- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Purification of Recombinant Human Mitochondrial RNA Polymerase (POLRMT) and the Initiation Factors TFAM and TFB2M
Published: Vol 13, Iss 23, Dec 5, 2023 DOI: 10.21769/BioProtoc.4892 Views: 2497
Reviewed by: Abhilash PadavannilJeremy BirdHassan RasouliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2159 Views
Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2361 Views
An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2428 Views
Abstract
Human mitochondrial DNA (mtDNA) encodes several components of oxidative phosphorylation responsible for the bulk of cellular energy production. The mtDNA is transcribed by a dedicated human mitochondrial RNA polymerase (POLRMT) that is structurally distinct from its nuclear counterparts, instead closely resembling the single-subunit viral RNA polymerases (e.g., T7 RNA polymerase). The initiation of transcription by POLRMT is aided by two initiation factors: transcription factor A, mitochondrial (TFAM), and transcription factor B2, mitochondrial (TFB2M). Although many details of human mitochondrial transcription initiation have been elucidated with in vitro biochemical and structural studies, much remains to be addressed relating to the mechanism and regulation of transcription. Studies of such mechanisms require reliable, high-yield, and high-purity methods for protein production, and this protocol provides the level of detail and troubleshooting tips that are necessary for a novice to generate meaningful amounts of proteins for experimental work. The current protocol describes how to purify recombinant POLRMT, TFAM, and TFB2M from Escherichia coli using techniques such as affinity column chromatography (Ni2+ and heparin), how to remove the solubility tags with TEV protease and recover untagged proteins of interest, and how to overcome commonly encountered challenges in obtaining high yield of each protein.
Key features
• This protocol builds upon purification methods developed by Patel lab (Ramachandran et al., 2017) and others with greater detail than previously published works.
• The protocol requires several days to complete as various steps are designed to be performed overnight.
• The recombinantly purified proteins have been successfully used for in vitro transcription experiments, allowing for finer control of experimental components in a minimalistic system.
Keywords: POLRMTBackground
The mitochondrial RNA polymerase (POLRMT) is a DNA-dependent RNA polymerase (RNAP) that transcribes the human mitochondrial DNA (mtDNA), which encodes several protein components of the complexes that carry out oxidative phosphorylation. POLRMT is structurally similar to other single-subunit RNAPs such as T7 phage RNAP (Schwinghammer et al., 2013; Sultana et al., 2017), and current models suggest that POLRMT transcribes short RNA primers used in mtDNA replication initiation (Tan et al., 2022). Considering the central role of POLRMT in cellular metabolism and maintenance of mtDNA, it is not surprising that mutations associated with mitochondrial dysfunction map to POLRMT, such as respiratory chain defects, hypotonia, and global developmental delays (Oláhová et al., 2021).
To start transcription de novo from a promoter sequence, POLRMT requires two initiation factors: transcription factor A (TFAM) and transcription factor B2 (TFB2M), which aid the melting of the promoter sequence and the formation of a stable initiation complex (Hillen et al., 2017; Ramachandran et al., 2017). TFAM is also the mtDNA packaging protein that compacts the mtDNA into structures called nucleoids. While too much TFAM can prevent proteins involved in transcription and other processes from accessing the mtDNA, TFAM is required to bend the upstream promoter region to recruit POLRMT to the promoter site; the underlying mechanisms of this process are yet to be worked out. Published work suggests that post-translational modifications (PTMs) to TFAM, such as acetylation or phosphorylation, do not compromise its function as a transcription factor but make it easier for POLRMT to remove TFAM from DNA, thus increasing transcription processivity through TFAM-compacted DNA (Reardon and Mishanina, 2022).
This protocol describes in detail how to isolate recombinant POLRMT, TFAM, and TFB2M from Escherichia coli in high purity and how to overcome the challenges in achieving high protein yield. These protocols build upon the procedures developed in the Patel lab and others (Yakubovskaya et al., 2014; Ramachandran et al., 2017) and describe the steps in greater detail than in any previously published work to provide access to these resources to a wider community. The resulting proteins can be used to reconstitute human mitochondrial transcription on promoter sequences in vitro, as in prior biochemical and structural studies (Bird et al., 2018; Tan et al., 2022). These proteins can be post-translationally modified to probe the impact of PTMs on their function to fine-tune mitochondrial transcription output (Reardon and Mishanina, 2022). Finally, the protocols can also be applied to express and purify mutant proteins to study the underlying basis of mutations associated with mitochondrial dysfunction.
Materials and reagents
4× Laemmli sample buffer (Bio-Rad, catalog number: 1610747)
10× Tris/Glycine/SDS (Bio-Rad, catalog number: 1610732)
Dithiothreitol (DTT) (Fisher BioReagents, catalog number: BP172-5), see Recipes for preparation
Ethanol (EtOH) (Fisher Scientific, catalog number: 2818500)
Glycerol (Fisher Scientific, catalog number: BP229-1)
HiTrap heparin column (GE Healthcare, catalog number: 17-0407-03)
Hydrochloric acid (HCl) (Fisher Scientific, catalog number: LC149502)
Imidazole (molecular biology grade, > 99%) (Fisher Scientific, catalog number: O3196-500)
Isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Fisher Scientific, catalog number: BP1755100), see Recipes for preparation
LB agar, Miller (Fisher Scientific, catalog number: BP1425-2)
LB broth (LB), Miller, granulated (Fisher Scientific, catalog number: BP9723-2)
Lysozyme (Fisher Scientific, catalog number: BP535-5)
Precision Plus ProteinTM All Blue Prestained Protein Standards (Bio-Rad, catalog number: 1610373)
Protease inhibitor cocktail (PIC) powder (used in this protocol as 1,000× stock and prepared following instructions online: https://www.sigmaaldrich.com/US/en/product/sigma/p2714) (Sigma-Aldrich, catalog number: P2714-1BTL)
Sodium hydroxide (NaOH) pellets (Fisher Scientific, catalog number: S318-500)
β-mercaptoethanol (BME) (Fisher Scientific, catalog number: AC125472500)
Part I: POLRMT purification materials and reagents
Amicon Ultra-15 centrifugal filter units (50K MWCO, 15 mL) (Millipore Sigma, catalog number: UFC905024)
Ammonium sulfate (AS) (Fisher Scientific, catalog number: A702-3)
Ampicillin (Fisher Scientific, catalog number: BP1760-25), see Recipes for preparation
Cytiva CaptoTM DEAE chromatography media (Fisher Scientific, catalog number: 45-002-960) or, if using an FPLC, HiTrap DEAE Fast Flow (Cytiva, catalog number: 17-5154-01)
HisTrap FPLC column (Cytiva, catalog number: 17-3712-06)
Polyethyleneimine (PEI), MN 60,000 50% (Fisher Scientific, catalog number: AC178571000), see Recipes for preparation
Poly-Prep® chromatography columns (Bio-Rad, catalog number: 7311550)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: BP358-212, CAS number: 7647-14-5)
Tris base (Fisher BioReagents, catalog number BP152-1)
Tween 20 (Fisher BioReagents, catalog number: BP337-100)
Part II: TFAM purification materials and reagents
1 kb DNA ladder (NEB, catalog number: N3232S)
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Fisher Scientific, catalog number: BP310-1)
Amicon Ultra-15 centrifugal filter units (3K MWCO, 15 mL) (Millipore Sigma, catalog number: UFC900324)
Ethidium bromide solution, 10 mg/mL (Bio-Rad, catalog number: 1610433)
HisPur Ni-NTA resin (Thermo Scientific, catalog number: 88222)
Kanamycin (Fisher Scientific, catalog number: 611290050), see Recipes for preparation
Potassium chloride (KCl) (molecular biology grade) (Fisher Scientific, catalog number: P217-3)
Sodium chloride (Fisher Scientific, catalog number: BP358-212, CAS number: 7647-14-5)
Part III: TFB2M purification materials and reagents
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Fisher Scientific, catalog number: BP310-1)
Amicon Ultra-4 centrifugal filter units (10K MWCO, 4 mL) (Millipore Sigma, catalog number: UFC801008)
HisPur Ni-NTA resin (Thermo Scientific, catalog number: 88222)
Kanamycin (Fisher Scientific, catalog number: 611290050), see Recipes for preparation
Potassium chloride (KCl) (Fisher Scientific, catalog number: P217-3)
Potassium hydroxide (KOH) (Fisher Scientific, catalog number: P250 500)
Biological materials
Plasmids available upon request
Escherichia coli (E. coli) BL21-CodonPlus (DE3)-RIPL competent cells (Agilent, catalog number: 230280) or
E. coli BL21-CodonPlus (DE3)-RIL competent cells (Agilent, catalog number: 230245)
Optional: E. coli ArcticExpress (DE3) competent cells (Agilent, catalog number: 230192)
Tobacco Etch Virus (TEV) protease (Kapust et al., 2001; Raran-Kurussi et al., 2017)
POLRMT construct in pPROEXHTb. It contains residues 43–1230 [lacking its N-terminal mitochondrial targeting sequence (MTS)] with an N-terminal hexahistidine 6x-His-tag
TFAM construct in pPROEXHTb. It contains residues 43–246 (lacking its N-terminal MTS) with an N-terminal 6x-His-tag and a TEV protease site
TFB2M construct in pT7TEV-HMBP4. It contains residues 20–398 with an N-terminal 6x-His-tag, maltose binding protein (MBP) tag, and a TEV protease site
Solutions
See Recipes for all solutions.
Part I: POLRMT purification
5% Polyethylenimine (PEI) solution pH 7.0
POLRMT Lysis Buffer
POLRMT HisTrap Buffer A
POLRMT HisTrap Buffer B
POLRMT Heparin No Salt Buffer
POLRMT Heparin Buffer A
POLRMT Heparin Buffer B
Part II: TFAM purification and activity test
TFAM Lysis Buffer
TFAM Ni-NTA Buffer A
TFAM Ni-NTA Buffer B
TFAM TEV Cleavage Buffer
TFAM Heparin Buffer A
TFAM Heparin Buffer B
TFAM Storage Buffer (2×)
10× TFAM Binding Buffer
TFAM Electrophoretic Mobility Shift Assay (EMSA) Buffer
Part III: TFB2M Purification
TFB2M Lysis Buffer
TFB2M Ni-NTA Buffer A
TFB2M Ni-NTA Buffer B
TFB2M TEV Cleavage Buffer
TFB2M No Salt Heparin Buffer
TFB2M Heparin Buffer A
TFB2M Heparin Buffer B
Recipes
Common stock solutions
Note: Sterilize all stocks by filtering through a 0.22 μm syringe filter, aliquot into 1 mL portions, and store at -20 °C.
1 M IPTG Stock
2.38 g of IPTG
Dissolve in Milli-Q water and bring the final volume up to 10 mL.
*Add 200 μL of 1 M IPTG per 1 L of cell culture for a final concentration of 0.2 mM IPTG.
Amp stock (100 mg/mL)
1 g of ampicillin
Dissolve in Milli-Q water and bring the final volume up to 10 mL.
Kan stock (25 mg/mL)
0.25 g of kanamycin
Dissolve in Milli-Q water and bring the final volume up to 10 mL.
1 M DTT Stock
1.54 g of DTT
Dissolve in Milli-Q water and bring the final volume up to 10 mL.
Part I: POLRMT Purification
Note: All buffers (except for 5% PEI) are adjusted to pH 7.0 with HCl or NaOH, sterilized with a 0.22 μm filter, and stored at 4 °C. Add BME to the Lysis and HisTrap purification buffers and DTT to the HiTrap heparin purification buffers right before use. Prepare cold, filtered Milli-Q H2O and 20% EtOH to wash and store the FPLC columns after use.
5% PEI solution
Reagent (stock concentration) Final concentration Quantity PEI (50%) 5% 10 mL Milli-Q H2O n/a 90 mL Total 100 mL Adjust pH to 7.0 with HCl when the solution is cold or on ice. No need to filter.
POLRMT Lysis Buffer
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.9) 40 mM 10 mL NaCl (3 M) 400 mM 33.3 mL Glycerol (100%) 15% v/v 37.5 mL Tween 20 (100%) 0.1% v/v 0.25 mL EDTA (500 mM) 1 mM 0.5 mL BME* (14.3 M) 5 mM See Note Lysozyme* 1 mg/mL See Note PIC (1,000×)* 1× See Note Milli-Q H2O n/a 168 mL Total 250 mL *Add right before use. Pour 50 mL of the buffer into a conical tube and then add these components (5 mg of lysozyme, 17.5 μL of BME, and 50 μL of PIC) to the tube instead of the entire bottle of buffer. PIC was prepared following instructions online and is used as 1,000× stock: https://www.sigmaaldrich.com/US/en/product/sigma/p2714.
POLRMT HisTrap Buffer A
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.9) 40 mM 40 mL NaCl (3 M) 300 mM 100 mL Glycerol (100%) 15% v/v 150 mL Tween 20 (100%) 0.1% v/v 1 mL Imidazole 20 mM 1.36 g BME* (14.3 M) 5 mM 350 μL Milli-Q H2O n/a 707 mL Total 1,000 mL *Add BME right before use.
POLRMT HisTrap Buffer B
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.9) 40 mM 20 mL NaCl (3 M) 300 mM 50 mL Glycerol (100%) 15% v/v 75 mL Tween 20 (100%) 0.1% v/v 0.5 mL Imidazole 500 mM 17.02 g BME* (14.3 M) 5 mM 175 μL Milli-Q H2O n/a 353 mL Total 500 mL *Add BME right before use.
POLRMT Heparin No Salt Buffer
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.9) 40 mM 4 mL NaCl (3 M) 0 mM n/a Glycerol (100%) 15% v/v 15 mL Tween 20 (100%) 0.1% v/v 0.1 mL EDTA (500 mM) 1 mM 0.2 mL DTT* (1 M) 1 mM 100 μL Milli-Q H2O n/a 81.6 mL Total 100 mL This buffer is used to adjust the salt concentration of the elution from the HisTrap column. *Add DTT right before use.
POLRMT Heparin Buffer A
Reagent (Stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.9) 40 mM 40 mL NaCl (3 M) 150 mM 50 mL Glycerol (100%) 15% v/v 150 mL Tween 20 (100%) 0.1% v/v 1 mL EDTA (500 mM) 1 mM 2 mL DTT* (1 M) 1 mM 1,000 μL Milli-Q H2O n/a 756 mL Total 1,000 mL *Add DTT right before use.
POLRMT Heparin Buffer B
Reagent (Stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.9) 40 mM 20 mL NaCl (3 M) 1,000 mM 167 mL Glycerol (100%) 15% v/v 75 mL Tween 20 (100%) 0.1% v/v 0.5 mL EDTA (500 mM) 1 mM 1 mL DTT* (1 M) 1 mM 500 μL Milli-Q H2O n/a 236 mL Total 500 mL *Add DTT right before use.
Part II: TFAM purification
Note: All buffers are adjusted to pH 7.5 with HCl or NaOH, sterilized with a 0.22 μm filter, and stored at 4 °C. Prepare cold, filtered Milli-Q H2O and 20% EtOH to wash and store the FPLC columns after use.
TFAM Lysis Buffer
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 20 mM 4 mL NaCl (3 M) 200 mM 13.33 mL Glycerol (100%) 15% v/v 30 mL Imidazole 10 mM 0.136 g BME* (14.3 M) 5 mM See Note Lysozyme* 1 mg/mL See Note Protease inhibitor cocktail* (1,000×) 1× See Note Milli-Q H2O n/a 152.47 mL Total 200 mL *Add right before use. We only need 50 mL of the buffer in a conical tube; then, add these components (5 mg of lysozyme, 17.5 μL of BME, and 50 μL of PIC) to the tube instead of the entire bottle of buffer.
TFAM Ni-NTA Buffer A*
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 20 mM 20 mL NaCl (3 M) 200 mM 66.65 mL Glycerol (100%) 15% v/v 150 mL Imidazole 10 mM 0.681 g BME* (14.3 M) 5 mM 350 μL Milli-Q H2O n/a 762.35 mL Total 1,000 mL *This buffer is the same as the TFAM Lysis Buffer without lysozyme and PIC. Add BME right before use.
TFAM Ni-NTA Buffer B
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 20 mM 20 mL NaCl (3 M) 200 mM 66.65 mL Glycerol (100%) 15% v/v 150 mL Imidazole 500 mM 34.04 g BME* (14.3 M) 5 mM 350 μL Milli-Q H2O n/a 762.35 mL Total 1,000 mL *Add BME right before use.
TFAM TEV Cleavage Buffer
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 20 mM 40 mL NaCl (3 M) 300 mM 200 mL Glycerol (100%) 15% v/v 300 mL BME* (14.3 M) 5 mM 700 μL Milli-Q H2O n/a 1,459 mL Total 2,000 mL *Add BME right before use.
TFAM Heparin Buffer A
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 20 mM 20 mL NaCl (3 M) 200 mM 66.67 mL Glycerol (100%) 15% v/v 150 mL DTT* (1 M) 1 mM 1,000 μL Milli-Q H2O n/a 762.33 mL Total 1,000 mL *Add DTT right before use.
TFAM Heparin Buffer B
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 30 mM 30 mL NaCl (3 M) 1500 mM 500 mL Glycerol (100%) 15% v/v 150 mL DTT* (1 M) 1 mM 1,000 μL Milli-Q H2O n/a 319 mL Total 1,000 mL *Add DTT right before use.
TFAM Storage Buffer (2×)
Reagent (stock concentration) Final concentration Quantity Tris-HCl (1 M, pH 7.5) 80 mM 16 mL NaCl (3 M) 200 mM 13.33 mL DTT* (1 M) 2 mM 400 μL Milli-Q H2O n/a 170.27 mL Total 200 mL *Add DTT right before use.
10× TFAM Binding Buffer
500 mM Tris-HCl pH 8.0
1 M KCl
50 mM MgCl2
1 mM DTT (add right before use)
TFAM EMSA Running Buffer
50 mM Tris-acetate pH 8.0
2.5 mM EDTA
Part III: TFB2M purification
Note: All buffers are adjusted to pH 8.0 with HCl or KOH, sterilized with a 0.22 μm filter, and stored at 4 °C. Use BME for the Lysis and HisTrap buffers and DTT for the HiTrap Heparin column and Storage buffers. Prepare cold, filtered Milli-Q H2O and 20% EtOH to wash and store the FPLC columns after use.
TFB2M Lysis Buffer
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8.0) 20 mM 6 mL KCl (3 M) 1 M 100 mL Glycerol (100%) 5% v/v 15 mL Imidazole 10 mM 0.204 g BME* (14.3 M) 5 mM See Note Lysozyme* 1 mg/mL See Note Protease inhibitor cocktail* (PIC, 1,000×) 1× See Note Milli-Q H2O n/a 179 mL Total 300 mL *Add right before use. We only need 50 mL of the buffer in a conical tube; then, add these components (5 mg of lysozyme, 17.5 μL of BME, and 50 μL of PIC) to the tube instead of the entire bottle of buffer.
TFB2M Ni-NTA Buffer A
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8.0) 20 mM 20 mL KCl (3 M) 300 mM 100 mL Glycerol (100%) 5% v/v 50 mL Imidazole 20 mM 1.36 g BME* (14.3 M) 5 mM 350 μL Milli-Q H2O n/a 830 mL Total 1,000 mL *Add BME right before use.
TFB2M Ni-NTA Buffer B
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8.0) 20 mM 10 mL KCl (3 M) 300 mM 50 mL Glycerol (100%) 5% v/v 25 mL Imidazole 500 mM 17.02 g BME* (14.3 M) 5 mM 175 μL Milli-Q H2O n/a 415 mL Total 500 mL *Add BME right before use.
TFB2M TEV Cleavage Buffer
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8.0) 20 mM 40 mL KCl (3 M) 300 mM 200 mL Glycerol (100%) 5% v/v 100 mL BME (14.3 M) * 5 mM 699.3 μL Milli-Q H2O n/a 1,659.3 mL Total 2,000 mL *Add BME right before use.
TFB2M No Salt Heparin Buffer
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8.0) 20 mM 4 mL Glycerol (100%) 5% v/v 10 mL EDTA (500 mM) 1 mM 0.4 mL DTT (1 M) * 1 mM 0.2 mL Milli-Q H2O n/a 185.4 mL Total 200 mL *Add DTT right before use.
TFB2M Heparin Buffer A
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8) 20 mM 20 mL KCl (3 M) 200 mM 66.67 mL Glycerol (100%) 5% v/v 50 mL EDTA (500 mM) 1 mM 2 mL DTT (1 M) * 1 mM 1 mL Milli-Q H2O n/a 860.33 mL Total 1,000 mL *Add DTT right before use.
TFB2M Heparin Buffer B
Reagent (stock concentration) Final concentration Quantity HEPES-KOH (1 M, pH 8.0) 20 mM 10 mL KCl (3 M) 1.5 M 250 mL Glycerol (100%) 5% v/v 25 mL EDTA (500 mM) 1 mM 1 mL DTT (1 M) * 1 mM 0.5 mL Milli-Q H2O n/a 213.5 mL Total 500 mL *Add DTT right before use.
SDS-PAGE sample recipes
Note: These recipes make 20 μL of each SDS-PAGE sample. Boil samples for 5 min at 95 °C before running and load 9–12 μL per lane. All POLRMT samples can be run on 8%–10% SDS-PAGE. For TFAM and TFB2M, 10%–12% SDS-PAGE would be suitable. We run the gels in 1× Tris/Glycine/SDS buffer and stain with Coomassie.
Uninduced, induced, pellet, lysate, PEI pellet, PEI supernatant, AS pellet, Ni-NTA or HisTrap flowthrough, and wash samples
1.5 μL of sample or scrape a small amount of pellet directly into the microfuge tube
13.5 μL of Milli-Q water
5 μL of 4× Laemmli buffer
Ni-NTA Elution samples, AS supernatant
7.5 μL of sample or small piece of pellet
7.5 μL of Milli-Q water
5 μL of 4× Laemmli buffer
TEV-cleaved sample, HisTrap, and Heparin column fractions
Note: Load as much as possible (at least 12 μL per lane) to visualize very diluted samples
10 μL of sample or small piece of pellet
5 μL of Milli-Q water
5 μL of 4× Laemmli buffer
Laboratory supplies
17 mm × 100 mm culture tubes, 14 mL (VWR, catalog number: 60818-689)
BD disposable syringe, 50 mL (Fisher Scientific, catalog number: 13-689-8)
Biotix 10 μL tips (Fisher Scientific, catalog number: 12-111-021)
Fernsbach culture flasks, 2.8 L (Fisher Scientific, catalog number: 09-552-39)
Membrane filters, 0.22 μm pore size, 47 mm diameter (Fisher Scientific, catalog number: 09-719-2B)
Petri dishes (Fisher Scientific, catalog number: FB0875713)
Pipette tips, microfuge tubes, and low-retention tubes (Genesee Scientific, catalog numbers: 24-282, 23-150RL, 23-165RL, 28-103, 22-281LR)
Serological pipettes, 5, 10, and 25 mL (VWR, catalog numbers: 75816-094, 75816-100, 75816-090)
Syringe filters (0.22 μm pore size, 25 mm diameter) (VWR, catalog number: 76479-024)
Equipment
1 L centrifuge bottles with sealing closure (Thermo Scientific, catalog number: 31411006)
4 L capacity centrifuge (Thermo Scientific, Sorvall LYNX 4000 Superspeed Centrifuge, catalog number: 75006580)
Benchtop centrifuges (Eppendorf, 5910 R, 5425)
Benchtop incubator shaker (Eppendorf, model: New Brunswick Benchtop Incubator Shaker I24)
BioSpectrometer Basic (Eppendorf)
Dialysis tubing, 14,000 MWCO (Sigma-Aldrich, catalog number: D9777-100FT)
Fast protein liquid chromatography (FPLC) system (Bio-Rad, NGC Quest 10 Chromatography System, catalog number: 7880001)
Gel imager (Bio-Rad, model: Gel Doc EZ Imager)
Glass column for Ni-NTA resin (Bio-Rad, Econo-Column Chromatography Column, catalog number: 7371512)
Hot plate stirrer (VWR, catalog number: 97042-674)
Magnetic stir bar (Fisher Scientific, catalog number: 14-513-51)
Reusable bottle top filter unit (Thermo Scientific, Nalgene, catalog number: DS0320-2533)
SDS-PAGE running system (Bio-Rad, Mini-PROTEAN Tetra System)
Sonicator (Qsonica, CL-18 Sonicator Probe)
Software and datasets
Bio-Rad Image Lab (https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z)
ImageJ version 1.53e (https://imagej.net/ij/index.html)
Procedure
Part I: POLRMT Purification
Overexpression of recombinant POLRMT in E. coli
Transform the plasmid into BL21-CodonPlus (DE3)-RIPL and plate on LB agar plates containing ampicillin (or the appropriate antibiotic). Incubate inverted overnight at 37 °C.
Note: This transformation can also be done with BL21-CodonPlus-RIL. This step can be skipped if glycerol stocks of cells carrying the POLRMT-coding plasmid have been made previously.
Get an autoclaved flask with 50 mL of autoclaved LB media and add 50 μL of Amp stock (100 mg/mL, see Recipes).
Pick five separate colonies from the plate with a sterile pipette tip or stick for inoculation (or add a small piece of glycerol stock).
Once inoculated, start incubating the cultures in the afternoon around 5 pm at 28 °C and shake overnight (14–17 h) at 200 rpm (see General note 2 for temperature and oversaturation of cells).
In four flasks (2.8 L Fernsbach culture flasks), prepare 1 L each of autoclaved LB media and add 1 mL of Amp stock per liter of media.
Add 10 mL of inoculum per liter of LB media with ampicillin (see General note 3 for how to make glycerol stocks).
Incubate the inoculated flasks at 37 °C with shaking at 200 rpm. Check cell density by measuring the OD600 after 3–4 h to see if it is between 0.4 and 0.6.
Once in the above OD range, lower the shaker temperature to 18 °C. Withdraw a 1 mL sample (label as “Uninduced”) into a microfuge tube and tape it on the side of the flask to let it continue shaking.
Cool down the flasks to 18 °C from 30 min to 1 h before adding IPTG (0.2 mM final concentration) to induce POLRMT expression. To do this, add 200 μL of 1 M IPTG stock (see Recipes) per 1 L of LB (see General note 4 for cooling flasks).
Continue growing overnight (16–20 h) at 18 °C at 200 rpm.
Withdraw a 1 mL sample at the end of the induction period (“Induced”) and remove the “Uninduced” sample tube off the side of the flask (see Recipes for SDS-PAGE samples). Run out samples on a 10% SDS-PAGE gel and Coomassie stain to visualize POLRMT overexpression.
Harvest the cells by centrifugation at 2,991× g for 30 min at 4 °C in 1 L centrifuge bottles.
Safely dispose of the supernatant with 10% bleach and scrape the pellets into a 50 mL conical tube. Store the combined pellet at -80 °C (flash freezing is not necessary) or proceed with the next step.
Lysis of POLRMT overexpression pellet by sonication
Note: All steps below should be done at 4 °C or on ice unless noted otherwise.
Thaw the pellet by putting the tube in tap water. Scrape out the cells with a spatula into a metal beaker and add 50 mL of cold Lysis Buffer with BME, lysozyme, and PIC. Optional: Stir on a stir plate with a stir bar at 4 °C for 1 h to let the lysozyme break down cells.
Put the beaker in an ice bucket and sonicate cell suspension for 30 min at 4 °C (30 s on and off, 60% amplitude). Check that the suspension has become less viscous and slightly darker at this point.
Centrifuge the suspension at 30,000× g for 1 h at 4 °C.
Pour supernatant (“Lysate”) into a cold beaker on ice and record the volume.
Take samples to run on an SDS-PAGE gel: Pellet and Lysate (see Recipes). There will be more POLRMT in the pellet than in the lysate, but we will use the soluble POLRMT in the lysate for purification (Figure 1B).
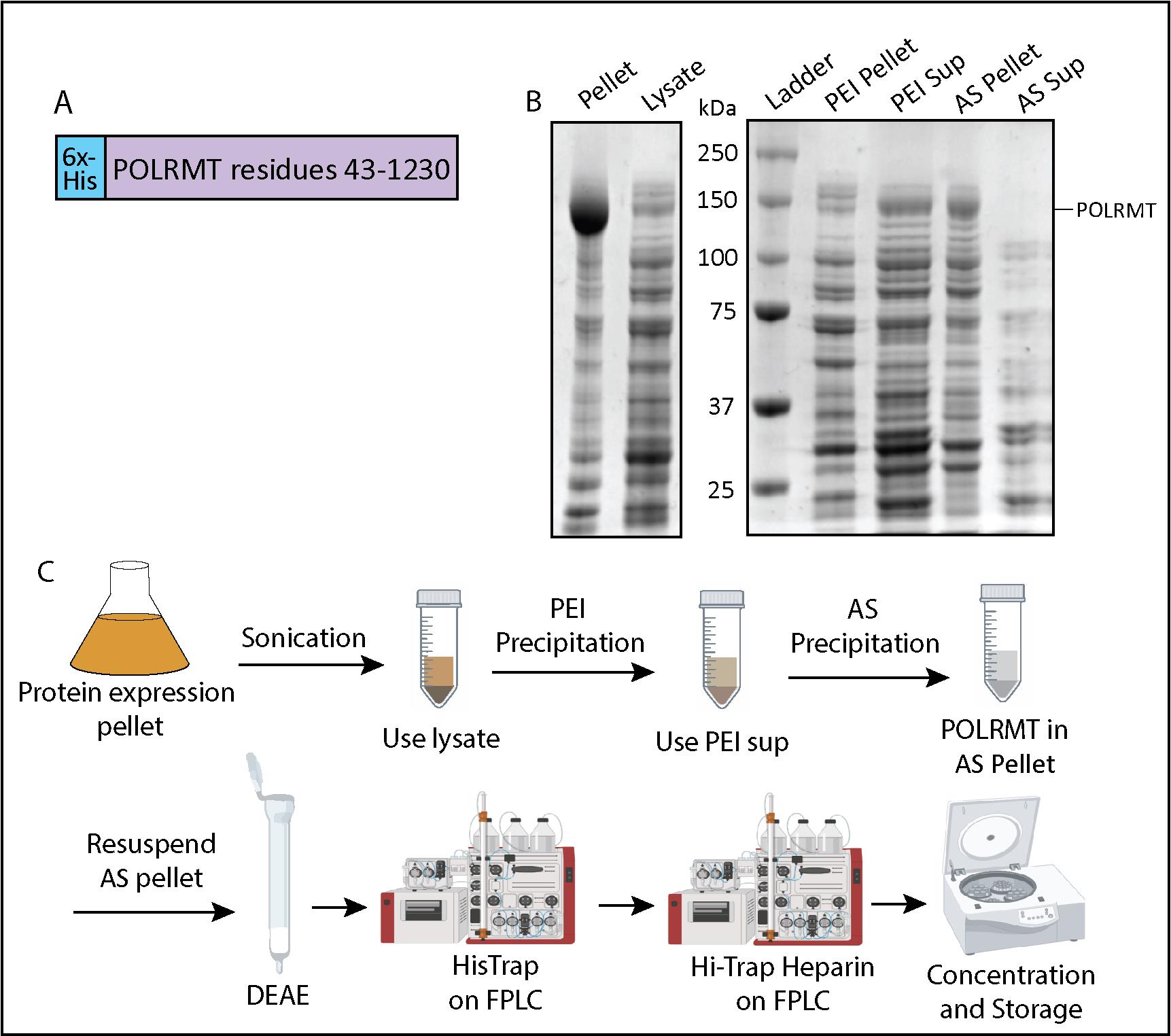
Figure 1. Mitochondrial RNA polymerase (POLRMT) purification flowchart and polyethyleneimine (PEI) and ammonium sulfate (AS) precipitation. (A) Expression construct containing the nucleotide sequence for POLRMT residues 43–1,230 (Uniprot: O00411) and an N-terminal 6x-His-tag. (B) 10% SDS-PAGE analysis of samples from the sonication step, PEI precipitation, and AS precipitation steps. (C) Schematic of POLRMT purification including sonication, PEI precipitation, AS precipitation, a DEAE column, a HisTrap column, a HiTrap heparin column, and concentration.
POLRMT PEI precipitation
With the lysate in a cold glass beaker on ice or at 4 °C, add a stir bar and check that it gently stirs the solution while on a stir plate.
Slowly add cold 5% PEI (see Recipes) dropwise into the lysate to get 0.3% PEI final concentration. To calculate how much 5% PEI we need, find 6% of the volume of the lysate:
______ mL of 5% PEI to be added = ______ mL of lysate × 0.06
Cover the top of the beaker with aluminum foil and keep stirring gently at 4 °C for an additional 30 min.
Centrifuge at 30,000× g for 10 min at 4 °C.
Take out samples for gel analysis [PEI pellet, PEI supernatant (see Recipes)] and run them out on a 10% SDS-PAGE gel. POLRMT should be in the supernatant (Figure 1B).
POLRMT AS precipitation
Add AS powder slowly (for a period of 30 min) to the supernatant to get 55% saturation.
For example, 55% saturation of 100 mL of lysate requires 35.1 g of AS.
______mL lysate × 0.351 g/mL = _______ grams AS to be added
Cover the top of the beaker with aluminum foil and gently stir at 4 °C for 1 h (or overnight) to dissolve AS and precipitate proteins.
Centrifuge at 30,000× g for 10 min at 4 °C. The POLRMT protein is in the pellet.
Take out samples for gel analysis: AS pellet, AS supernatant (see Recipes). On an SDS-PAGE gel, you should see POLRMT (~140 kDa) in the AS pellet and not in the AS supernatant (Figure 1B).
Record the weight of the pellet in grams and store it at -80 °C.
Pause point: The AS pellet can be stored for a few months before moving on to FPLC purification.
POLRMT DEAE gravity column and overnight HisTrap column
Note: One column volume (CV) corresponds to 5 mL for the HisTrap column (Cytiva). The DEAE gravity column can be replaced by using a HiTrap DEAE Fast Flow column (Cytiva) attached in tandem with the HisTrap column.
Start by attaching the 5 mL HisTrap column and loading POLRMT HisTrap buffers (see Recipes) onto the FPLC.
Wash and equilibrate the column (2 mL/min for five CV each of filtered Milli-Q water, HisTrap Buffer B, and then HisTrap Buffer A).
Resuspend the AS pellet in the HisTrap Buffer A by adding 4 mL of cold buffer per gram of pellet. Stir gently until dissolved.
Note: The following steps with the DEAE column can be done at room temperature as long as all the buffers are cold and the samples are kept on ice.
Prepare the DEAE by placing 10 mL of the DEAE resin suspension into a clean gravity column. Once the resin settles, you should get approximately 5 mL of bed resin.
Wash the DEAE with 5× bed volume of cold filtered water and cold HisTrap Buffer A.
Note: This DEAE column can be washed with water and stored in 20% EtOH for reuse.
With the stopper on the gravity column closed, pour the dissolved AS pellet into the column and gently stir with a spatula. Optional: Incubate for 15 min at 4 °C with gentle rocking.
Elute into a clean container on ice and then add 10 mL of HisTrap Buffer A to ensure all POLRMT is washed out.
Filter the eluent with a 0.22 μm syringe filter and prepare to load it on the HisTrap column on the FPLC. Use these settings for the FPLC run while monitoring eluent absorbance at 280 nm (A280) and collect all flowthrough/eluent for a later SDS-PAGE analysis (Figure 2B):
Sample application: 0.3 mL/min (for running overnight).
Column wash with Buffer A: 0.5 mL/min (for running overnight). Wash the column until the A280 trace returns to baseline (approximately five CV or 25 mL). After reaching baseline, continue to wash for at least 20 mL.
Protein elution: 1 mL/min, collect 5 mL fractions.
Elution program:
0–50% Buffer B gradient for 40 CV (200 mL). Optional: Wash the column with Buffer B. No need to collect samples from these steps.
70% Buffer B for five CV (25 mL)
100% Buffer B for five CV (25 mL)
Wash the column with five CV of water and five CV of 20% EtOH (use a lower flow rate of 1 mL/min) and store the columns.
Note: POLRMT elutes in a small broad peak and is very dilute. Do not let the fractions sit for longer than one day, as the more concentrated fractions of POLRMT will start precipitating (see General note 5).
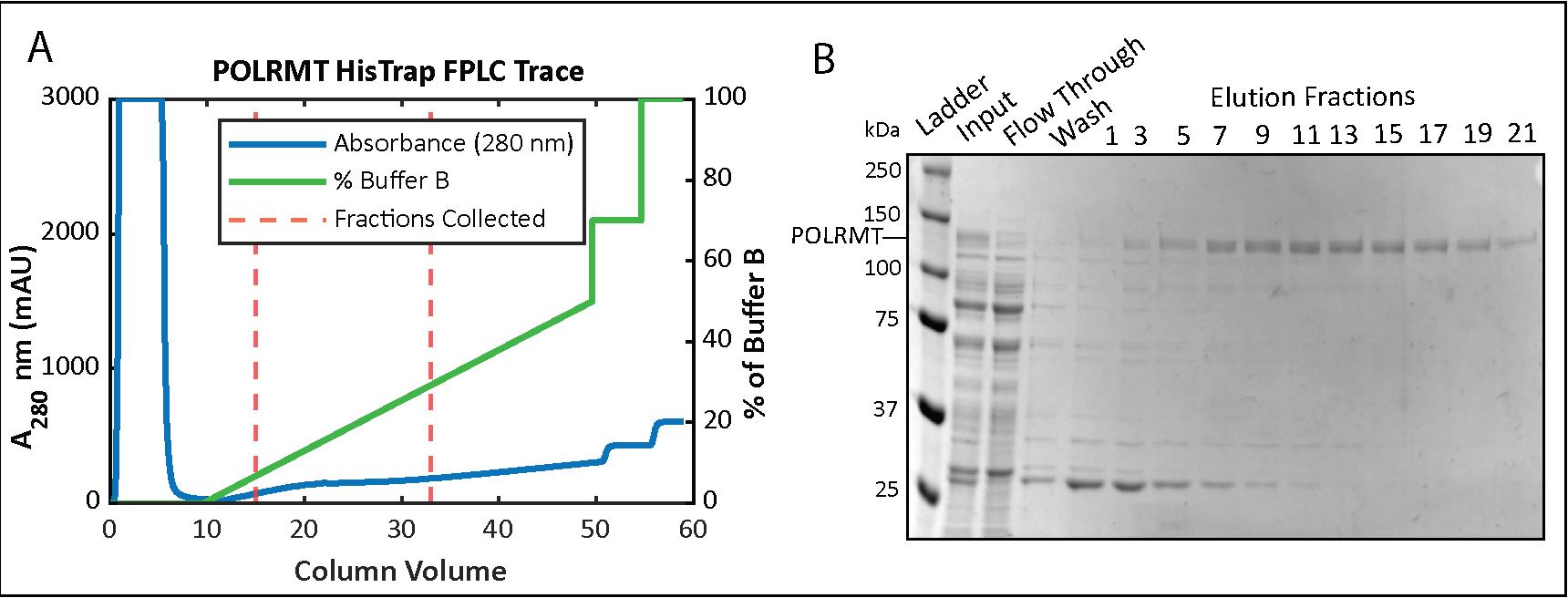
Figure 2. Mitochondrial RNA polymerase (POLRMT) HisTrap chromatography. (A) Chromatogram for the elution phase of the HisTrap column run on the FPLC and (B) 8% SDS-PAGE analysis of the collected fractions.Take out samples of the HisTrap fractions and run them on a 10% SDS-PAGE gel alongside a protein ladder. POLRMT appears around 140 kDa on an SDS-PAGE gel and is more concentrated roughly around fractions #4–22 (Figure 2B). (To save time, start preparing the filters and concentrate while the gel is running; see step below.)
Equilibrate two 50K MWCO Amicon filters at 4,000× g and 4 °C.
Concentrate POLRMT by centrifuging the Amicon filtration units at 1,300× g. Pipette solution gently every 5–10 min to avoid precipitation. Stop when the total volume becomes less than 25 mL (see General note 6 about this concentration step).
Add an equal volume of No Salt Heparin Buffer to the concentrate to bring the final NaCl concentration to 150 mM (see General note 7).
POLRMT overnight HiTrap heparin column
Wash and equilibrate the 5 mL HiTrap heparin column in Buffer A on the FPLC (2 mL/min for five CV each of filtered Milli-Q water, Heparin Buffer B, and then Heparin Buffer A).
Load your protein sample that has been diluted with the No Salt Heparin buffer.
Run the following FPLC program:
Sample application: 0.3 mL/min (for running overnight).
Column wash with Buffer A: 0.5 mL/min (for running overnight). Wash the column until the A280 trace returns to baseline (approximately five CV or 25 mL). After reaching baseline, continue to wash for at least 20 mL.
Protein elution: 2 mL/min, collect 3 mL fractions.
0–50% Buffer B gradient for 20 CV (100 mL).
55% Buffer B for five CV or 25 mL (POLRMT elutes here!).
70% Buffer B for five CV or 25 mL.
100% Buffer B for five CV or 25 mL.
Wash the column with five CV of water and five CV of 20% EtOH (use a lower flow rate of 1 mL/min) for storage of the columns.
Concentration and storage of POLRMT
Run a 10% SDS-PAGE gel of the fractions (see Recipes) and pool together the fractions with POLRMT (Figure 3B).
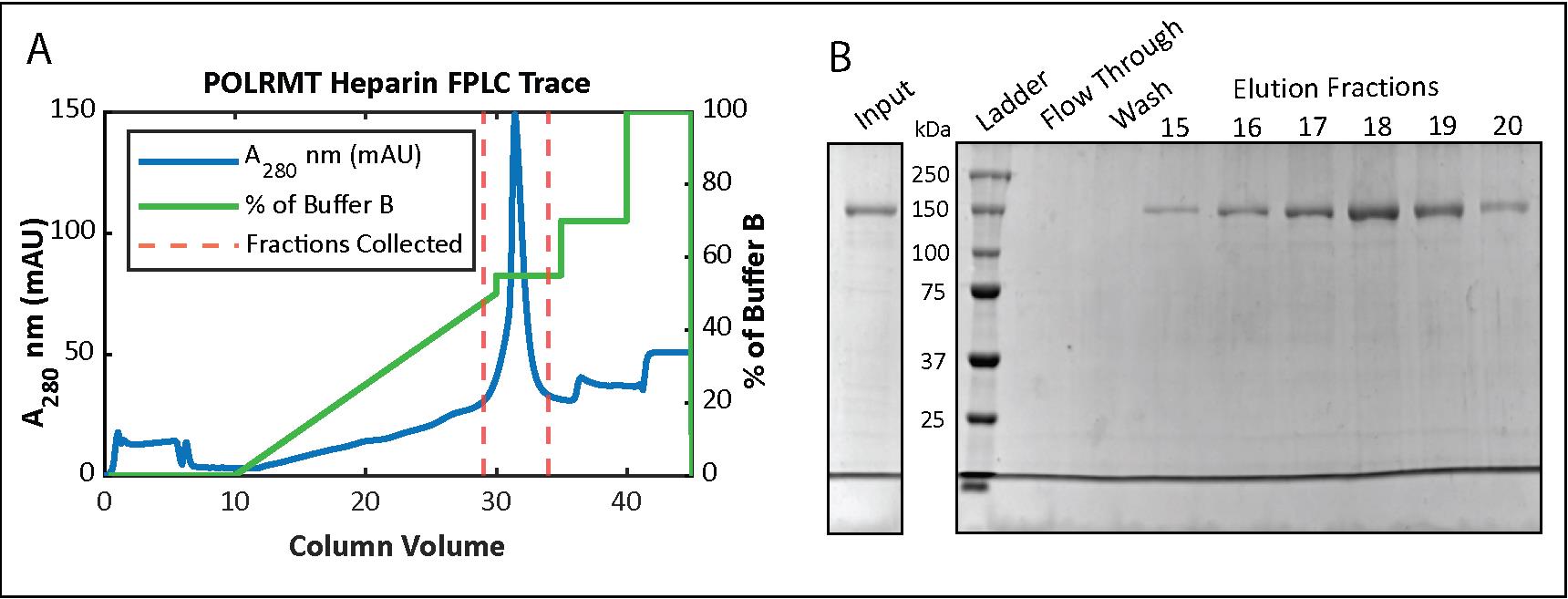
Figure 3. Mitochondrial RNA polymerase (POLRMT) HiTrap Heparin chromatography. (A) Chromatogram for the elution phase of the HiTrap Heparin column run on the FPLC and (B) 10% SDS-PAGE analysis of the collected fractions.Equilibrate the membrane of a 50K MWCO Amicon filtration unit with 150 mM NaCl Heparin Buffer by spinning in the centrifuge for 5 min. Add pooled POLRMT fractions to the unit.
Start centrifuging at 2,500× g at 4 °C and gradually decrease the speed to 1,900× g, pipetting gently from the bottom of the membrane and checking concentration frequently until at least 7 μM. POLRMT will easily precipitate out, so if you notice precipitate forming or if the concentration in the solution is not increasing, stop concentrating (see General note 8).
Measure and calculate protein concentration using these parameters by the Protparam tool (https://web.expasy.org/protparam/):
6x-His-POLRMT: 137.7 kDa or 137,700 mg/mole
Molar extinction coefficient: 143,700 L (cm*M)-1 (assuming all cys reduced)
Abs 0.1% = 1.043 (assuming all cys reduced)
Add sterile glycerol to get a final concentration of 25% glycerol.
Aliquot 10 μL into low-retention tubes, flash freeze in liquid nitrogen, and store at -80 °C.
Part II: TFAM purification
Overexpression of recombinant TFAM in E. coli
Transform BL21-CodonPlus(DE3)-RIPL cells using ~200 ng of ProEX_Htb_TFAM plasmid and plate on LB agar plates containing ampicillin. Incubate inverted overnight at 37 °C.
Grow 5 × 5 mL starter cultures overnight in LB supplemented with ampicillin, one colony per tube.
In the morning, spin down the 5 mL starter cultures at 2,782× g for 6 min and pour off the media. Carefully resuspend the cells in a total of 1 mL of LB, transferring the 1 mL to resuspend all the cells from the five tubes.
Add ampicillin to LB media (1 mL of 100 mg/mL ampicillin stock per 1 L of LB). Evenly split the 1 mL cell suspension between the 1 L LB flasks.
Incubate at 37 °C at 200 rpm. Check the OD600 after 4–5 h to see if it is between 0.4 and 0.6.
Once in the above OD range, withdraw a 1 mL sample (“uninduced”) into a microfuge tube, tape the tube on the side of the flask, and lower the shaker temperature to 16 °C.
Cool down the flasks before inducing with IPTG (0.2 mM final concentration; add 200 μL of 1 M IPTG stock per 1 L of LB).
Continue shaking overnight at 16 °C (~16–20 h) at 200 rpm.
Take a sample at the end of the induction period (“Induced”) and run it alongside the “Uninduced” sample (see Recipes) on an SDS-PAGE gel to visualize protein expression.
Harvest the cells by centrifuging at 2,991× g for 15 min at 4 °C.
Dispose of the supernatant and scrape the pellets into a 50 mL conical tube. Store the combined pellet at -80 °C or proceed with the cell lysis.
Cell lysis: lysozyme treatment and sonication
Note: All steps below should be done at 4 °C or on ice unless noted otherwise.
Thaw cells if frozen. Add 40 mL of TFAM Lysis Buffer (with lysozyme, BME, and PIC; see Recipes) to the pellet (10 mL of lysis buffer per liter of expressed culture) and resuspend the pellet until homogenous.
Incubate for 1 h at 4 °C.
Sonicate cell suspension for 20 min at 4 °C (20 s on and off, 60% amplitude).
Centrifuge at 30,000× g for 1 h at 4 °C.
Save ~10 μL of “Cell Pellet” and “Lysate” samples (see Recipes) for SDS-PAGE gel analysis.
TFAM Ni-NTA resin #1
Note: This is Ni-NTA by centrifugation method at 4 °C.
Prepare a 5 mL Ni-NTA resin bed in a 50 mL conical tube according to the manufacturer’s instruction and wash three times with Ni-NTA Buffer A by resuspending the resin in the buffer, spinning it down at 363× g for 3 min at 4 °C, carefully decanting the buffer, and repeating the process.
Add the supernatant and incubate overnight at 4 °C with gentle agitation.
Spin down the resin, collect the supernatant as “Sample Application” into a conical tube, and begin resin washing steps by adding 5 mL of Ni-NTA Buffer A to the resin, gently mixing, and spinning the sample at 1,932× g at 4 °C.
Collect the supernatant as “Wash 1” and repeat washes to wash #10.
Beginning with wash #10, check the absorbance of the sample at 280 nm and continue the washes until the absorbance is close to 0.
Elute the proteins by adding 5 mL of Ni-NTA Buffer B and spinning as before. Collect the sample as “Elution 1” and repeat this 3–5 times, keeping “Elution” supernatants separate.
Run a 10% or 12% SDS-PAGE gel with collected samples from the cell lysis, sample application, washing, and elution (see Recipes). A TFAM band just above 25 kDa should be obvious but some contaminants will still be present (Figure 4B).
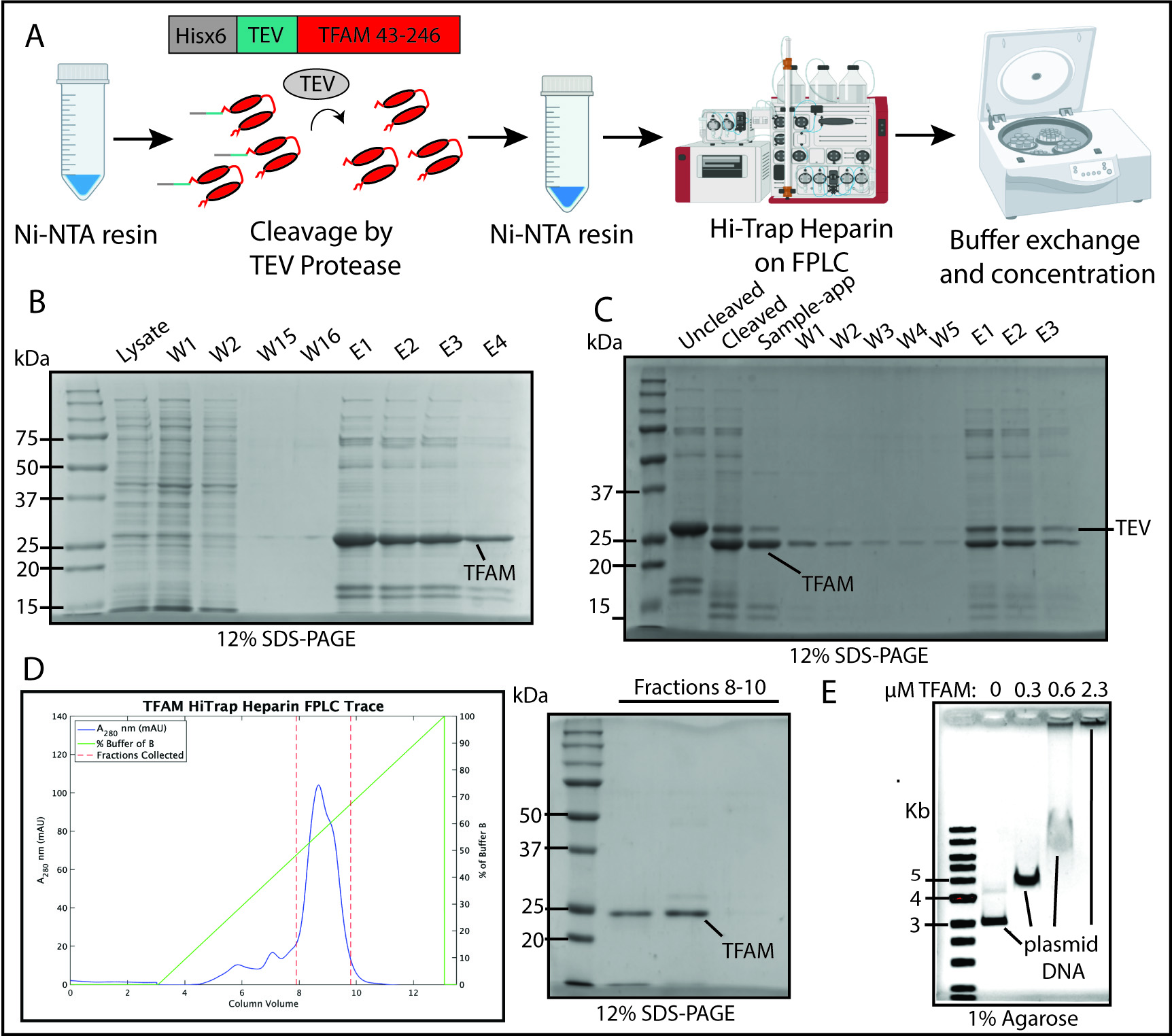
Figure 4. Transcription factor A, mitochondrial (TFAM) purification flowchart, Ni-NTA and HiTrap Heparin chromatography, and TFAM DNA-compaction test. (A) Schematic of TFAM purification including Ni-NTA Resin purification, cleavage by TEV protease, a second Ni-NTA resin step, a HiTrap Heparin column, and concentration. (B) 12% SDS-PAGE analysis of Ni-NTA #1 results including washes (W) and elution steps (E). (C) 12% SDS-PAGE analysis of TEV protease cleavage of TFAM and Ni-NTA resin step. (D) Chromatogram for the elution phase of the HiTrap Heparin column run on the FPLC and the SDS-PAGE analysis of the collected fractions. (E) Electrophoretic mobility shift assay (EMSA) to test TFAM ability to compact plasmid DNA.
Dialysis and TEV cleavage of the His tag on TFAM
Add 2 L of ice-cold TFAM TEV Cleavage Buffer to a large beaker.
Place the elution samples with significant TFAM content into dialysis tubing and add TEV protease at a 1:30 TEV:TFAM protein molar ratio. (Estimate TFAM content using the absorbance of the pooled elution samples at 280 nm. See step G for protein mass and extinction coefficient.)
Allow the sample to dialyze in Cleavage Buffer and cleave overnight at 4 °C with gentle stirring.
Check the success of TEV cleavage on a 12% SDS-PAGE gel by comparing the sample before and after dialysis. If cleavage is complete, continue to the next step. If not, add more TEV protease and continue dialysis (Figure 4C).
Note: The version of TEV protease used in our lab is ~26 kDa in size. This means it will look a lot like uncleaved TFAM on the SDS-PAGE gel.
TFAM Ni-NTA resin #2
Prepare a 5 mL resin bed of Ni-NTA resin as before (see step C1).
Add the dialyzed sample to the resin and incubate overnight at 4 °C as before with gentle agitation.
Spin down the resin as before (1,932× g at 4 °C) and collect the supernatant [“Sample application” (this will have the TEV-cleaved, now untagged TFAM)].
Add 5 mL of Ni-NTA Buffer B to the resin, mix, and spin to collect “Elution 1.” You want to collect this to be sure the TEV protease was bound by the Ni resin.
Repeat the elution 2–3 times.
Run a 10% or 12% SDS-PAGE gel to be sure that TFAM is in the “Sample application” (Figure 2C).
TFAM concentration and HiTrap heparin column
Note: An alternative to the first concentration step is to adjust the salt concentration by adding a version of the Ni-NTA Buffer A without any salt. Adjust the salt to <200 mM for use on the HiTrap heparin column.
Wash and equilibrate the 5 mL HiTrap heparin column (Cytiva) on the FPLC with five CV of heparin Buffer B and then Buffer A.
Add elution fractions from Ni-NTA resin #2 to a 50 mL conical tube and begin buffer exchanging the sample by adding it in 15 mL volumes to a 3K MWCO Amicon filter and spinning at 3,434× g at 4 °C. Stop concentrating when the buffer exchange sample is down to 2 mL.
When the entire protein sample has been added and spun down to approximately 2 mL, fill the 3K MWCO Amicon filter with Heparin Buffer A and continue to spin. Continue to add Heparin Buffer A and spin until ~30 mL has flown through the filter and you are left with 2–3 mL. This lowers the salt concentration from 300 mM to 200 mM to ensure binding to the HiTrap Heparin column.
Load the buffer-exchanged protein sample onto the prepared HiTrap Heparin column.
Run using these settings:
Sample application: 1.0 mL/min.
Column wash: 1.0 mL/min until A280 reaches ~0.
Elution: 2.5 mL/min with 2 mL fractions.
0–100% Buffer B gradient over 10 CV (50 mL).
Wash the column with five CV of water and five CV of 20% EtOH (1 mL/min) for storage.
Concentration and storage of TFAM
Run a 10% or 12% SDS-PAGE gel of the fractions and pool together the fractions with TFAM (Figure 4D)
Concentrate as before using a fresh 3K MWCO Amicon filter.
When all the pooled fractions have been concentrated to a small volume (0.5–1 mL), add 2× Storage Buffer to fill the filter and continue to centrifuge. Repeat this process to pass two full volumes of Storage Buffer through the filter to exchange the buffer. Stop when the second full pass of 2× Storage buffer has reached ~2 mL and begin measuring the absorbance at 280 nm for TFAM concentration while continuing to centrifuge.
Measure and calculate protein concentration using these parameters from the protein sequence by the Protparam tool (https://web.expasy.org/protparam/):
TFAM after TEV cleavage: 24.4 kDa or 24,400 mg/mole.
Molar extinction coefficient: 35,410 L (cm*M)-1 (assuming all cys reduced).
Abs 0.1% = 1.45 (assuming all cys reduced).
When the desired concentration has been reached (100–200 mM), move the sample from the Amicon filter to a conical tube and add an equal volume of filtered 50% glycerol for a final concentration of 25% glycerol.
Aliquot 20 mL per tube, flash freeze, and store at -80 °C.
Testing TFAM DNA-compaction activity with electrophoretic mobility shift assay (EMSA)
Note: To know if the purified TFAM is active, it is best to run an electrophoretic mobility shift assay. TFAM can bind DNA non-specifically, so any plasmid DNA can be used. If the bands shift upwards in the assay, the TFAM is active and will work with transcription assays. In our assays, we use the Puc19 plasmid.
Prepare a 1% agarose EMSA gel using TFAM EMSA Running Buffer
Prepare reactions with 1× TFAM Binding Buffer, add ~200 ng of plasmid DNA, and add TFAM in increasing amounts to the plasmid. These reactions are typically done in total volumes of 20–30 mL.
Incubate samples at 37 °C for 30 min.
Add filtered glycerol to 10% in the samples and load onto the EMSA gel along with a 1 kb DNA ladder.
Run the EMSA gel at 70 V for 65 min at room temperature in TFAM EMSA Running Buffer.
Stain the gel with gentle agitation using ethidium bromide for 30 min at room temperature and image using UV light on a Bio-Rad Gel Doc EZ Imager system or equivalent (Figure 4E).
Part III: TFB2M purification
Expression of TFB2M
Transform the plasmid containing TFB2M (Figure 5) into BL21-CodonPlus (DE3)-RIPL and spread on Kan-containing (25 μg/mL, see Recipes) plates. Incubate inverted overnight at 37 °C.
Note: The transformation step can be skipped if glycerol stocks have been previously made. E. coli strain ArcticExpress (DE3) could also be used for transformation.
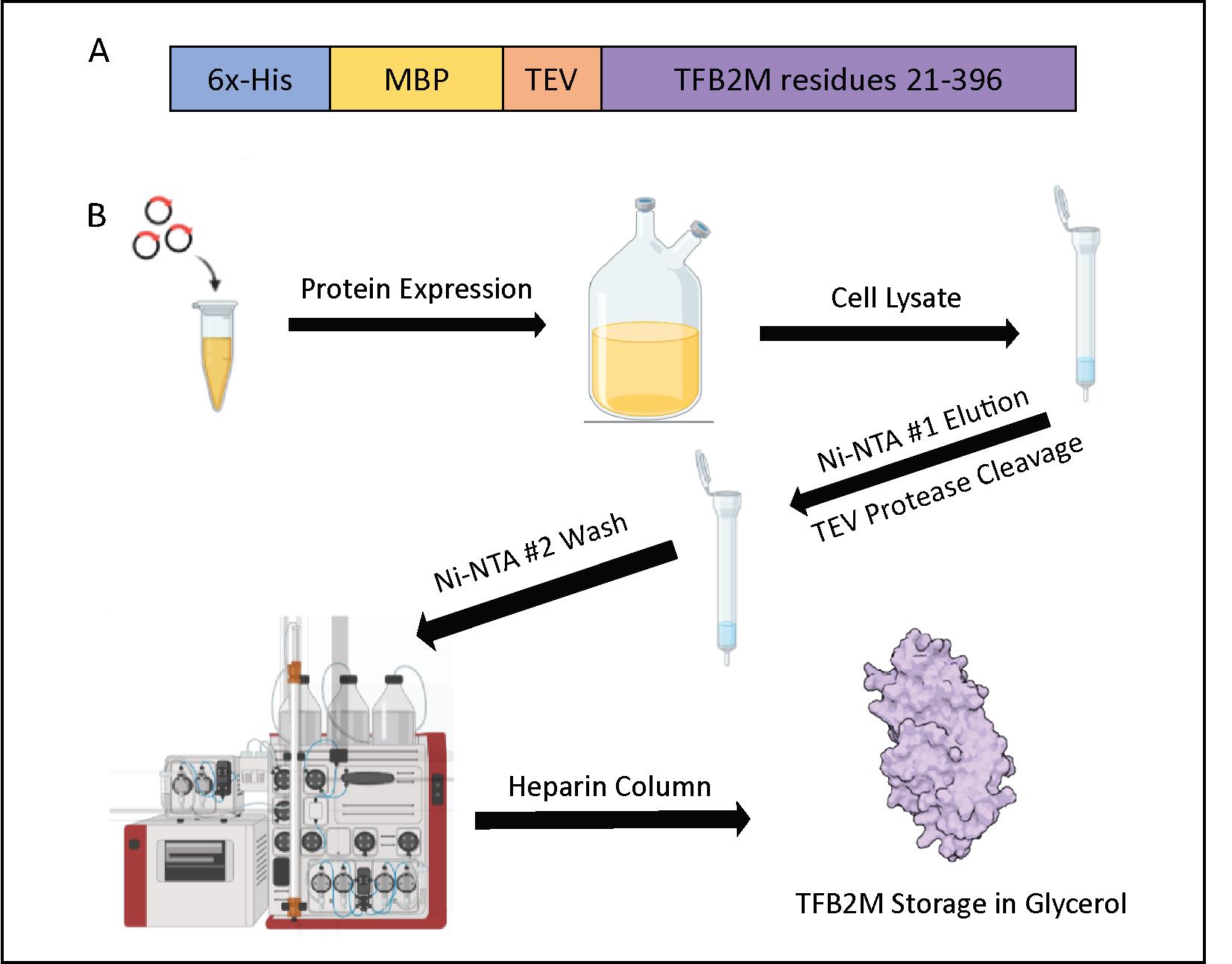
Figure 5. Transcription factor B2, mitochondrial (TFB2M) purification flowchart. (A) Expression construct containing the nucleotide sequence for TFB2M, corresponding to residues 21–396 (Uniprot: Q9H5Q4). This plasmid has an N-terminal 6x-His-tagged Maltose-binding protein (MBP) solubility tag connected to TFB2M with a TEV-cleavage site. (B) Schematic of TFB2M purification including protein expression, Ni-NTA #1 Resin purification, cleavage by TEV protease, Ni-NTA #2 Resin step, a HiTrap Heparin column, and concentration.Add five colonies or a small chunk of the frozen glycerol stock to 5–10 mL of LB culture with Kan (25 μg/mL). Grow overnight while shaking at 28–37 °C; avoid letting the starter culture become oversaturated (see General note 2 for POLRMT purification).
In the morning, add primary culture (2.5–5 mL) to 1 L of LB culture.
Grow at 37 °C at 200 rpm until OD600 reaches 0.7–0.8 (approximately 3–5 h).
Once in the OD range, cool down the shaker to 14 °C and withdraw a 1 mL sample (“uninduced”) before inducing the rest of the culture with 0.2 mM final IPTG (refer to Recipes). Continue to shake for 16–20 h at 14 °C.
Take a 1 mL sample (“Induced”) at the end of the induction period to run on an SDS-PAGE gel.
Harvest the cells by centrifuging at 2,991× g for 15 min at 4 °C.
Store cell pellet at -80 °C.
Sonication
Note: All steps below should be done at 4 °C or on ice unless noted otherwise.
Add 40 mL of Lysis Buffer (with lysozyme, BME, and PIC, see Recipes) to 4 L-worth of cell pellet (10 mL of lysis buffer per liter of expressed culture) and stir the pellet at 4 °C for 1 h.
In the cold room, sonicate for 20 min at 60% amplitude, 30 s on and off.
Centrifuge at 30,000× g for 1 h at 4 °C to remove cell debris.
Ni-NTA Resin #1
Set up a gravity column by clamping it onto a stand and pipette 10 mL of Ni-NTA resin solution into the column to get 5 mL bed volume.
Wash the resin with filtered, cold Milli-Q water three times and then wash once to equilibrate with your Ni Buffer A.
Note: The volume of each wash should be five times the bed volume of the resin.
Pour the cell lysate onto Ni-NTA resin and gently stir with a metal spatula to promote the binding of your His-tagged protein. Securely close the column/vial and rock gently for 1 h at 4 °C.
Clamp the column again on the stand and collect your sample application flowthrough. Label the sample as “Ni flowthrough” for SDS-PAGE.
Prepare the following mixtures of Buffer A and B to perform the wash and elution steps:
One CV = 5 mL bed resin.Wash 1, 2, and 3: 0% B, five CV: prepare three tubes containing 50 mL of Buffer A each.
Wash 4: 5% B, five CV: one tube containing 47.5 mL of Buffer A and 2.5 mL of Buffer B.
Elution 1: 40% B, two CV: one tube containing 6 mL of Buffer A and 4 mL of Buffer B.
Elution 2: 50% B, two CV: one tube containing 5 mL of Buffer A and 5 mL of Buffer B.
Elution 3: 100% B, two CV: one tube with 10 mL of Buffer B.
Keep them on ice or at 4 °C before use.
Note: The washes and elution can be done at room temperature if all the buffers are cold and samples are kept on ice. Avoid drying out the resin.
Wash the resin with Wash 1. Collect and label the flowthrough as “W1.”
Repeat with Wash 2 and Wash 3.
Wash a final time with Wash 4 (which contains 5% v/v of Buffer B in Buffer A). Save the flowthrough as “W4.”
Elute TFB2M with Elution 1. Collect and label the elution as “E1.”
Repeat with Elution 2 and 3.
Pool elution 1, 2, and 3 together and estimate the protein concentration with the spectrophotometer:
6x-His-MBP-TFB2M: 87.7 kDa or 87,691 mg/mole.
Molar extinction coefficient: 117,230 L (cm·M)-1 (assuming all cys reduced).
Abs 0.1% = 0.00134 (assuming all cys reduced).
Overnight TEV cleavage
Estimate the amount of TFB2M using the absorbance of the pooled elution samples at 280 nm:
Amount of total TFB2M (spectrometer reading) =_____mg/mL
Total volume = ____ mL
Total amount of protein = ____mg (assume it is all TFB2M)
We need a molar ratio of 1:25 TEV:TFB2M
Amount of TEV to be added =_____mg
Concentration of your TEV protease stock = _____mg/mL
Volume of TEV to be added = _____mL.
Add 2 L of ice-cold TEV Cleavage Dialysis Buffer to a large beaker with a large stir bar.
Add TEV to the same tube as your Ni-resin eluted TFB2M, gently mix, and then transfer into the dialysis tubing (see General note 9).
Allow the sample to dialyze in the TEV Cleavage Buffer for 19–20 h overnight at 4 °C with gentle stirring.
Check the completion of TEV cleavage of TFB2M on a 10% or 12% SDS-PAGE gel by comparing the TFB2M band size before (~88 kDa) and after dialysis (~43 kDa). If cleavage is complete, continue to the next step. If not, add more TEV protease and continue dialysis (Figure 6).
Note: The MBP tag (42 kDa) will be similar in size to TFB2M, and the band may be more intense. The version of purified TEV protease used in our lab is approximately 26 kDa in size.
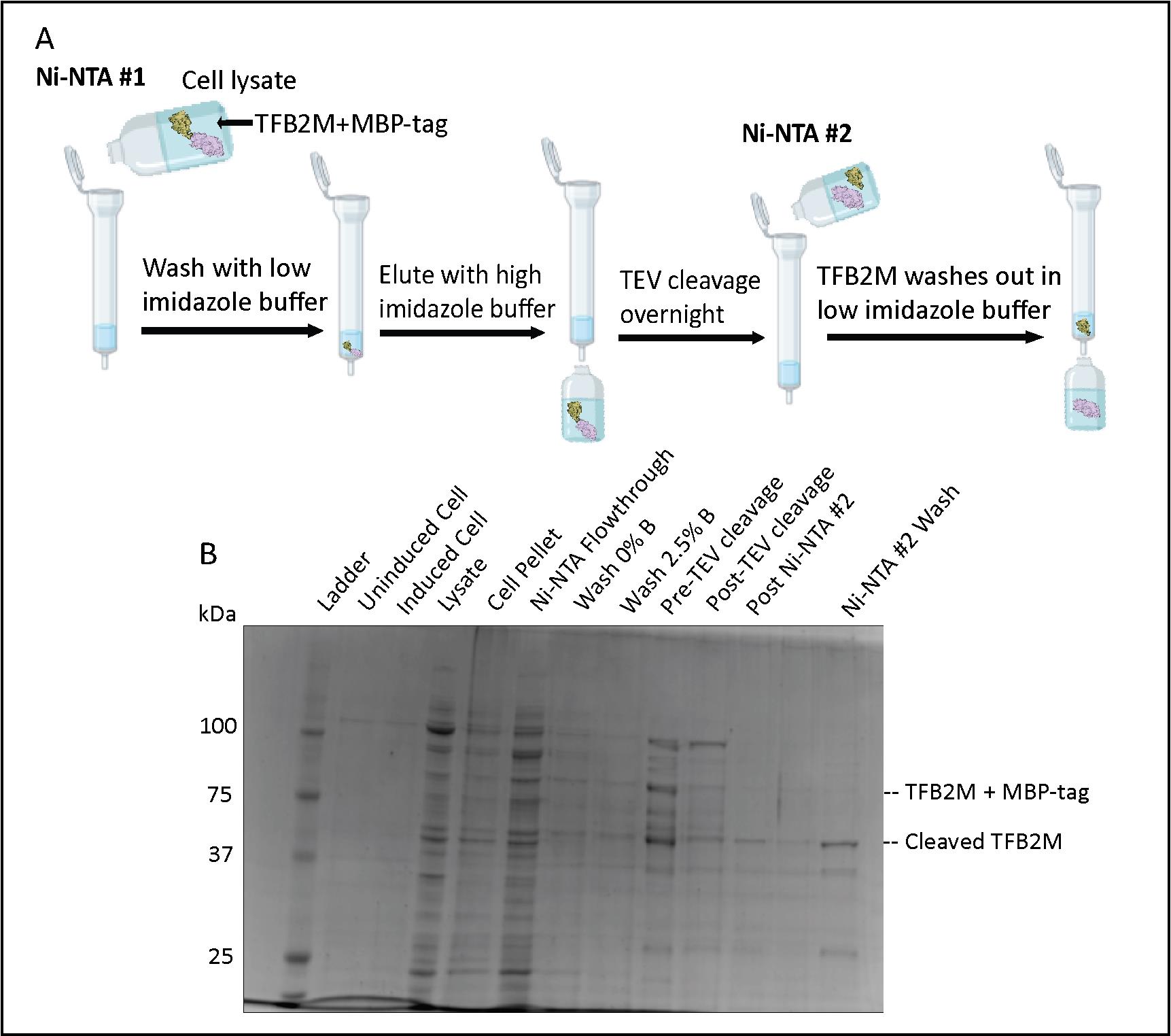
Figure 6. Transcription factor B2, mitochondrial (TFB2M) Ni-NTA chromatography. (A) Flowchart of the Ni-NTA #1, TEV cleavage, and Ni-NTA #2 steps. (B) SDS-PAGE of TFB2M expression, the first Ni-NTA purification step, the TEV cleavage process, and TFB2M presence in Ni-NTA #2 wash.
Ni-NTA Resin #2
Note: This is to remove His-tagged TEV protease and other contaminants from the cleaved TFB2M, which is now missing its His-tag. TFB2M will be in the flowthrough instead of the elution (Figure 6A).
Using the same Ni-NTA column from the previous step, wash the column with 5× bed volume of Buffer B to remove all the bound proteins, and then wash with Buffer A to equilibrate the column.
Before applying your cleaved TFB2M sample onto the Ni-NTA resin, add Buffer B to the TFB2M solution to bring the imidazole concentration to 20 mM (matching that in Buffer A):
(20 mM imidazole needed) (____ mL of cleaved TFB2M sample) = (500 mM imidazole in Buffer B) (x mL Buffer B needed)
Note: Introducing a small (20 mM) amount of imidazole ensures that most of TFB2M does not bind to the resin. The other protein contaminants like the His-MBP-tag and His-tagged TEV protease will stay bound to the Ni-NTA.
Add the sample to the Ni resin and rock gently for 30 min at 4 °C.
Collect the flowthrough (or “wash”) containing TFB2M and run a 10% or 12% SDS-PAGE gel to be sure that TFB2M is present in the flowthrough (Figure 6B).
Note: What remains bound to the column are the His-MBP tag and His-tagged TEV protease. Be aware that the His-MBP tag is similar in size to untagged TFB2M (approximately 42 kDa), and its band is often more intense on a gel.
Overnight heparin column
Wash and equilibrate a 5 mL HiTrap heparin column (Cytiva) on the FPLC.
Dilute the TFB2M sample (currently in 300 mM KCl buffer) with the No Salt Heparin Buffer to get a final salt concentration between 75 and 150 mM KCl.
Note: The lower the salt, the more TFB2M binds to the heparin column. Do not concentrate TFB2M before loading it on the column or it will precipitate out.
Run using these settings:
Note: One column volume (CV) is 5 mL for the HiTrap Heparin column (Cytiva).
Sample application: 1 mL/min.
Column wash: 0.5 mL/min (for running overnight) for five CV.
Protein elution: 0.5 mL/min, collect 2 mL fractions. Elution program:
0–70% gradient for 10 CV (50 mL total)
100% for one CV
Wash the column with five CV of water and five CV of 20% EtOH (1 mL/min) for storage.
Collect fractions and run a sample on a 10%–12% SDS-PAGE to determine which fractions contain TFB2M (Figure 7B).
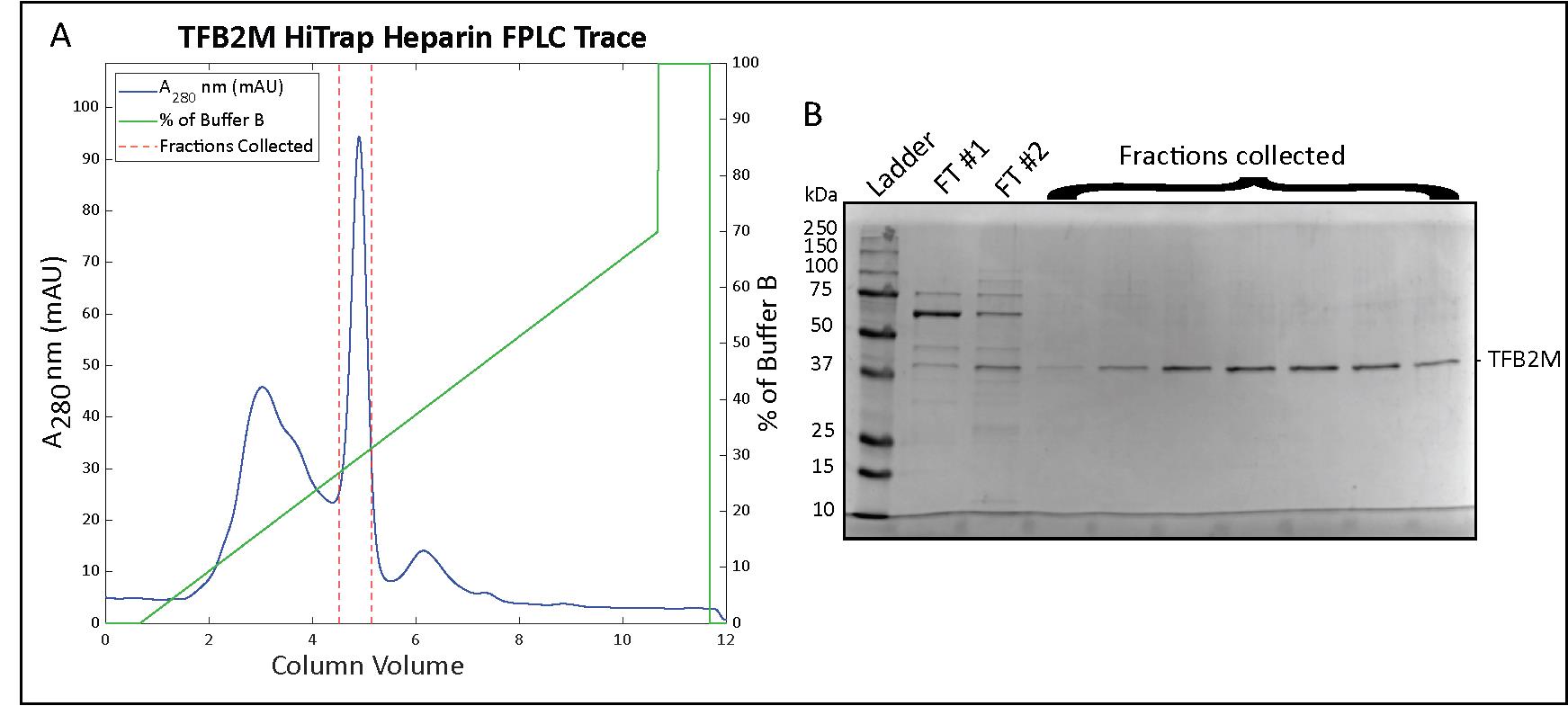
Figure 7. Transcription factor B2, mitochondrial (TFB2M) HiTrap Heparin chromatography. (A) Chromatogram for the elution phase of the HiTrap Heparin column run on the FPLC and (B) a 12% SDS-PAGE analysis of the collected fractions.
TFB2M concentration
Pool together the fractions containing TFB2M.
Equilibrate a 10K MWCO Amicon membrane in a centrifuge for 5 min with the Heparin Buffer A.
Start centrifuging the TFB2M sample at 1,300× g. Stop the centrifuge every 10 min and gently pipette TFB2M solution up and down with a pipette to mix. If you notice precipitate forming or if the concentration in the solution is not increasing, stop concentrating.
Measure and calculate the protein concentration using these parameters from the protein sequence by the Protparam tool (https://web.expasy.org/protparam/):
TFB2M after TEV cleavage: 43.405 kDa or 43,405 mg/mole.
Molar extinction coefficient: 49,390 L (cm·M)-1 (assuming all cys reduced).
Abs 0.1% = 1.138 (assuming all cys reduced).
Add sterile glycerol to get a final concentration of 25%, aliquot 15 μL into low-retention tubes, flash freeze in liquid nitrogen, and store at -80 °C (see General note 10 for estimating salt concentration in TFB2M samples).
Data analysis
To assess the purity of the proteins, run the purified samples on an SDS-PAGE gel (see Figure 8A below). Protein band intensity can be analyzed using Bio-Rad Image Lab and ImageJ software.
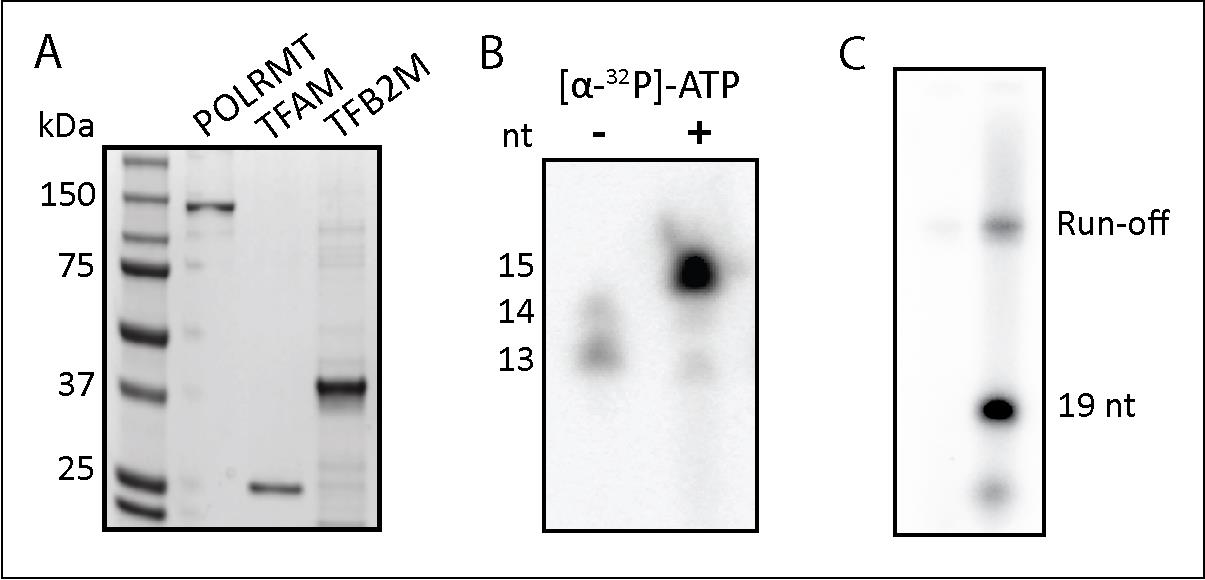
Figure 8. Validation of POLRMT, TFAM, and TFB2M purification. (A) SDS-PAGE analysis of purified POLRMT, TFAM, and TFB2M. (B) Validation of POLRMT activity on a nucleic acid scaffold. The size of a weakly radiolabeled RNA was increased by POLRMT-catalyzed incorporation of [a-32P]NTP to produce a 15-nt RNA. (C) TFB2M activity was validated in a promoter (LSP)-initiated transcription assay. The reaction was stopped at 19 nt with the incorporation of 3'-dCTP. The two lanes are the same sample loaded in increasing amounts.
To evaluate the expression of the proteins, a western blot can be run using an anti-His antibody. POLRMT can be identified this way after the purification as it still retains its His-tag. For TFAM and TFB2M, which will lack their His-tag after purification, performing the TFAM functional assay described above and performing an in vitro transcription assay will confirm the activity of the proteins.
Validation of protocol
The protocol was used to purify the POLRMT, TFAM, and TFB2M used for an in vitro transcription initiation assay in Figure 5D of our paper (Reardon and Mishanina, 2022).
POLRMT activity was validated by performing an in vitro transcription assay (Figure 8B) on a nucleic acid scaffold designed by Schwinghammer et al., 2013 (Supplementary Figure 1 in the paper). This assay indicated that POLRMT was able to add the incoming nucleotide to the starting RNA.
TFB2M activity was validated by performing an in vitro transcription initiation assay (Figure 8C) following the protocol by Ramachandran et al., 2017 (Figure 1 in the paper). The template contained the light strand promoter (LSP) sequence of mtDNA, which requires TFB2M and TFAM for transcription to occur.
General notes and troubleshooting
This protocol describes how to lyse cells with a sonicator, but a French press could be used instead. Similarly, a peristaltic pump could be used to generate buffer gradients instead of an FPLC.
For POLRMT expression: the goal is to prevent oversaturation of the cells. The lower the temperature, the longer the colonies can be left growing overnight, so any temperature between 25 °C and 37 °C will work. If colonies are oversaturated, the growth will be extremely slow, most likely because the leaky expression of POLRMT is toxic to E. coli cells.
Glycerol stocks could also be prepared at this time by storing 1 mL of the inoculum in 25% glycerol at -80 °C. We usually vortex 500 μL of inoculum with 500 μL of 50% sterile glycerol before freezing.
Lower temperatures will slow down translation and promote proper folding of the protein. To cool down the flasks, we leave them shaking on the incubator while they cool or put the flasks in large buckets of ice. The OD600 can be checked again but inducing in the range of 0.6–1.0 works fine.
We have tried to change the method to get more concentrated POLRMT in fewer fractions, but POLRMT ended up precipitating out very quickly, so it is not recommended.
We concentrate the HisTrap elution in the Amicon filter down to 25 mL (step 10) because 50 mL is a convenient volume that we can load onto our Bio-Rad NGC FPLC via a sample pump. If your system can handle a larger loading volume, you can add an equal volume of the No Salt Heparin buffer and skip the concentration step.
Because the HisTrap Buffer B has 300 mM NaCl, we need to bring the salt concentration down to the same as in the Heparin Buffer A to have POLRMT bind to the Heparin column. The Heparin column does not greatly increase purity but removes endogenous bound nucleic acids from the protein.
Previous attempts at buffer exchange resulted in increased POLRMT precipitation and are thus not recommended. We usually concentrate POLRMT to 6 or 7 μM before we start losing protein to precipitation.
The goal of the dialysis is to remove imidazole from the buffer, which inhibits TEV protease activity. It is also important to keep the salt concentration above 300 mM KCl to prevent TEV protease from crashing out. There is no EDTA in the TEV buffer because EDTA inhibits TEV activity.
Optional for downstream transcription experiments where salt concentration could be an issue: we can estimate the salt concentration of the purified TFB2M by the % Buffer B line at the time of TFB2M elution on the FPLC trace and calculate based on the known salt concentrations of Buffers A and B:
% of buffer B: _____
% of buffer A: 100% - % buffer B = ____
Volume of that fraction collected: ____mL
Volume of buffer B: Vol of fraction * % buffer B = _____mL
Volume of buffer A: Vol of fraction * % buffer A = _____mL
Moles of B = Vol Buffer B (1.5 mol)/(1,000 mL)=x mol KCl
Moles of A = Vol Buffer A (0.200 mol)/(1,00 0mL)= x mol KCl
Salt concentration of fraction = (Moles A+Moles B)/(Volume of fraction collected (mL)) × (1,000 mL)/(1 L) = x M salt concentration
Acknowledgments
We greatly appreciate Dr. Smita Patel (Rutgers University) and her lab members for providing the expression plasmids for POLRMT and TFAM and the protocols used in this work. We would like to thank Dr. Miguel Garcia-Diaz (Stony Brook University) for providing the expression plasmid for TFB2M. This work was supported by UCSD institutional support, Yinan Wang Memorial Chancellor’s Endowed Junior Faculty Fellowship, and R35GM142785 to T.V.M., R35GM118086 to S.S.P.; NIH Molecular Biophysics Training Grant (T32 GM008326) to A.H.H. and J.H.M-C.; NIH Multi-Scale Analysis of Biological Structure and Function Training Grant (T32 EB009380) to S.D.R. This protocol was adapted from and validated in previous works (Ramachandran et al., 2017; Reardon and Mishanina, 2022).
Competing interests
The authors declare no competing interests.
References
Bird, J. G., Basu, U., Kuster, D., Ramachandran, A., Grudzien-Nogalska, E., Towheed, A., Wallace, D. C., Kiledjian, M., Temiakov, D., Patel, S. S., et al. (2018). Highly efficient 5' capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase. eLife 7: e42179.
- Hillen, H. S., Morozov, Y. I., Sarfallah, A., Temiakov, D. and Cramer, P. (2017). Structural Basis of Mitochondrial Transcription Initiation. Cell 171(5): 1072–1081.e10.
- Kapust, R. B., Tözsér, J., Fox, J. D., Anderson, D., Cherry, S., Copeland, T. D. and Waugh, D. S. (2001). Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. Des. Sel. 14(12): 993–1000.
- Oláhová, M., Peter, B., Szilagyi, Z., Diaz-Maldonado, H., Singh, M., Sommerville, E. W., Blakely, E. L., Collier, J. J., Hoberg, E., Stránecký, V., et al. (2021). POLRMT mutations impair mitochondrial transcription causing neurological disease. Nat. Commun. 12(1): e1038/s41467-021-21279-0.
- Ramachandran, A., Basu, U., Sultana, S., Nandakumar, D. and Patel, S. S. (2017). Human mitochondrial transcription factors TFAM and TFB2M work synergistically in promoter melting during transcription initiation. Nucleic Acids Res. 45(2): 861–874.
- Raran-Kurussi, S., Cherry, S., Zhang, D., Waugh, D. S. (2017). Removal of Affinity Tags with TEV Protease. In: Burgess-Brown, N. (Ed.) Heterologous Gene Expression in E. coli (pp. 221–230). Methods in Molecular Biology. Humana Press, New York, NY.
- Reardon, S. D. and Mishanina, T. V. (2022). Phosphorylation and acetylation of mitochondrial transcription factor A promote transcription processivity without compromising initiation or DNA compaction. J. Biol. Chem. 298(4): 101815.
- Schwinghammer, K., Cheung, A. C. M., Morozov, Y. I., Agaronyan, K., Temiakov, D. and Cramer, P. (2013). Structure of human mitochondrial RNA polymerase elongation complex. Nat. Struct. Mol. Biol. 20(11): 1298–1303.
- Sultana, S., Solotchi, M., Ramachandran, A. and Patel, S. S. (2017). Transcriptional fidelities of human mitochondrial POLRMT, yeast mitochondrial Rpo41, and phage T7 single-subunit RNA polymerases. J. Biol. Chem. 292(44): 18145–18160.
- Tan, B. G., Mutti, C. D., Shi, Y., Xie, X., Zhu, X., Silva-Pinheiro, P., Menger, K. E., Díaz-Maldonado, H., Wei, W., Nicholls, T. J., et al. (2022). The human mitochondrial genome contains a second light strand promoter. Mol. Cell 82(19): 3646–3660.e9.
- Yakubovskaya, E., Guja, K. E., Eng, E. T., Choi, W. S., Mejia, E., Beglov, D., Lukin, M., Kozakov, D. and Garcia-Diaz, M. (2014). Organization of the human mitochondrial transcription initiation complex. Nucleic Acids Res. 42(6): 4100–4112.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Hsieh, A., Reardon, S. D., Munozvilla-Cabellon, J. H., Shen, J., Patel, S. S. and Mishanina, T. V. (2023). Expression and Purification of Recombinant Human Mitochondrial RNA Polymerase (POLRMT) and the Initiation Factors TFAM and TFB2M. Bio-protocol 13(23): e4892. DOI: 10.21769/BioProtoc.4892.
- Reardon, S. D. and Mishanina, T. V. (2022). Phosphorylation and acetylation of mitochondrial transcription factor A promote transcription processivity without compromising initiation or DNA compaction. J. Biol. Chem. 298(4): 101815.
Category
Molecular Biology > Protein > Expression
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









