- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Development of Recombinase Polymerase Amplification–Lateral Flow Dipstick (RPA–LFD) as a Rapid On-Site Detection Technique for Fusarium oxysporum
Published: Vol 14, Iss 1, Jan 5, 2024 DOI: 10.21769/BioProtoc.4915 Views: 1912
Reviewed by: Shweta PanchalMichael EnosSoumya Moonjely

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fast and Sustainable Thermo-osmotic DNA Extraction Protocol for Trans-spectrum Contingency and Field Use
Stavroula Goudoudaki [...] Yiannis Manoussopoulos
Sep 5, 2023 1636 Views
Novel Workflows for Separate Isolation of Pathogen RNA or DNA From Wastewater: Detection by Innovative and Conventional qPCR
Kristina M. Babler [...] Ayaaz Amirali
Feb 20, 2025 2993 Views

Improved Extraction Methods to Isolate High Molecular Weight DNA From Magnaporthaceae and Other Grass Root Fungi for Long-Read Whole Genome Sequencing
Michelle J. Grey [...] Mark McMullan
Mar 20, 2025 3106 Views
Abstract
Fusarium oxysporum can cause many important plant diseases worldwide, such as crown rot, wilt, and root rot. During the development of strawberry crown rot, this pathogenic fungus spreads from the mother plant to the strawberry seedling through the stolon, with obvious characteristics of latent infection. Therefore, the rapid and timely detection of F. oxysporum can significantly help achieve effective disease management. Here, we present a protocol for the recombinase polymerase amplification– lateral flow dipstick (RPA–LFD) detection technique for the rapid detection of F. oxysporum on strawberry, which only takes half an hour. A significant advantage of our RPA–LFD technique is the elimination of the involvement of professional teams and laboratories, which qualifies it for field detection. We test this protocol directly on plant samples with suspected infection by F. oxysporum in the field and greenhouse. It is worth noting that this protocol can quickly, sensitively, and specifically detect F. oxysporum in soils and plants including strawberry.
Key features
• This protocol is used to detect whether plants such as strawberry are infected with F. oxysporum.
• This protocol has potential for application in portable nucleic acid detection.
• It can complete the detection of samples in the field within 30 min.
Graphical overview
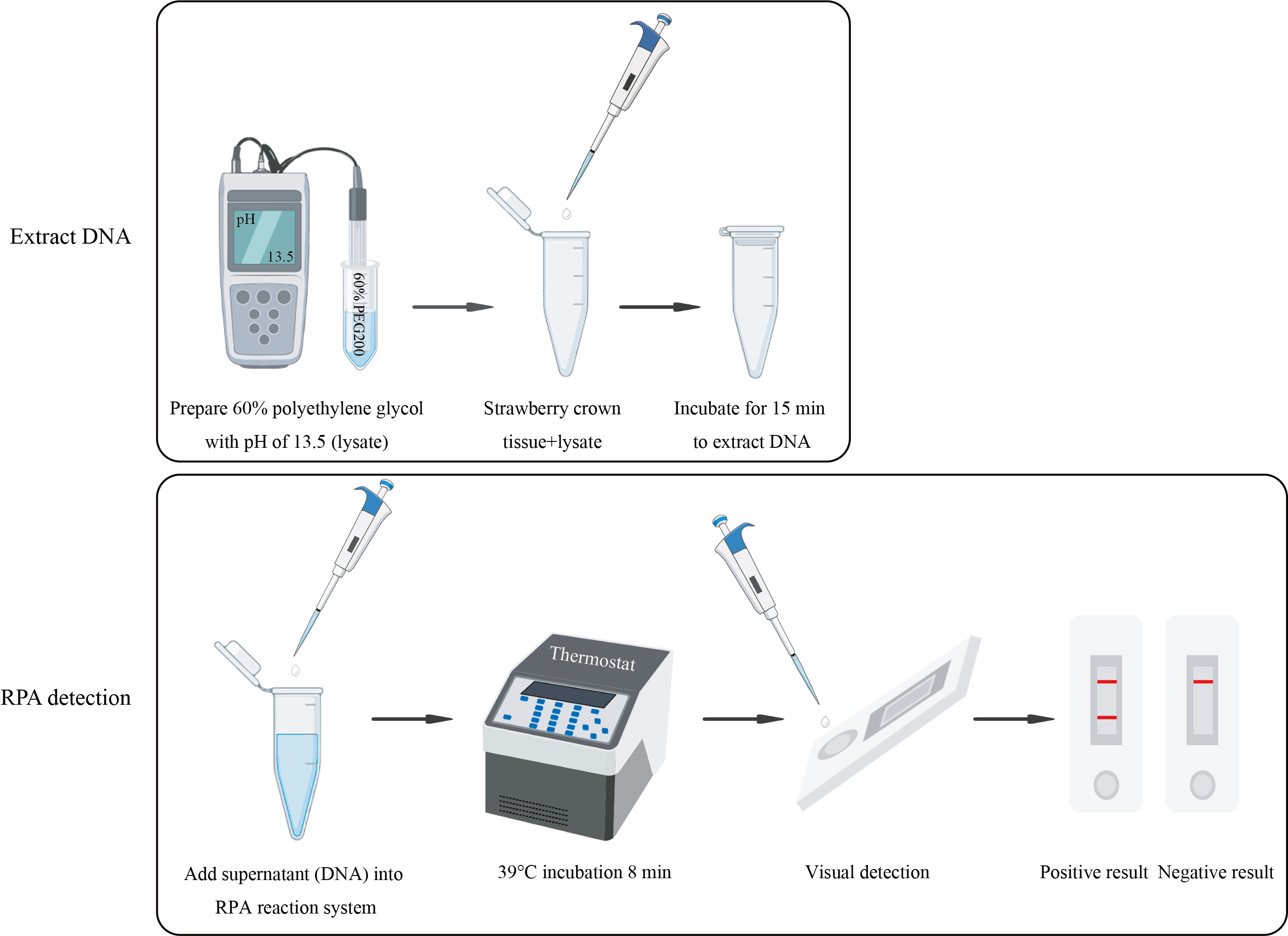
Background
Strawberry crown rot caused by Fusarium oxysporum has seriously affected strawberry yield [1]. The pathogen is transmitted from the mother plant to the strawberry seedling through the stolon [2]. Due to the particularities of pathogen transmission, the control of this disease is difficult. Therefore, rapid and accurate detection of F. oxysporum in the early stages is of utmost importance for preventing and controlling crown rot disease.
Koch’s postulates are the traditional method for pathogen detection through isolation, culture, and pathogenicity testing, being time-consuming and requiring professional training. Molecular detection and identification of the pathogen are highly accurate and sensitive alternative methods for disease diagnosis. Polymerase chain reaction (PCR), quantitative PCR, and other molecular detection techniques have been invented to detect pathogens [3, 4]. However, these techniques require professional experience in performing the tests and expensive equipment, which may not be possible in on-site detection. Altogether, these create a bottleneck for early-disease diagnosis. Fortunately, other molecular biological methods can overcome these limitations, such as the recombinase polymerase amplification (RPA) detection technique [5]. RPA is performed at constant temperature, and its most significant advantage is that the detection results can be visualized by lateral flow dipstick (LFD) [6]. This method is simple to operate and requires little laboratory equipment. Therefore, it has potential for application in portable nucleic acid detection.
Here, we develop and evaluate the RPA–LFD detection technique that can quickly, sensitively, and specifically detect F. oxysporum infection in strawberry [7]. By comparing the genomes of F. oxysporum with other strains common in soil and strawberry plants, we found that the sequence of CYP51C gene was unique and highly variable in Fusarium spp.; therefore, we chose the CYP51C gene to design primers and probes for F. oxysporum detection. We demonstrate how F. oxysporum can be accurately detected on-site. This protocol requires no professional experience, is easy to operate, and the results can be observed within 8 min at 39 °C. This advantage makes it a valuable diagnostic tool, especially in the absence of experienced microscopists. The sensitivity of this protocol is consistent with PCR, and it is accurate enough to detect F. oxysporum. In summary, this protocol is validated on the early field diagnosis of strawberry crown rot. Additionally, it can be applied to detect F. oxysporum in soils and other plants that are damaged by this fungus and can be adjusted to detect other pathogens, only by redesigning specific primers and probes according to the genome sequence of the pathogen to be detected.
Materials and reagents
Biological materials
DNA of F. oxysporum, F. tricinctum, F. proliferatum, F. fujikuroi, F. graminearum, F. solani, F. equiseti, Colletotrichum siamense, C. gloeosporioides, C. aenigma, C. fructicola, Botrytis cinerea, Phoma sp., and Stagonosporopsis sp.
Strawberry plant
Potato
Reagents
KOH (SangonBiotech, catalog number: A610441)
Polyethylene glycol (PEG200) (SangonBiotech, catalog number: A601780-0500)
EDTA disodium salt dihydrate (SangonBiotech, catalog number: A610185-050)
CTAB, cetyltrimethylammonium bromide (SangonBiotech, catalog number: A600108-0500)
Sodium chloride (NaCl) (SangonBiotech, catalog number: A610476-0001)
Tris (SangonBiotech, catalog number: A610195-0500)
Ethanol absolute (SangonBiotech, catalog number: A500737-0500)
Isopropanol (Shanghai Hushi, catalog number: 180109218)
Trichloromethane (Shanghai Hushi, catalog number: 10006818)
Dextrose (SangonBiotech, catalog number: A610219-0500)
Agar (SangonBiotech, catalog number: A505255-025)
Primer and probe:
F91:5′-TCAACTGGCATCGTCAACATCACCGAAGTAA-3′
R91:5′ Biotin-CCAGGCATGACGAAGTTGATAGGTTGAAAGC-3′
Probe:5′ 6-FAM-AGGCTCCCTCCTCGGTAACGAAGTCCGCTCCA/idSp/GTTTGACAGCACATT-3′ C3 Spacer
Solutions
CTAB extracting solution (see Recipes)
70% Ethanol absolute (see Recipes)
60% PEG200 with pH 13.2–13.5 (see Recipes)
2 M KOH (see Recipes)
Potato dextrose agar (PDA) (see Recipes)
Recipes
CTAB extracting solution
*Note: The solute is first mixed and then ddH2O is added to the constant volume of 1,000 mL.
Reagent Final concentration Quantity or Volume CTAB
Tris
EDTA
NaCl
H2O
2%
100 mM
20 mM
1.4 M
n/a
20 g
12.114 g
7.448 mL
81.816 g
to 1,000 mL
Total n/a 1,000 mL 70% ethanol absolute
Reagent Final concentration Quantity or Volume Ethanol absolute
H2O
70% (v/v)
n/a
70 mL
30 mL
Total n/a 100 mL 2 M KOH
*Note: Store at 4 °C.
Reagent Final concentration Quantity or Volume KOH
H2O
2 M
n/a
1.12 g
10 mL
Total n/a 10 mL 60% PEG200 with pH 13.2–13.5
*Note: This solution needs to be prepared and used immediately.
Reagent Final concentration Quantity or Volume KOH (Recipe 3)
PEG200
H2O
n/a
60% (v/v)
n/a
0.33 mL
8 mL
11.67 mL
Total n/a 20 mL Potato dextrose agar (PDA)
*Note: Boil the potato in 1,000 mL of H2O using an induction cooker and pan and filter. Then, add the dextrose and agar to the filtrate.
Reagent Final concentration Quantity or Volume Potato
Dextrose
Agar
H2O
200 g/L
20%
15%
n/a
200 g
20 g
15 g
1,000 mL
Total n/a 1,000 mL
Laboratory supplies
1.5 mL microcentrifuge tubes (Axygen, catalog number: MCT-150-C)
HybriD DNA thermostat rapid amplification kit (APWL, catalog number: WLN8203KIT)
HybriDetect lateral flow dipstick (APWL, catalog number: WLFS8201)
Forceps (Shanghai Leigu, catalog number: W-002903)
Sterile blade (VWR International, catalog number: 55411-050)
Beakers (Shanghai Leigu, catalog number: B-000103)
Stir bars (Shanghai Leigu, catalog number: B-040315)
Graduated cylinders (Shanghai Leigu, catalog number: B-031603)
Glass bottles (Shanghai Leigu, catalog number: B-W00323)
90 mm Petri dish (Nantong Baiyao, catalog number: YJ-90-12g)
Inoculating needle (Shanghai Leigu, catalog number: W-006001)
Forceps (Shanghai Leigu, catalog number: W-002806)
Quartz sand (SangonBiotech, catalog number: A500823-0001)
Equipment
Micropipettes 0.1–2.5 µL, 2–20 µL, 20–200 µL (Eppendorf, model: 3123000217, 3123000233, 3123000250)
Incubator, 25 °C (Shanghai Boxun, model: MJX-250B-Z)
4 °C refrigerator (Haier, model: SC-339JN)
Grinding machine (Shanghai Jingxin, model: JXFSTPRP-24L)
Ultramicro accounting protein analyzer (BioDrop, model: BioDrop µlite+)
Centrifuge (ThermoFisher, model: 75002440)
Thermostat (Hangzhou Aosheng, model: 028-14484-20110001)
Balance (Shanghai Haosheng, model: JA303A)
Induction cooker (Midea, model: RT21E0105)
Pan (Supor, model: ST22P1)
Vortex shaker (SangonBiotech, model: WH-861)
Software and datasets
DNAMAN v6.0.3.99
Snapgene v.5.2.4
Procedure
Design of primers and probes
The sequence of the CYP51C gene was unique and highly variable in Fusarium spp. Therefore, we used DNAMAN v6.0.3.99 to design primers based on the CYP51C sequence of the common Fusarium species in soil and plants, such as F. tricinctum, F. proliferatum, F. fujikuroi, F. graminearum, F. solani, and F. equiseti. The specific sequence of F. oxysporum was found by CYP51C gene sequence to design specific primers (Figure 1).
Recommended primers length was between 31 and 32 bp. Shorter primers can affect amplification speed and detection sensitivity, while longer primers can form secondary structures that affect amplification. The pairs of primers will be as follows:
F91: 5′-TCAACTGGCATCGTCAACATCACCGAAGTAA-3′
R91: 5′-CCAGGCATGACGAAGTTGATAGGTTGAAAGC-3′
Biotin label was applied to the 5′ end of the screened downstream primers R91. Tailed primers will be as follows:
5′ Biotin-CCAGGCATGACGAAGTTGATAGGTTGAAAGC-3′.
The probe was designed with an antigen marker (6-FAM) modification at the 5′ end, a modified group (C3 Spacer) at the 3′ end, and a dSpacer (tetrahydrofuran, THF) labeled at the middle of the 5′ and 3′ ends as the recognition site for NFO. C3 Spacer was blocking groups that prevent the DNA strand from extending, FAM was a fluorophore. Exonuclease NFO (buffer) recognized dSpacer (tetrahydrofuran, THF) and cut it to form a free hydroxyl end extending at the 3′ end. Finally, the amplification product of 5′ FAM-RPA-3′ Biotin was obtained. Gold particle of biotin antibody and FAM antibody fixed on LFD (T line), 5′ FAM-RPA-3′ biotin will be captured to appear bands.
The labeled probe will be as follows:
5′ 6-FAM-AGGCTCCCTCCTCGGTAACGAAGTCCGCTCCA/idSp/GTTTGACAGCACATT-3′ C3 Spacer.
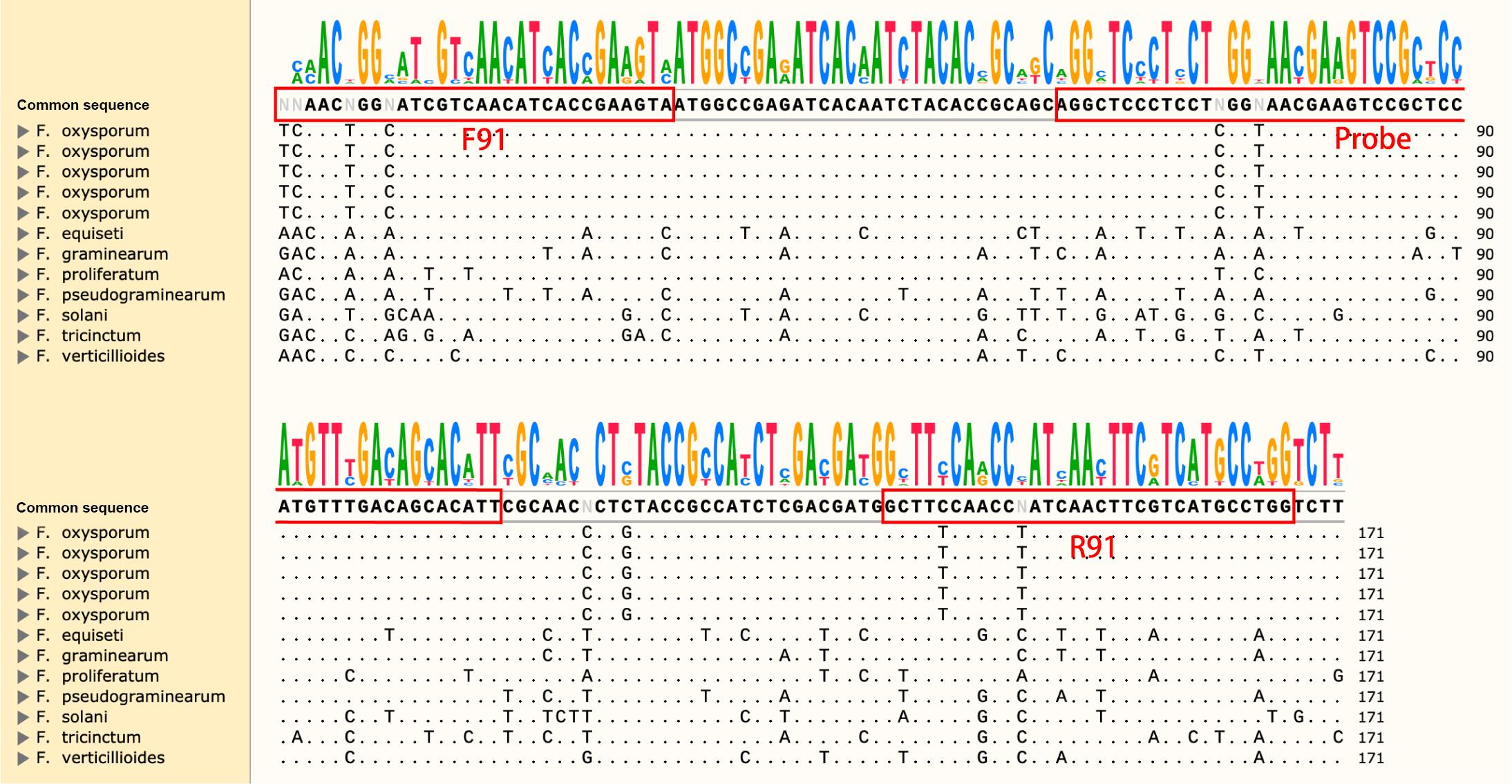
Figure 1. Screenshot of Snapgene with Fusarium sp. genome sequence comparison. Specific primers were designed at locations with large sequence differences. The red boxes indicated the locations of the primers and probes.
DNA extraction
Culture the F. tricinctum, F. proliferatum, F. fujikuroi, F. graminearum, F. solani, F. equiseti, C. siamense, C. gloeosporioides, C. aenigma, C. fructicola, Botrytis cinerea, Phoma sp., and Stagonosporopsis sp. isolates, stored in a 4 refrigerator, in a PDA plate using an inoculating needle. Culture at 28 until colony diameter reaches 6 cm.
Scrape the mycelium into a 1.5 mL tube, add 0.1 g of quartz sand and 300 μL of CTAB extracting solution, and grind for 120 s at 85 Hz in a grinding machine.
Add 400 μL of CTAB extracting solution and shake for 15 s using the vortex.
Add 700 μL of trichloromethane, shake for 15 s using the vortex, and centrifuge at 13,800× g for 10 min.
Remove 500 μL of supernatant and transfer it to a new 1.5 mL tube. Add 500 mL of isopropanol and centrifuge at 13,800× g for 10 min.
Discard the supernatant, add 500 μL of 70% ethanol, and centrifuge at 13,800× g for 5 min.
Discard the supernatant, invert the tube for 2 h to dry, and add 50 μL of ddH2O. Store at 4°C.
Specific detection of primers and probes
Set up the reaction system as follows:
Buffer A 29.4 μL
10 μM upstream F91 2 μL
10 μM downstream R91 2 μL
10 μM probe 0.6 μL
ddH2O 11.5 μL
Template DNA 2 μL
Buffer B 2.5 μL
Buffer B needs to be added last. Buffer A and Buffer B are supplied with the HybriD DNA thermostat rapid amplification kit.
Incubate at 38 °C for 10 min.
Dilute the reaction product 20 times using ddH2O and add 50 μL of the diluted reaction product to the sample hole of HybriDetect LFD. Interpretation of test results is available within 5 min. If the test results are not read within 5 min, the test is not successful.
If the control line is visible and the detection line is not, this means a negative result. If both the detection and control lines are visible, this means a positive result. If neither the detection and control lines are visible, this means that the experimental operation was performed incorrectly or that the kit was damaged (Figure 2).
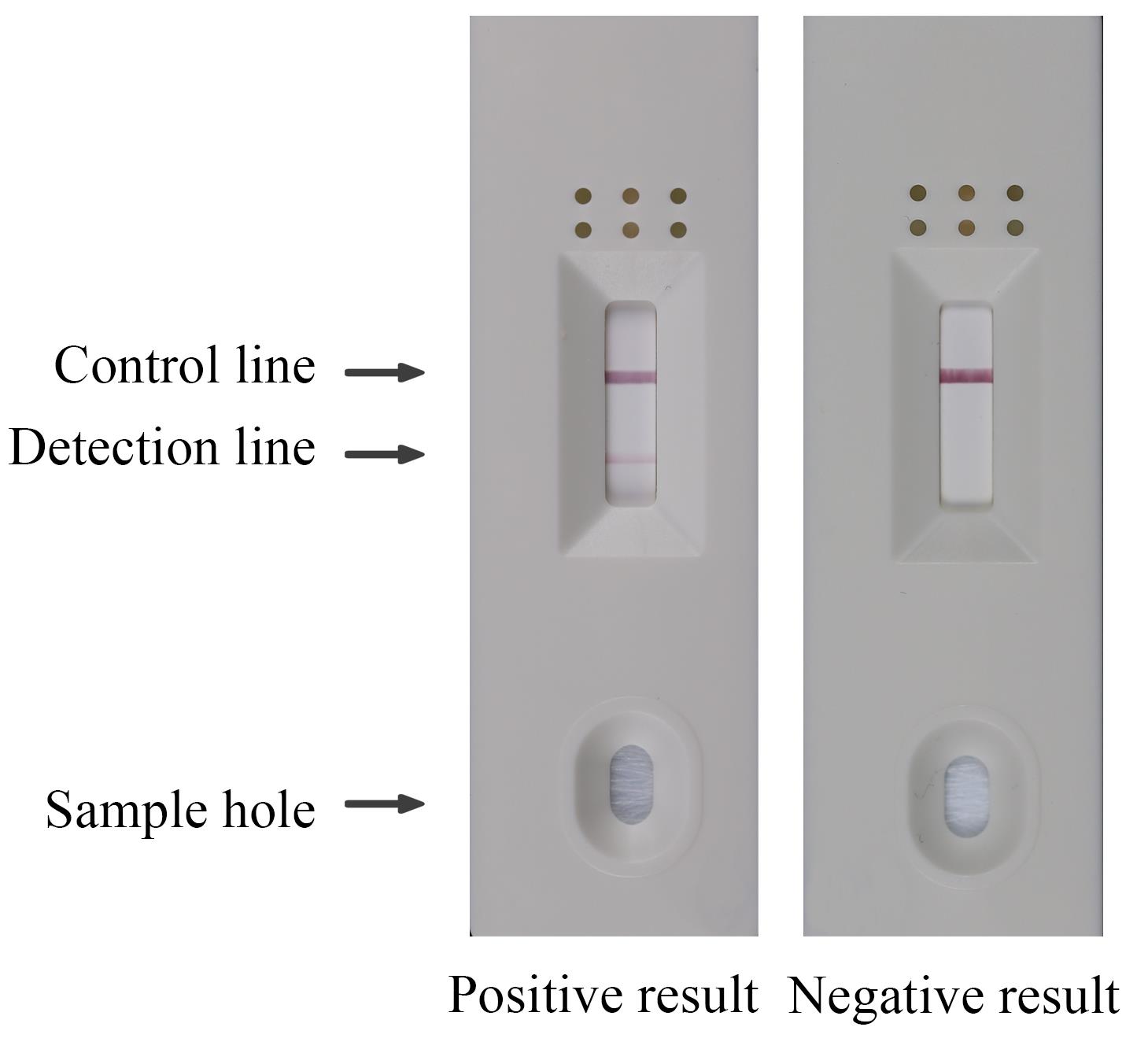
Figure 2. Assessment of specificity of primers pair F91/R91 and probe for recombinase polymerase amplification–lateral flow dipstick (RPA–LFD) detection of F. oxysporumRPA–LFD reaction condition optimization
According to kit instructions, set the reaction temperature to 33, 36, 39, 42, and 45 °C and the reaction time to 10 min.
After determining the optimal reaction temperature, set the reaction time to 4, 6, 8, 10, and 12 min.
Visualize the amplification results according to the above method.
The best reaction condition was determined according to the color depth of the detection line (Figure 3).
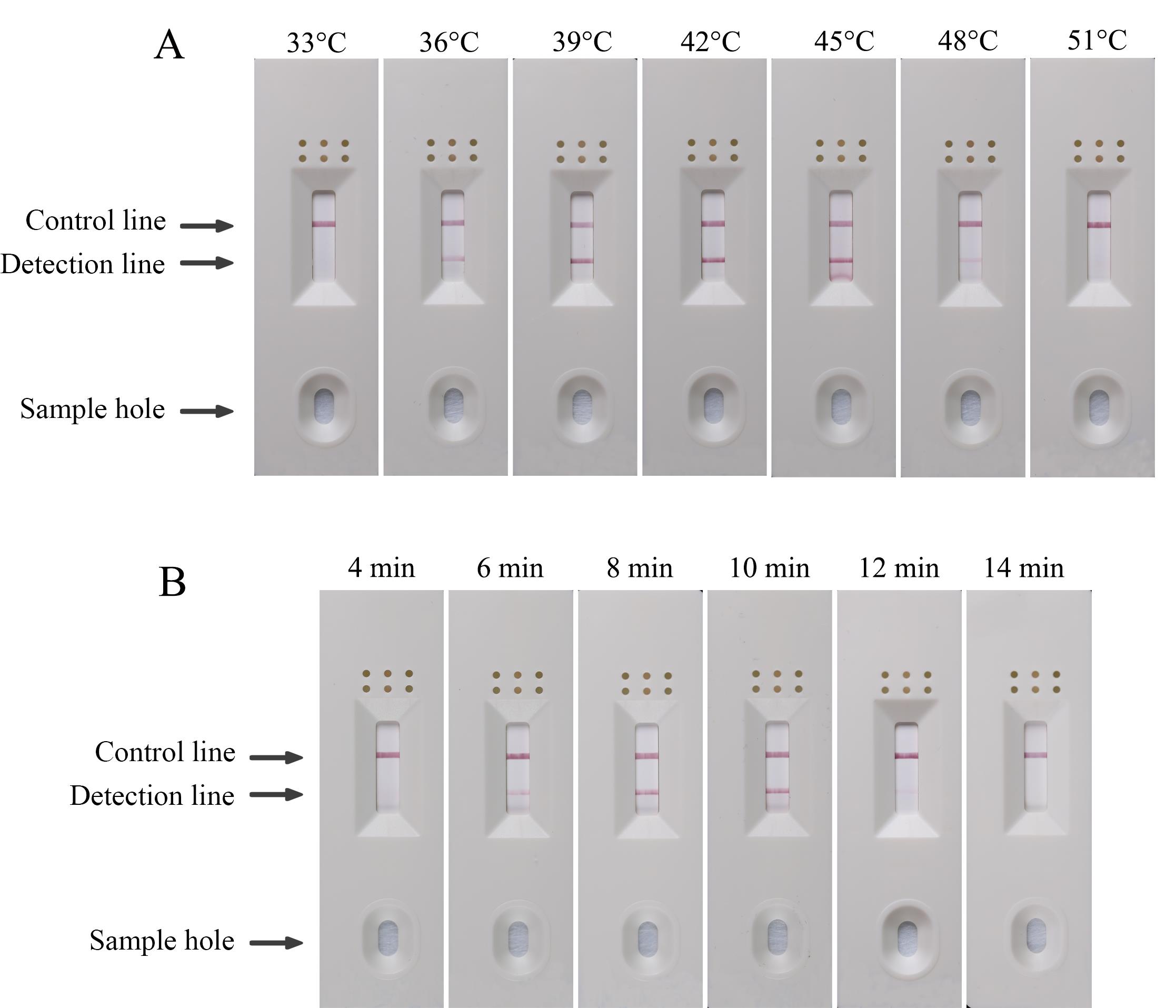
Figure 3. Optimization of recombinase polymerase amplification–lateral flow dipstick (RPA–LFD) detection for F. oxysporum. (A) Optimization of the RPA reaction temperature (33, 36, 39, 42, 45, 48, and 51 °C). (B) Optimization of the RPA reaction time (4, 6, 8, 10, 12, and 14 min).
Detection system sensitivity verification
Set the DNA concentration to 10 ng/μL, 1 ng/mL, 100 pg/μL, 10 pg/μL, 1 pg/μL, and 100 fg/μL. DNA concentration was determined using Ultramicro accounting protein analyzer.
Add different concentrations of DNA templates to the reaction system.
Incubate at 39 °C for 8 min.
Visualize the amplification results according to step C3.
The detection limit was determined by the presence or absence of the detection line.
Field sample detection
Take 100 mg of strawberry crown tissue (approximately 125 mm3 of crown tissue) and place in a 1.5 mL tube.
Add 100 μL of 60% PEG200 .
Incubate at room temperature for 15 min to lyse plant tissue and release DNA.
Add the lysate supernatant as a template into the RPA reaction system. Set up the reaction system as follows:
Buffer A 29.4 μL
Upstream F91 2 μL
Downstream R91 2 μL
Probe 0.6 μL
ddH2O 11.5 μL
Template DNA 2 μL
Buffer B 2.5 μL
Buffer B needs to be added last. Buffer A and Buffer B are supplied with the kit.
Incubate at 39 °C for 8 min.
Visualize the amplification results according to step C3.
If the control line is visible and the detection line is not, this indicates that the strawberry was not infected by F. oxysporum. If both the detection and control lines are visible, this indicates that the strawberry was infected by F. oxysporum (Figure 4).
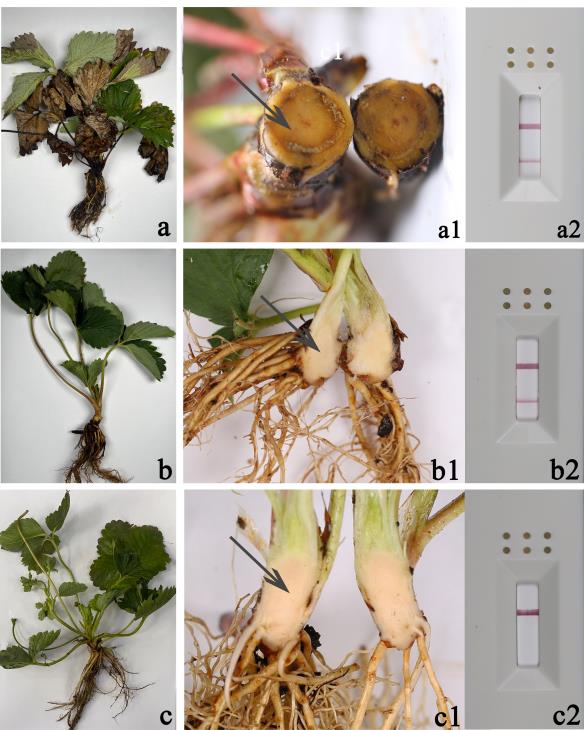
Figure 4. Symptoms of strawberry crown rot and the result of recombinase polymerase amplification–lateral flow dipstick (RPA–LFD) detection. (a–a2) Plant with crown rot symptoms and positive test results. (b–b2) Plant without crown rot symptoms and positive test results. (c–c2) Plant without crown rot symptoms and negative test results. The tissue indicated by the arrow is used for detection.
Data analysis
Determination of primers and probes specificity:
Judge the specificity of primers and probes by the presence or absence of detection lines of HybriDetect LFD. The detection line was visible only for F. oxysporum and the detected result is positive; the detection lines were not visible for other genomic DNA, and the detected result is negative (Table 1). These results indicate that the primers F91/R91 and probe had good specificity for F. oxysporum.
Table 1. Results of other isolates detected by RPA-LFD
Species Detection result F. oxysporum + F. tricinctum - F. proliferatum - F. fujikuroi - F. graminearum - F. solani - F. equiseti - C. siamense - C. gloeosporioides - C. aenigma - C. fructicola - Botrytis cinerea - Phoma sp. - Stagonosporopsis sp. - “+” represents positive result, “-” represents negative result.
Determination of optimal reaction conditions:
According to the manufacturer’s instructions, the optimal reaction temperature was determined with 10 min as the reaction time. The result showed that 39 °C is the optimal reaction temperature, and the optimal reaction time was 8 min.
Determination of detection limits:
The detection limit was determined by the presence or absence of the detection line. The HybriDetect LFD still had bands in the detection line at a template concentration of 1 pg/μL, and we concluded that the detection limit of the RPA-LFD detection was 1 pg/μL.
Field application of RPA-LFD detection:
The appearance of the test line indicated a positive result, suggesting that the plant was infected with F. oxysporum.
Validation of protocol
Hu et al. (2023) [7]. Establishment of the recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) detection technique for Fusarium oxysporum. Plant Disease. (Figure 4).
General notes and troubleshooting
General notes
The length of primers for RPA detection was 31–32 bp; too-short primers will affect the amplification speed and detection sensitivity.
Before the positive test, the negative test was performed to rule out the formation of false positives due to the primers probe.
Care should be taken to avoid cross contamination during the experimental operation.
This operation was effective only when the HybriDetect LFD control line is striped.
HybriDetect LFD interpretation results were completed within 5 min.
Troubleshooting
Problem 1: The combination of primers and probe could not specifically detect the target fungus.
Possible cause(s): Poor specificity of primers and probe.
Solution(s): The specific primers need to be screened again, and the probes need to be screened after the specific primers are obtained.
Problem 2: The test line of negative control showed banding.
Possible cause(s): The primers bound to the probe itself.
Solution(s): Re-screen primers and probe.
Problem 3: The color of the detection line was weak.
Possible cause(s): The reaction conditions were not optimal, or the DNA concentration is low.
Solution(s): Optimize reaction conditions and increase DNA concentration.
Acknowledgments
This research was funded by Agriculture and Social Development Research Project of Hangzhou (202203A07), Guizhou Research academy [110202101048(LS-08)], and Key Research and Development Project of Zhejiang Province, China (No. 2020C02005).
The protocols described here are adapted from our previous work [7].
Competing interests
The authors declare no competing interests.
References
- Hassan, O. and Chang, T. (2021). Morphological and molecular characteristics of fungal species associated with crown rot of strawberry in South Korea. Mol. Biol. Rep. 49(1): 51–62. doi: 10.1007/s11033-021-06841-9
- Freeman, S. and Katan, T. (1997). Identification of Colletotrichum Species Responsible for Anthracnose and Root Necrosis of Strawberry in Israel. Phytopathology 87(5): 516–521. doi: 10.1094/phyto.1997.87.5.516
- Lévesque, C. A. (2001). Molecular methods for detection of plant pathogens—What is the future? Can. J. Plant. Pathol. 23(4): 333–336. doi: 10.1080/07060660109506953
- Cao, X., Yao, D., Zhou, Y., West, J. S., Xu, X., Luo, Y., Ding, K., Fan, J. and Duan, X. (2016). Detection and quantification of airborne inoculum of Blumeria graminis f. sp. tritici using quantitative PCR. Eur. J. Plant Pathol. 146(1): 225–229. doi: 10.1007/s10658-016-0908-8
- Phoulivong, S., Cai, L., Chen, H., McKenzie, E. H. C., Abdelsalam, K., Chukeatirote, E. and Hyde, K. D. (2010). Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity 44(1): 33–43. doi: 10.1007/s13225-010-0046-0
- Ghosh, D. K., Kokane, S. B. and Gowda, S. (2020). Development of a reverse transcription recombinase polymerase based isothermal amplification coupled with lateral flow immunochromatographic assay (CTV-RT-RPA-LFICA) for rapid detection of Citrus tristeza virus. Sci. Rep. 10(1): e1038/s41598–020–77692–w. doi: 10.1038/s41598-020-77692-w
- Hu, S., Yan, C., Yu, H., Zhang, Y. and Zhang, C. Q. (2023). Establishment of the Recombinase Polymerase Amplification–Lateral Flow Dipstick Detection Technique for Fusarium oxysporum. Plant Disease 107(9): 2665–2672. doi: 10.1094/pdis-12-22-2841-re
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Hu, S., Yu, H. and Zhang, C. (2024). Development of Recombinase Polymerase Amplification–Lateral Flow Dipstick (RPA–LFD) as a Rapid On-Site Detection Technique for Fusarium oxysporum. Bio-protocol 14(1): e4915. DOI: 10.21769/BioProtoc.4915.
Category
Microbiology > Pathogen detection
Molecular Biology > DNA > DNA extraction
Biological Sciences > Microbiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









