spectroflurometry
Manish Bhuwan [...] Seyed E. Hasnain
Jan 20, 2017 12676 Views
Snapshots of the Signaling Complex DesK:DesR in Different Functional States Using Rational Mutagenesis and X-ray Crystallography
Juan Andres Imelio [...] Alejandro Buschiazzo
Aug 20, 2017 8210 Views
Abstract
The human pathogenic yeast Candida albicans can attach to epithelial cells or indwelling medical devices to form biofilms. These microbial communities are highly problematic in the clinic as they reduce both sensitivity to antifungal drugs and detection of fungi by the immune system. Amyloid structures are highly organized quaternary structures that play a critical role in biofilm establishment by allowing fungal cells to adhere to each other. Thus, fungal amyloids are exciting targets to develop new antifungal strategies. Thioflavin T is a specific fluorescent dye widely used to study amyloid properties of target proteins in vitro (spectrophotometry) and in vivo (epifluorescence/confocal microscopy). Notably, thioflavin T has been used to demonstrate the ability of Als5, a C. albicans adhesin, to form an amyloid fiber upon adhesion. We have developed a pipeline that allows us to study amyloid properties of target proteins using thioflavin T staining in vitro and in vivo, as well as in intact fungal biofilms. In brief, we used thioflavin T to sequentially stain (i) amyloid peptides, (ii) recombinant proteins, (iii) fungal cells treated or not with amyloid peptides, (iv) fungal amyloids enriched by cell fractionation, and (v) intact biofilms of C. albicans. Contrary to other methods, our pipeline gives a complete picture of the amyloid behavior of target proteins, from in vitro analysis to intact fungal biofilms. Using this pipeline will allow an assessment of the relevance of the in vitro results in cells and the impact of amyloids on the development and/or maintenance of fungal biofilm.
Key features
• Study of amyloid properties of fungal proteins.
• Visualization of the subcellular localization of fungal amyloid material using epifluorescence or confocal microscopy.
• Unraveling of the amyloid properties of target proteins and their physiological meaning for biofilm formation.
• Observation of the presence of amyloid structures with live-cell imaging on intact fungal biofilm using confocal microscopy.
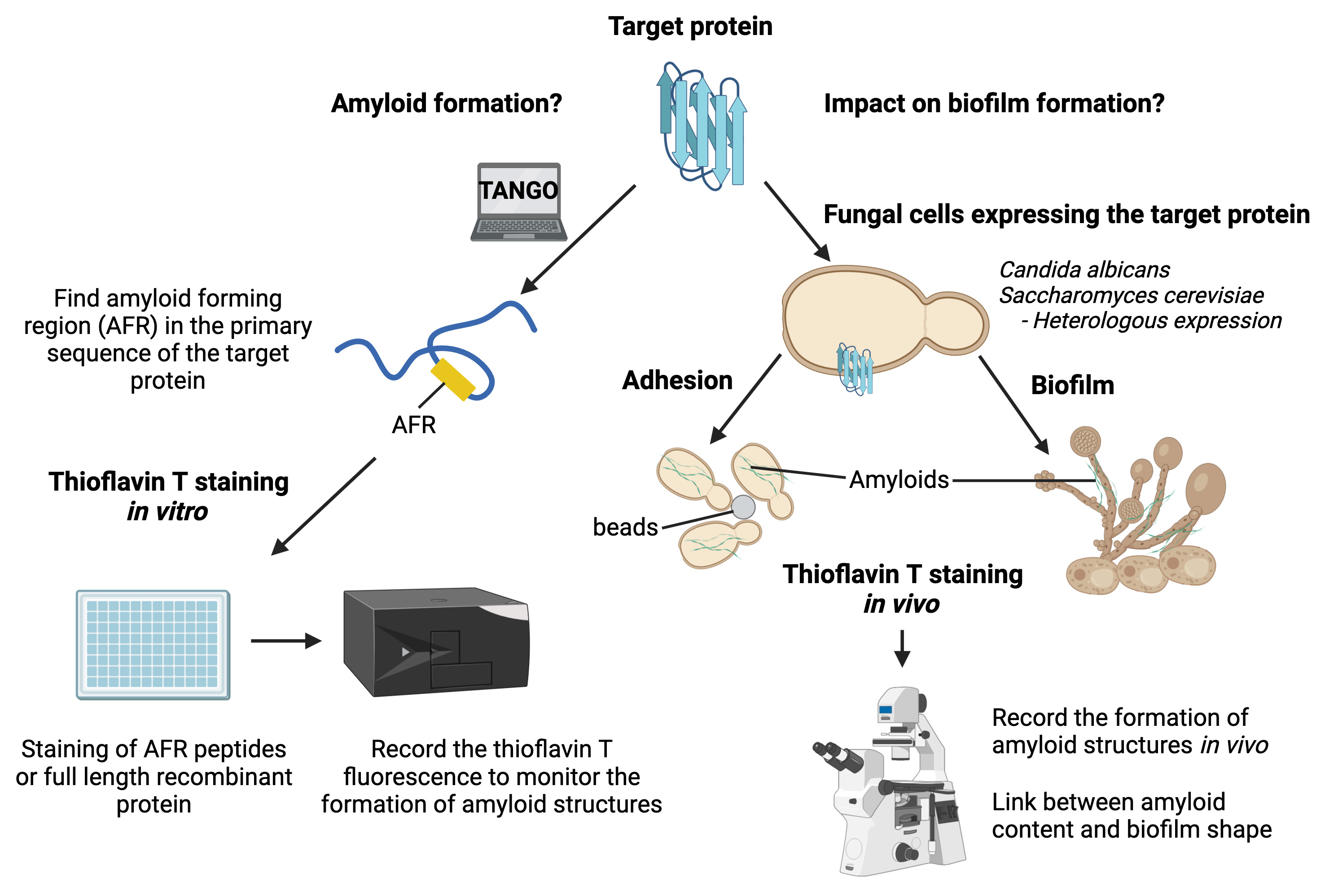
Graphical overview
Background
Under certain circumstances (e.g., broad-spectrum antibiotics, neutropenia, abdominal surgery, or central venous catheter), the human fungal pathogen Candida albicans can be translocated through the gastrointestinal mucosa to reach the bloodstream and colonize organs such as kidneys or the brain (Pfaller and Diekema, 2007). Upon host colonization, C. albicans can form a highly structured microbial community named biofilm (Cabral et al., 2014). Biofilm formation starts with adhesion of cells on biotic or abiotic surfaces (Nobile and Johnson, 2015). Then, hyphae are extruded and elongated from the mother cells before the synthesis of the extracellular matrix. Once the matrix covers the fungal community, yeast cells are released from the biofilm, allowing new areas to be colonized. C. albicans biofilms are difficult to cure, as such structures reduce both sensitivity to antifungal agents and pathogen recognition by the host immune system (Brown et al., 2012). The cell wall plays a critical role in fungal biofilm establishment by allowing C. albicans cells to interact with their environment as well as maintaining the biofilm integrity. Over the past decade, multiple reports have shown that amyloid structures present in the microorganisms’ cell wall create adhesion forces between cells that promote biofilm formation (Ho et al., 2019). We have recently uncovered that Pga59, a cell wall protein, forms amyloid structures to promote cell–cell adhesion forces and hence contribute to biofilm formation in C. albicans (Mourer et al., 2023). To draw these conclusions, we took advantage of the amyloid-specific fluorescent dye thioflavin T (ThT). Specific binding of ThT to amyloid structures results in a shift of its emission wavelength and in an increase of its quantum yield. Hence, ThT is a powerful tool used to detect amyloid components in biological specimens. Regarding fungal biofilms, some cell wall proteins have been positively stained by ThT, thus demonstrating their ability to adopt the shape of amyloid structure (Mourer et al., 2023; Ramsook et al., 2010). However, the link between in vitro experiments and the fact that amyloid properties of proteins are important for biofilm formation or establishment is often missing. We developed a pipeline to explore the amyloid properties of target proteins as well as their relevance for fungal cell adhesion and biofilm formation. We used ThT to successively stain octapeptides, recombinant proteins, amyloid structures enriched from fungal cells, adherent cells treated or not with amyloid peptides, and finally intact biofilms. Using this pipeline can formally confirm or reject that 1) the target protein is able to form an amyloid structure and 2) this amyloid is physiologically relevant for biofilm formation. This protocol has the potential to be applied to other microorganisms that can form biofilms like the bacteria Pseudomonas aeruginosa or Staphylococcus epidermidis and hence, create new knowledge regarding the impact of amyloid proteins on biofilm formation.
Materials and reagents
Biological materials
Candida albicans BWP17 (Wilson et al., 1999)
Saccharomyces cerevisiae BY4742 (Winston et al., 1995)
Reagents
Thioflavin T (Sigma-Aldrich, catalog number: T3516-5g)
Dimethylsulfoxide (DMSO) (Sigma-Aldrich, catalog number: 276855-250 mL)
M-280 Dynabeads, tosyl-activated magnetic beads (Thermo Fisher Scientific, catalog number: 14204)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9418)
Concanavalin A, Alexa FluorTM 594 conjugate (Thermo Fisher Scientific, catalog number: C11253)
RNase A solution (Promega, catalog number: A7973)
TritonTM X-100 (Sigma-Aldrich, catalog number: X100-100ML)
Sucrose (Sigma-Aldrich, catalog number: S0389-500G)
10% CriterionTM XT Bis-Tris protein gel (Bio-Rad, catalog number: 3450112)
Bio-Rad protein assay dye reagent concentrate (Bradford) (Bio-Rad, catalog number: 5000006)
Poly-L-lysine solution (Sigma-Aldrich, catalog number: P8920)
Aclar® embedding film (TED PELLA, INC, catalog number: 10501-10)
GibcoTM RPMI 1640 medium (Thermo Fisher Scientific, catalog number: 10379144)
Dulbecco’s phosphate buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 11590476)
Ethanol absolute (Merck, catalog number: 1009831011)
Magnesium chloride (Sigma-Aldrich, catalog number: 208337-100G)
Nonidet P-40 (Sigma-Aldrich, catalog number: I8896)
cOmplete, EDTA-free protease inhibitor tablets (Roche, catalog number: 11873580001)
Phenylmethylsulfonyl fluoride (PMSF) (Thermo Fisher Scientific, catalog number: 36978)
Glycerol (Sigma-Aldrich, catalog number: G5516-100ML)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: 62862)
Dithiothreitol (Sigma-Aldrich, catalog number: D0632-1G)
Tris hydrochloride (Thermo Fisher Scientific, catalog number: 15893661)
Sodium chloride (Sigma-Aldrich, catalog number: S9888-500G)
Bromophenol blue (Sigma-Aldrich, catalog number: B0126-25G)
Bradford reagent (Bio-Rad, catalog number: 500205)
BactoTM yeast extract (Thermo Fisher Scientific, catalog number: 212750)
BactoTM peptone (Thermo Fisher Scientific, catalog number: 211677)
Dextrose (Sigma-Aldrich, catalog number: D9434-1KG)
Yeast nitrogen base without amino acids (Sigma-Aldrich, catalog number: Y0626-1KG)
Glucose (Sigma-Aldrich, catalog number: G8270-1KG)
Arginine (Sigma-Aldrich, catalog number: A5006-100G)
Uridine (Sigma-Aldrich, catalog number: U3750-25G)
Histidine (Merck, catalog number: H3911)
Yeast synthetic drop-out medium supplements without uracil (Sigma-Aldrich, catalog number: Y1501-20G)
Solutions
Thioflavin T solution (ThT) (see Recipes)
Thioflavin T buffer (see Recipes)
Lysis buffer (see Recipes)
Amyloid-Prion resuspension buffer (see Recipes)
Amyloid-Prion buffer R (see Recipes)
YPD medium (see Recipes)
SD complete medium (see Recipes)
SC-Ura medium (see Recipes)
Recipes
Thioflavin T solution (3.3 mM)
Reagent Final concentration Quantity Thioflavin T 3.3 mM 10.53 mg Absolute ethanol n/a 10 mL Total n/a 10 mL Protect the thioflavin T solution from light with aluminum foil and store the solution at -20 °C up to six months.
Thioflavin T buffer
Reagent Final concentration Quantity Tris hydrochloride (1 M, pH 8.0) 20 mM 200 μL Sodium chloride (5 M) 150 mM 300 μL Thioflavin T (3.3 mM) 40 μM 121 μL Distilled H2O n/a 9.4 mL Total n/a 10 mL Prepare the thioflavin T buffer freshly before each experiment.
Lysis buffer
Reagent Final concentration Quantity Tris hydrochloride (1 M, pH 7.5) 50 mM 750 μL Sodium chloride (5 M) 150 mM 450 μL Magnesium chloride (1 M) 5 mM 75 μL Nonidet P-40 0.1% 15 μL PMSF (200 mM) 1 mM 75 μL cOmplete protease inhibitor n/a 2 tablets Distilled H2O n/a 13.6 mL Total n/a 15 mL Store the lysis buffer without protease inhibitors (PMSF and cOmplete protease inhibitor tablets) at 4 °C for two months maximum. Add protease inhibitors freshly before each experiment.
Amyloid-Prion resuspension buffer
Reagent Final concentration Quantity Tris hydrochloride (1 M, pH 7.5) 50 mM 350 μL Sodium chloride (5 M) 100 mM 140 μL SDS (20%) 2% 700 μL Dithiothreitol (1 M) 5 mM 35 μL Glycerol 5% 350 μL cOmplete protease inhibitor n/a 1 tablet Distilled H2O n/a 5.4 mL Total n/a 7 mL Store the Amyloid-Prion resuspension buffer for eight weeks at 4 °C without dithiothreitol and protease inhibitor. Both the dithiothreitol and the protease inhibitor tablet should be added just before the start of the experiment.
Amyloid-Prion buffer R
Reagent Final concentration Quantity Tris hydrochloride (1 M, pH 7.5) 10 mM 100 μL SDS (20%) 0.4% 200 μL Dithiothreitol (DTT, 1 M) 5 mM 50 μL Distilled H2O n/a 9.7 mL Total n/a 10 mL The amyloid-prion buffer R can be kept at room temperature for three months. Add the dithiothreitol just before starting the experiment.
YPD medium
Reagent Final concentration Quantity BactoTM yeast extract 1% 10 g BactoTM peptone
Dextrose
Distilled H2O
2%
2%
n/a
20 g
20 g
q.s. to 1 L
Total n/a 1 L SD complete medium
Reagent Final concentration Quantity Yeast nitrogen base without amino acids 0.67% 6.7 g Glucose
Arginine (2 mg/mL)
Uridine (4 mg/mL)
2%
20 mg/mL
40 mg/mL
20 g
10 mL
10 mL
Histidine (2 mg/mL) 20 mg/mL 10 mL Distilled H2O n/a q.s. to 1 L Total n/a 1 L SC-Ura medium
Reagent Final concentration Quantity Yeast nitrogen base without amino acids 0.67% 6.7 g Glucose 2% 20 g Yeast synthetic drop-out medium supplements without uracil 0.1% 1 g Distilled H2O n/a q.s. to 1 L Total n/a 1 L
Laboratory supplies
1.5 mL Eppendorf tubes (Eppendorf, catalog number: 0030125207)
50 mL FalconTM tubes (Fisher Scientific, catalog number: 10203001)
TPP® 96-well plate (Merck, catalog number: Z707902-108EA)
Culture plate 12 wells TPP (Dutscher, catalog number: 109212)
1.5 mL screw-cap micro tubes (Fisher Scientific, catalog number: 11549924)
Glass beads 0.5 mm, acidic wash (Sigma-Aldrich, catalog number: G8772-100 G)
Petri dishes 35 mm (Greiner, catalog number: P5112)
Ultracentrifuge polypropylene tubes (Beckman-Coulter, catalog number: 331372)
Glass slide (Merck, catalog number: S9027-1CS)
Coverslip (VWR, catalog number: 43210.KG)
Centrifuge bottles (Beckman-Coulter, catalog number: C31600)
Plastic cuvettes BRANDTM (Fisher Scientific, catalog number: 10566581)
Equipment
Microplate reader (TECAN, model: Infinite® 200 PRO)
Microcentrifuge (Eppendorf, catalog number: Z606235)
Ultracentrifuge OPTIMA XPN (Beckman-Coulter, catalog number: A94468)
Upright confocal microscope (Zeiss, model: LSM700)
ThermoMixer® C (Eppendorf, catalog number: 5382000015)
Sorvall RC-5B centrifuge (Thermo Fisher Scientific, catalog number: sorvall-RC-5B)
Vortex mixer (VWR, catalog number: 444-1372)
Centrifuge 5810 (Eppendorf, catalog number: 5810000010)
Microscope (Zeiss, model: Epifluorescence ApoTome)
Spectrophotometer (GE UltrospecTM 2100 pro, catalog number: GE80211221)
Bullet Blender Storm Pro (Next Advance, catalog number: 152081)
2 L Erlenmeyer flask (VWR, catalog number: 10545-844)
Electrophoresis power supply (Bio-Rad, model: 200/2.0)
Mini trans-blot electrophoretic transfer cell (Bio-Rad, catalog number: 1703930)
Water bath (Cole-Parmer, catalog number: WB-300-15)
Benchtop tube rotator RotoFlex (Cole-Parmer, catalog number: 120VAC)
Multitron shaker (Infors HT)
Software and datasets
Fiji ImageJ software (https://imagej.net/software/fiji/downloads)
Procedure
Identification and validation of amyloid-forming regions present in a protein of interest
Download the complete amino acid sequence of the target protein from the Uniprot database (https://www.uniprot.org/).
Copy and paste the primary sequence of the studied protein on the TANGO amyloid prediction tool (http://tango.crg.es/). The software will predict the presence of amyloid-forming regions in the primary sequence and, therefore, the potential of the protein to be assembled as an amyloid structure.
Note: TANGO provides a β-aggregation potential for every amino acid of the sequence. The higher the β-aggregation potential for an amino acid, the more likely that it is involved in the formation of an amyloid structure. This step will allow the identification of regions within the primary sequence of the protein that could be involved in amyloid formation. In our study, we have selected a region with a β-aggregation potential equal or better than 30%.
Order peptides corresponding to regions identified in step A2. All peptides were ordered from Thermo Fisher Scientific.
Note: All peptides must have acetyl and amide group at the N-terminus and C-terminus, respectively. These modifications avoid introduction of charged groups in the peptide as well as its degradation by exopeptidases.
Centrifuge the peptides at 18,000× g for 1 min to ensure that all powder is at the bottom of the tubes. Resuspend each peptide at a concentration of 100 μM in DMSO (stock solution).
Pause point: The protocol could be stopped at this step and the peptide solution stored at 4 °C for up to a month.
Note: Make sure that the powder is well resuspended either by pipetting or vortexing.
Dilute stock solutions of each peptide at a final concentration of 5 μM in the thioflavin T buffer. Distribute in triplicate 100 μL of diluted peptides in a 96-well plate. A solution composed of DMSO, 20 mM Tris-HCl, 150 mM NaCl, and 40 μM thioflavin T should be added to the plate as a negative control.
Note: Thioflavin T is resuspended at 3.3 mM in absolute ethanol.
Incubate the 96-well plate at 37 °C in a Tecan Infinite plate reader for 16 h.
Note: Set up the parameters to record thioflavin T fluorescence every hour with an excitation wavelength of 440 nm and emission wavelength of 496 nm.
Evaluation of amyloid structure assembly on recombinant full-length protein
Express and purify the recombinant protein of interest.
Critical point: Once purified, the recombinant protein should be used as soon as possible to avoid degradation of the polypeptide chain and/or random aggregation.
Notes:
As numerous methods exist to express and subsequently purify recombinant proteins, we cannot recommend a unique path to produce the protein of interest with high purity. Protein expression and purification correspond to a trial-and-error process. If the behavior of the protein is unknown, several expression systems (Escherichia coli, Pichia pastoris, Baculovirus expression system, Chinese hamster ovary mammalian cell line) should be tested for their ability to produce the required quantity (1 mg/mL) of recombinant protein. We used pET28a to express His6-Pga59 in E. coli SHuffle strain (Lobstein et al., 2012).
The appropriate chromatography strategy should be carefully designed to reach a protein purity as high as 95%. In our case, proteins were solubilized from inclusion bodies with a solution containing 6 M guanidium hydrochloride, refolded by dialysis, and purified by affinity chromatography (α-HIS tag). It is highly recommended to assess the purity of proteins using a MALDI-TOF mass spectrometry analysis.
Make a 10-time dilution of the recombinant protein in the dialysis buffer (final volume: 100 μL). Transfer the diluted protein in a quartz cuvette and measure its concentration using UV-spectrophotometry at 280 nm.
Caution: As the recombinant protein is purified under the monomeric form, the protein concentration must be kept low (5–10 mM) to avoid unwanted random aggregation.
Note: The concentration is determined by applying the Beer-Lambert’s law A = ϵlc, where A is the absorbance at 280 nm, ϵ the molar extinction coefficient, l the length of the optical path, and c the concentration. ϵ can be calculated by inserting the primary sequence of the protein in the Expasy ProtParam tool (https://web.expasy.org/protparam/). If the protein has no aromatic residues (tryptophan, histidine, tyrosine, and phenylalanine), the absorbance should be measured at 214 nm (absorbance of peptidic bonds).
Dilute recombinant proteins at 5 μM in 300 μL of thioflavin T buffer and distribute triplicates (3 × 100 μL) of each mix in 96-well plates.
Critical point: Before starting the thioflavin T staining assay, the recombinant protein must be present under the monomeric form. Starting the amyloidogenesis with preexisting aggregates in the mixture will impair amyloid assembly and hence the results of the thioflavin T staining.
As described in step A6, incubate the plate at 37 °C in a Tecan Infinite plate reader for 16 h. Measure the fluorescence every hour to monitor amyloidogenesis.
Thioflavin T staining of amyloid structures on intact fungal cells upon adhesion
Isolated colonies of C. albicans are inoculated in 4 mL of YPD medium and incubated under shaking (220 rpm) overnight at 30 °C.
Note: According to the protein of interest, the appropriate genetic background should be constructed to perform experiments for section C (e.g., knockout cells).
The same day as C. albicans precultures, wash 200 μL of magnetic beads twice with PBS.
Resuspend the beads in 1 mL of heat-denatured BSA (1 mg/mL) and incubate at 37 °C overnight on a benchtop rotator.
Note: To denature BSA, incubate the solution for 1 h in a water bath at 70°C.
The day after, dilute 20 μL of the culture in 980 μL of YPD. Transfer in a 1 mL plastic cuvette and measure the optical density OD at 600 nm (OD600).
Caution: If OD600 of cell dilution is above 1, it is likely that it is outside of the linearity zone of the spectrophotometer. Therefore, to be precise about cells’ quantity, samples should be diluter further to have an OD600 between 0.1 and 1.
Dilute the overnight culture to an OD600 of 0.3 in 5 mL of fresh YPD medium. Then, incubate cells for four additional hours under shaking (220 rpm) at 30 °C.
Note: Cells should be at mid-logarithmic phase before starting the ThT staining (OD600 around 0.8 or 1).
Centrifuge 108 yeast cells at 956× g for 4 min and wash them twice with sterile PBS.
Note: The quantity of cells is determined by measuring the OD600 with a spectrophotometer. One milliliter of C. albicans culture at OD600 of 1 corresponds to 2 × 107 fungal cells.
Resuspend fungal cells in 1 mL of SD complete medium containing 25 μM of ThT and 106 magnetic beads treated with heat-denatured BSA. Incubate the cells at room temperature on a benchtop rotator for 1 h.
Note: At this step, the impact of amyloid peptides on fungal adhesion can be assessed on a wild-type strain of C. albicans. For this, proceed exactly as described in steps C1–C5, but at step C6 add 5 μM of the peptide of interest in the SD medium.
Drop 10 μL of fungal cells on a glass slide and add a coverslip on top of the droplet. Observe the presence of amyloid structure by recording the ThT signal with the blue filter of an epifluorescence microscope.
Heterologous expression of amyloidogenic proteins in S. cerevisiae
Note: If too much redundancy arises between the protein of interest and other proteins from the host organism, it could be very difficult to observe phenotypes using ThT staining. One alternative would be to heterologously express the protein in another microorganism such as S. cerevisiae.
Inoculate S. cerevisiae from isolated colonies in 4 mL of SC-Ura medium and incubate overnight at 30 °C. Prepare magnetic beads as described in step C2.
The day after, dilute 20 μL of the overnight culture in 980 μL of YPD, transfer the entire volume in a plastic cuvette, and measure the OD600 nm.
Dilute the preculture of S. cerevisiae cells to an OD600 of 0.5 in 4 mL of fresh SC-Ura medium. Incubate cells with agitation (220 rpm) at 30 °C for 4 h.
Centrifuge yeast cells at 956× g for 3 min and resuspend the resulting pellet in 1 mL of SC-Ura medium.
Then, treat the cell suspension simultaneously with 106 BSA-coated magnetic beads and 25 μM of ThT. Incubate the mixture on a benchtop rotator at room temperature for 1 h.
Note: At this step, fluorescent dyes targeting specific cellular compartments can be added to perform colocalization experiments with the signal generated by the ThT. As our amyloids of interest are assembled in the cell wall, we used Concanavalin A Alexa Fluor 594-conjugate as cell wall marker.
Centrifuge cells at 956× g for 3 min and wash the pellet twice with 1 mL of PBS.
Add a droplet of fungal cells (10 μL) on a glass slide and cover with a coverslip. Monitor the ThT signal using epifluorescence microscopy with the blue filter.
Isolation of amyloid material from fungal cells
Note: To study the effect of various stimuli on amyloidogenesis in fungal cells, whole amyloid material could be isolated by cell fractionation (Kryndushkin et al., 2017) and subsequently stained with thioflavin T. We used this approach to show the formation of amyloids in C. albicans upon adhesion.
Inoculate fungal cells in 50 mL of YPD medium and incubate the flasks at 30 °C overnight under agitation (220 rpm).
Note: Prepare a solution of beads with heat-denatured BSA as described in step C2.
The day after, measure the OD600 of the preculture as described in step C3.
Dilute overnight culture to an OD600 of 0.3 in 1 L of YPD medium. Incubate cells at 30 °C for 4 h with shaking (220 rpm).
Caution: To ensure good oxygenation of fungal cells, the 1 L YPD culture should be performed in two flasks of 2 L (2 × 500 mL).
Transfer the cultures in 500 mL centrifuge bottles and centrifuge (Sorvall centrifuge) fungal cells at 956× g for 10 min.
Resuspend the pellet in 50 mL of SD medium and transfer the cell suspension in a 50 mL Falcon tube. Add 1.25 × 107 BSA-coated beads to the mixture, except for the negative control, and insert Falcon tubes on a benchtop rotator. Incubate cells for 1 h at room temperature.
Centrifuge fungal cells at 956× g for 4 min (Eppendorf 5810 R centrifuge). Discard the supernatant and store pellets at -80 °C until needed.
Pause point: The frozen pellet can be stored at -80 °C for months before being processed for amyloids extraction.
Thaw pellets on ice and wash once with cold PBS. Centrifuge at 956× g for 4 min and discard the supernatant.
Resuspend pellets with 4 mL of lysis buffer and distribute the entire volume in 1.5 mL screw-cap tubes (600 μL per tube). To each screw-cap tube, add 200 μL of 0.5 mm glass beads.
Mechanically disrupt yeast cells with a Bullet Blender bead beater using a cycle of six times for 1 min at full speed. Centrifuge the protein mixture at 800× g for 5 min at 4 °C in a tabletop centrifuge. Take out the supernatant that contains amyloid materials and transfer it in a clean 1.5 mL Eppendorf tube.
Note: Between each cycle, incubate the lysates on ice for 1 min.
Incubate protein extracts with RNase A (0.1 mg/mL) at room temperature for 10 min. Add Triton X-100 at a final concentration of 0.5% and incubate on ice for 10 min. Centrifuge the lysate at 2,000× g for 10 min at 4 °C. Discard the pellet and transfer the supernatant in a new 1.5 mL Eppendorf tube.
Load the supernatant from the previous step on a 2 mL sucrose layer (40%). Centrifuge samples at 200,000× g for 2 h at 4 °C.
Note: Make sure to use an ultracentrifuge tube for this step to avoid plastic collapsing at 200,000× g.
Resuspend the pellet in 200 μL of amyloid-prion resuspension buffer by pipetting up and down with a pipette. Incubate the mix for 10 min at 37 °C in a ThermoMixer device without shaking.
Centrifuge the solution at 5,000× g for 10 min in a tabletop centrifuge. Save the resulting supernatant in a 1.5 mL Eppendorf tube and discard the pellet.
For each 100 μL of amyloid preparation, add 0.01% of bromophenol blue and 5% of glycerol. Mix well and load the solution on a 10% polyacrylamide gel. Run the gel first for 15 min at 70 V and then switch to 200 V for 40 min. Use as many wells as required according to your sample volume.
Wash the polyacrylamide gel in distilled water once and cut out the top part (3 mm) of the stacking gel with a scalpel. Pool all gel sections per sample in a 1.5 mL Eppendorf tube and freeze gel squares at -20 °C for at least 20 min.
To each tube, subsequently add 200 μL of amyloid-prion buffer R, vortex for 10 s, and incubate for 15 min at 98 °C. Then, vortex and centrifuge at 4,000× g for 1 min. Take out the supernatant and drop it in a new 1.5 mL tube. Repeat this step twice.
Note: All supernatants must be pooled.
Quantify proteins in each sample using a classical Bradford assay.
Pause point: At this point, the protocol can be stopped, and amyloids stored at -20 °C until thioflavin T staining assays.
Dilute the amyloid-enriched fraction at a final concentration of 100 μM in 300 μL of thioflavin T buffer. Distribute 100 μL of amyloid fractions in a 96-well plate in triplicate. A solution composed of DMSO, 20 mM Tris-HCl, 150 mM NaCl, and 40 μM thioflavin T should be used as a negative control.
Incubate the 96-well plate at 37 °C in a Tecan Infinite plate reader for 16 h. Record ThT fluorescence every hour.
Track the presence of amyloid structures in intact biofilms of C. albicans
Note: C. albicans biofilms can be grown on a plastic surface (Aclar film) before being stained with ThT and then imaged with a confocal microscope to observe the presence of amyloid structures.
Inoculate C. albicans cells in 4 mL of YPD medium and incubate overnight at 30 °C.
Coat the Aclar film with a freshly made poly-L-lysine solution (0.1%) for 30 min at 37 °C. Wash the Aclar film twice with distilled water, cut it in small squares (1 cm × 1 cm), and finally sterilize each side under UV light for 15 min.
The day after, wash yeast cells twice with sterile PBS and dilute fungal cells at 1.106 cells/mL in RPMI medium.
Note: The RPMI medium should be prewarmed at 37 °C before use.
Place sterile Aclar film squares in wells from a 12-well tissue culture plate. Add 3 mL of diluted cells to each well.
Incubate fungal cells for 1 h at 37 °C with moderate shaking (110 rpm) in an Infors HT Multitron to allow adhesion of C. albicans to the plastic surface. Then, remove non-adherent cells by washing the wells twice with 1 mL of sterile PBS and fill up with 3 mL of prewarmed RPMI medium (37 °C).
Incubate the culture plates at 37 °C for 48 h at 110 rpm.
Wash mature biofilms once with PBS and stain with 2 mL of PBS containing 25 μM of ThT.
Caution: In order to prevent the biofilm to be detached from the Aclar film, the fungal community should be washed gently.
Wash biofilms two times with PBS and transfer them in a 35 mm Petri dish. Cover C. albicans biofilms with 2 mL of PBS and image using the blue channel of a confocal microscope.
Critical point: The objective of the microscope will be immersed in the culture medium. To avoid any damage, make sure to use an immersion objective.
Note: We record Z-stacks on a LSM700 upright using a 40× immersion objective. We then reconstruct the volume using Fiji ImageJ software.
Data analysis
For sections A–E, an example of each experiment has been published in Mourer et al. (2023). The step-by-step protocol has been summarized in Figure 1.
I) Regarding in vitro assays with peptides, recombinant proteins, or amyloid-enriched fractions from C. albicans (sections A, B, and E), the formation or the presence of amyloid structures is monitored by the ThT fluorescence over time. The higher the ThT fluorescence, the more amyloid structures are formed in the wells. If possible, add a peptide or a protein that is unable to form amyloids as an additional negative control to the blank. At the end of each experiment:
(i) Average the thioflavin T intensity value of triplicates for each sample.
(ii) Perform the statistical analysis to check if differences are relevant. Repeat this experiment three times, with three technical replicates for each peptide/protein. Samples are compared two by two using a Student’s t-test.
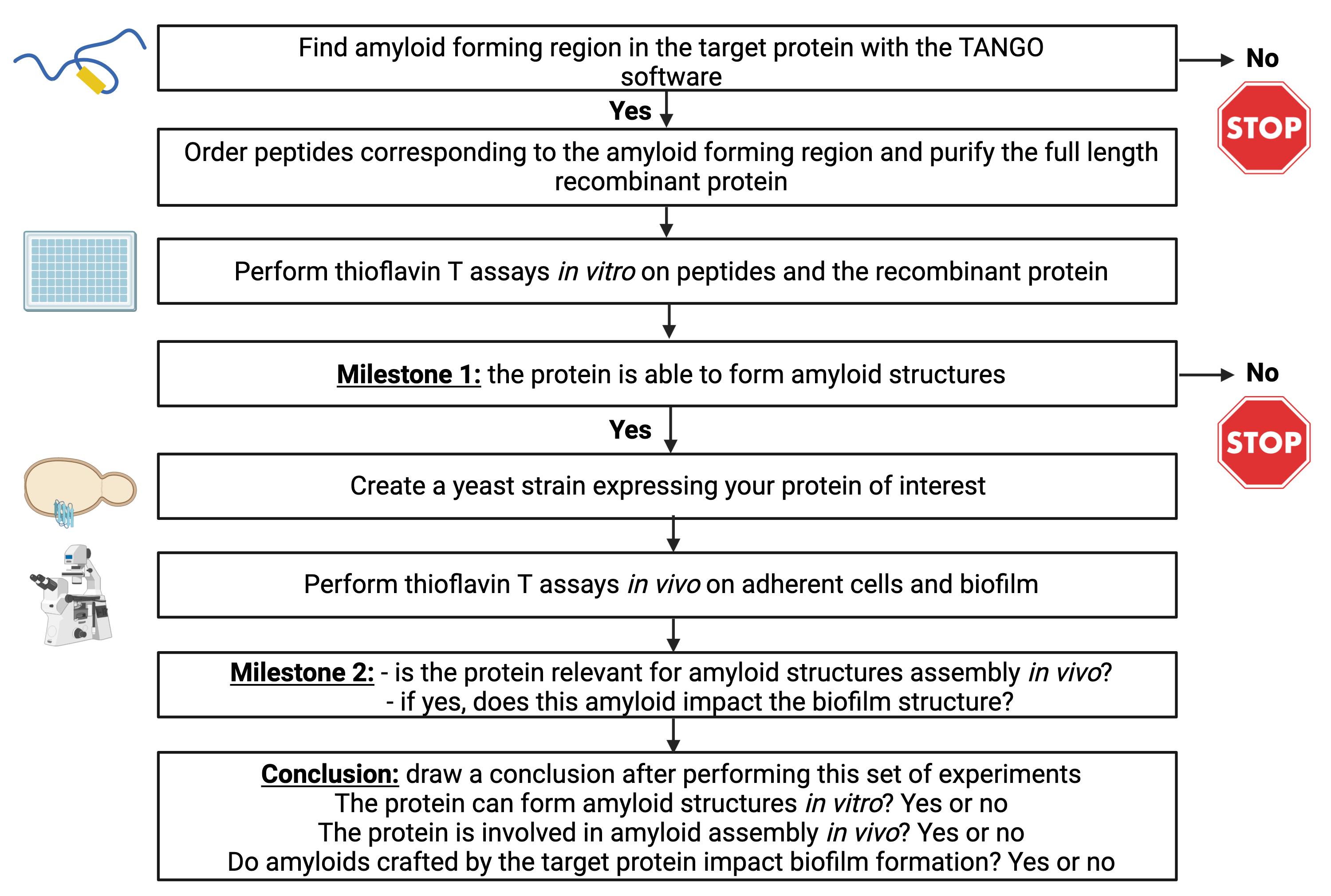
Figure 1. Summary of the protocol steps that must be followed successively to assess the propensity of a protein to form amyloids. The protocol will also assess the relevance of amyloids in vivo as well as their involvement in biofilm formation.
II) For in vivo staining (sections C, D, and F), the presence of amyloid structures is indicated by the blue fluorescence emitted by the ThT. According to the protein studied or the genotype of the yeast strains, the ThT pattern could change drastically. We suggest following this routine for data analysis:
(i) Observe fungal cells first at a rather low magnification using a 40× objective to evaluate the impact of the loss of certain proteins on amyloid formation, using ThT fluorescence intensity as a readout of amyloid content at the population level; indeed, the fluorescence levels correlate with the quantity of amyloids produced in the cells.
(ii) Record pictures of at least 100 cells at low magnification (40×).
(iii) Count the number of positive cells in each condition.
(iv) Repeat experiments at least three times in an independent manner and perform statistical analysis (compare two by two using a Student’s t-test).
(v) Draw a conclusion on the relevance of a target protein for amyloid formation in vivo and biofilm formation.
(vi) Then, switch to a 100× objective to observe the precise localization of amyloids within the cell. Fluorescent markers specific for cellular compartments could be used to assess colocalization with the ThT fluorescence as an indication of the presence of amyloid material in this specific compartment. Experiments must be carried out with three independent biological replicates.
III) Peptides encompassing the amyloid-forming region of candidate proteins can also be used for in vivo ThT staining on wild-type strains (section C). Follow the same routine as described in II.
The impact of wild-type and mutant peptides will be monitored by the ThT fluorescence intensity. If a peptide stimulates amyloid assembly, the ThT fluorescence will be higher in treated cells than in the untreated control. On the contrary, treating cells with peptides that inhibit amyloid assembly, for instance mutated peptides, will result in a lower ThT fluorescence than in the untreated control.
Finally, by using this protocol, we can also assess the relevance of amyloid structures for biofilm establishment. C. albicans biofilms can be easily stained with ThT and subsequently imaged with confocal microscopy to record Z-stacks of complete biofilms. According to the genotype of yeast cells, both the biofilm shape and thickness can be linked to the amyloid content of yeast cells that constitute the biofilm. Comparing the biofilm shape, thickness, and ThT fluorescence intensity to those of a wild-type strain biofilm will reveal if given mutations affect amyloid formation and hence biofilm establishment. An example of biofilm staining with ThT is presented in Figure 2, where we can see the shape, height, and amyloids (ThT staining in white). The experiments should be repeated three times with independent biological replicates. At this step, it is important to know how to record Z-stacks on the confocal microscope available in your institute or university. Also, users need to know how to reconstruct 2D images with Z-stacks using the Fiji ImageJ software.
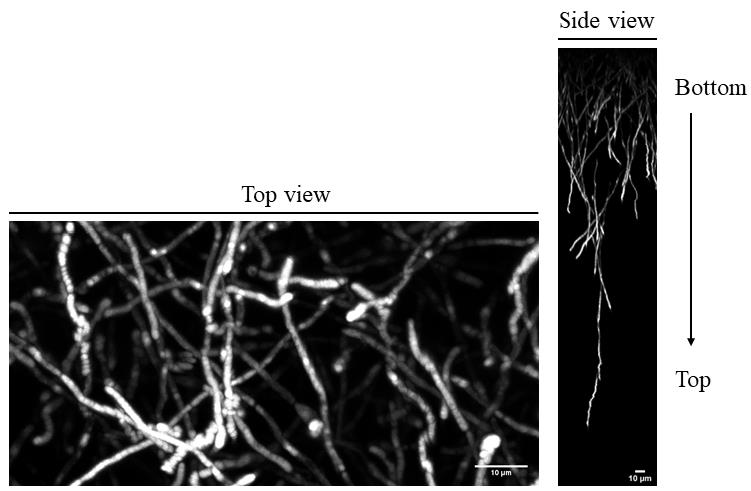
Figure 2. Thioflavin T (ThT) staining of intact fungal biofilm. C. albicans cells were allowed to attach on an Aclar film for 1 h. Fungal biofilm was then grown at 37 °C in RPMI under gentle shaking. After 48 h, amyloid structures were stained with a solution of PBS containing 25 mM of ThT. Z-stacks of biofilms were then recorded in the blue channel with a confocal microscope. Top view (left part) and side view (right part) of the biofilm are shown. Scale bars: 10 mm.
Validation of protocol
This protocol was developed to assess if a fungal candidate protein can be self-assembled to form an amyloid structure both in vivo and in vitro. If so, the physiological relevance of the protein for biofilm establishment or maintenance could be assessed by following section F of the present protocol. Except for ThT staining of intact biofilms, all experimental procedures were validated in Figure 1C, 2C, 3C, 4A and B, 5B, and 7 of Mourer et al., 2023 (doi: 10.1038/s41522-023-00371-x). For in vivo ThT staining of intact biofilm of C. albicans, see Figure 2 of the present manuscript.
General notes and troubleshooting
General notes
The culture conditions to perform ThT staining on fungal cells should be carefully determined. Indeed, C. albicans undergoing planktonic growth produces virtually no amyloid structures. Cells should be cultivated in conditions where amyloid assembly is active, like for instance cell adhesion triggered by magnetic beads. Otherwise, all attempts to stain amyloid fiber on fungal cells with ThT will be a failure.
The ThT fluorescence will be recorded in the blue channel either on epifluorescence or confocal microscopes. The signal of the resulting images will be weak and sometimes difficult to analyze. To visualize it better, use the Fiji ImageJ software to switch the color of images from blue to yellow using the channel tools option. Apply this change to all microscopic images.
Troubleshooting
Make sure that the buffer used to purify recombinant proteins is not autofluorescent in the blue channel (section B). This would generate erroneous results.
The concentration of recombinant proteins during amyloidogenesis should be kept low (section B). High concentration of proteins will interfere with the amyloid assembly and generate anarchic aggregates.
The purification of proteins that tend to aggregate under the monomeric form is a tedious process that could lead to project failure by lack of biological samples to perform experiments. If experimenters face difficulties to produce protein monomers, the purification product could be treated with hexafluoroisopropanol (HFIP). This compound will suppress the formation of amorphous aggregates and hence keep the protein in the monomeric form before starting in vitro thioflavin T assays (section B).
The ThT concentration required to stain amyloids in vivo can change depending on the cellular model (section C). According to the microorganism used, the amyloid content could change, and hence, the ThT concentration should be adjusted for efficient staining.
A low quantity of amyloid materials is extracted from fungal cells in section E. According to the growth conditions, C. albicans cells may be difficult to disrupt, and hence, a small amount of protein is extracted from cells during the lysis step. If this happens, do not skip the freezing step at -80 °C (step E6). The freezing will generate ice crystals in the membranes, which will positively impact cell lysis. Also, to increase cell lysis efficiency, add more glass beads on cells (400 μL instead of 200 μL).
Some amyloid structures can be sensitive to buffers that contain SDS, leading to a poor amount of amyloids recovery after the extraction protocol (section E). If amyloid structures are sensitive to SDS treatment, the detergent should be changed in amyloid-prion resuspension buffer as well as the amyloid-prion R buffer. We suggest replacing SDS by 1% Sarkosyl in both buffers.
C. albicans cells do not attach well on the plastic surface, making the imaging by confocal microscopy complicated. To solve this problem, two solutions are available: (i) increase the quantity of Poly-L-Lysine in step F2 or (ii) choose another extracellular matrix protein (e.g., Fibronectin) to coat the Aclar film (step F2).
Acknowledgments
T.M. was a recipient of the Pasteur-Cantarini postdoctoral fellowship from the Institut Pasteur. This work was supported by the French Government's Investissement d’Avenir program (Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases, ANR-10-LABX-62-IBEID). The graphical overview and Figure 1 were created with BioRender.com. This protocol has been used in Mourer et al. (2023), doi: 10.1038/s41522-023-00371-x.
Competing interests
The authors declare no competing interests.
References
- Brown, G. D., Denning, D. W., Gow, N. A. R., Levitz, S. M., Netea, M. G. and White, T. C. (2012). Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 4(165): e3004404.
- Cabral, V., Znaidi, S., Walker, L. A., Martin-Yken, H., Dague, E., Legrand, M., Lee, K., Chauvel, M., Firon, A., Rossignol, T., et al. (2014). Targeted Changes of the Cell Wall Proteome Influence Candida albicans Ability to Form Single- and Multi-strain Biofilms. PLoS Pathog. 10(12): e1004542.
- Ho, V., Herman-Bausier, P., Shaw, C., Conrad, K. A., Garcia-Sherman, M. C., Draghi, J., Dufrene, Y. F., Lipke, P. N. and Rauceo, J. M. (2019). An Amyloid Core Sequence in the Major Candida albicans Adhesin Als1p Mediates Cell-Cell Adhesion. mBio 10(5): e01766–19.
- Kryndushkin, D., Pripuzova, N. and Shewmaker, F. P. (2017). Isolation and Analysis of Prion and Amyloid Aggregates from Yeast Cells. Cold Spring Harb. Protoc. 2017(2): pdb.prot089045.
- Lobstein, J., Emrich, C. A., Jeans, C., Faulkner, M., Riggs, P. and Berkmen, M. (2012). SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 11(1): e1186/1475–2859–11–56.
- Mourer, T., El Ghalid, M., Pehau-Arnaudet, G., Kauffmann, B., Loquet, A., Brûlé, S., Cabral, V., d’Enfert, C. and Bachellier-Bassi, S. (2023). The Pga59 cell wall protein is an amyloid forming protein involved in adhesion and biofilm establishment in the pathogenic yeast Candida albicans. npj Biofilms Microbiomes 9(1): e1038/s41522–023–00371–x.
- Nobile, C. J. and Johnson, A. D. (2015). Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 69(1): 71–92.
- Pfaller, M. A. and Diekema, D. J. (2007). Epidemiology of Invasive Candidiasis: a Persistent Public Health Problem. Clin. Microbiol. Rev. 20(1): 133–163.
- Ramsook, C. B., Tan, C., Garcia, M. C., Fung, R., Soybelman, G., Henry, R., Litewka, A., O'Meally, S., Otoo, H. N., Khalaf, R. A., et al. (2010). Yeast Cell Adhesion Molecules Have Functional Amyloid-Forming Sequences. Eukaryotic Cell 9(3): 393–404.
- Wilson, R. B., Davis, D. and Mitchell, A. P. (1999). Rapid Hypothesis Testing with Candida albicans through Gene Disruption with Short Homology Regions. J. Bacteriol. 181(6): 1868–1874.
- Winston, F., Dollard, C. and Ricupero‐Hovasse, S. L. (1995). Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11(1): 53–55.
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Mourer, T., d’Enfert, C. and Bachellier-Bassi, S. (2024). Use of the Fluorescent Dye Thioflavin T to Track Amyloid Structures in the Pathogenic Yeast Candida albicans. Bio-protocol 14(3): e4932. DOI: 10.21769/BioProtoc.4932.
Category
Microbiology > Microbial biochemistry > Protein > Structure
Biochemistry > Protein > Fluorescence
Microbiology > Microbial biofilm
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








