- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Three-dimensional Co-culture Model for Live Imaging of Pancreatic Islets, Immune Cells, and Neurons in Agarose Gel
(§ Technical contact) Published: Vol 13, Iss 20, Oct 20, 2023 DOI: 10.21769/BioProtoc.4852 Views: 2612
Reviewed by: Mohan BabuAndrea GramaticaShun Yu Jasemine YangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Cryopreservation of Bulk-Produced Primary Rat Oligodendrocyte Progenitor Cells
Hanki Kim [...] Jun Young Choi
Jun 20, 2025 1410 Views

Protocol for 3D Bioprinting a Co-culture Skin Model Using a Natural Fibrin-Based Bioink as an Infection Model
Giselle Y. Díaz [...] Stephanie M. Willerth
Jul 20, 2025 3752 Views
A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1249 Views
Abstract
During the onset of autoimmune diabetes, nerve–immune cell interactions seem to play an important role; however, there are currently no models to follow and interfere with these interactions over time in vivo or in vitro. Two-dimensional in vitro models provide insufficient information and microfluidics or organs on a chip are usually challenging to work with. We present here what we believe to be the first simple model that provides the opportunity to co-culture pancreatic islets with sympathetic nerves and immune cells. This model is based on our stamping device that can be 3D printed (STL file provided). Due to the imprint in the agarose gel, sympathetic neurons, pancreatic islets, and macrophages can be seeded in specific locations at a level that allows for confocal live-cell imaging. In this protocol, we provide the instructions to construct and perform live cell imaging experiments in our co-culture model, including: 1) design for the stamping device to make the imprint in the gel, 2) isolation of sympathetic neurons, pancreatic islets, and macrophages, 3) co-culture conditions, 4) how this can be used for live cell imaging, and 5) possibilities for wider use of the model. In summary, we developed an easy-to-use co-culture model that allows manipulation and imaging of interactions between sympathetic nerves, pancreatic islets, and macrophages. This new co-culture model is useful to study nerve– immune cell– islet interactions and will help to identify the functional relevance of neuro-immune interactions in the pancreas.
Key features
• A novel device that allows for 3D co-culture of sympathetic neurons, pancreatic islets, and immune cells
• The device allows the capture of live interactions between mouse sympatheticneurons, pancreatic islets, and immune cells in a controlled environment after six days of co-culturing.
• This protocol uses cultured sympathetic neurons isolated from the superior cervical ganglia using a previously established method (Jackson and Tourtellotte, 2014) in a 3D co-culture.
• This method requires 3D printing of our own designed gel-stamping device (STL print file provided on SciLifeLab FigShare DOI: 10.17044/scilifelab.24073062).
Graphical overview
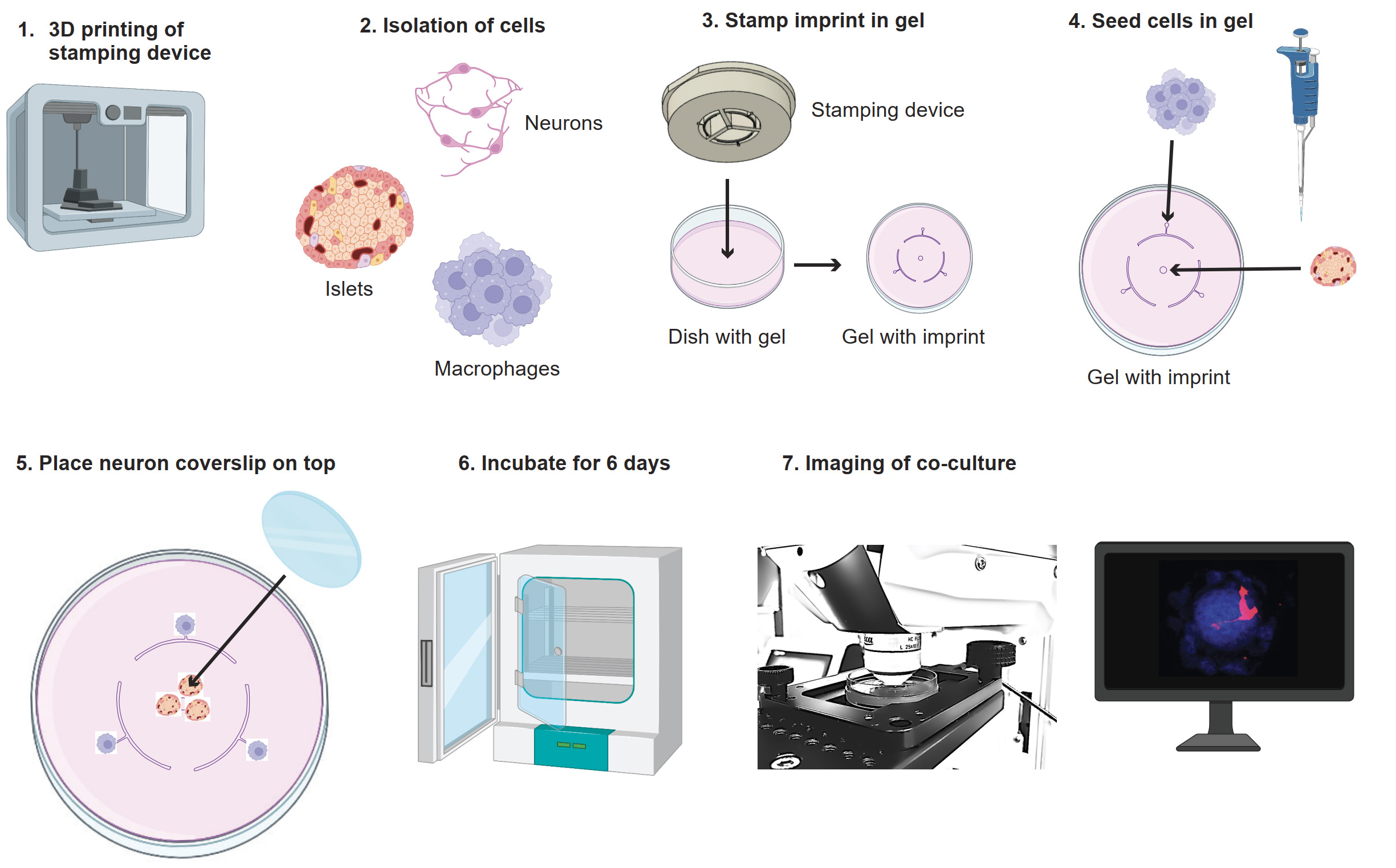
Graphical overview of co-culture model. 1) Print the stamp with a 3D printer. 2) Isolate neurons, islets, and macrophages. 3) Use the stamp to make the imprint in the agarose gel. 4) Seed the macrophages and islets in the agarose gel on their seeding points. 5) Place the coverslip with neurons on top. 6) Incubate the culture for six days. 7) Image the co-culture. Images adapted from BioRender.
Background
It is becoming increasingly clear that the sympathetic nervous system and its interactions with immune cells play an important role in maintaining homeostasis(Martinez-Sanchez et al., 2022). In line with this, type 1 diabetes (T1D) onset in mice halts after depletion of macrophages or sympathetic nerves, via surgical denervation or chemical ablation of nerves (Christoffersson et al., 2020). This shows that sympathetic nerves and macrophages play important roles in T1D onset. Moreover, less severe autoimmune diabetes onset was observed in non-obese diabetic mice (NOD) when the pancreatic nerve was stimulated (Guyot et al., 2019). This suggests that nerve signal modulation might be beneficial for T1D patients in the future and needs to be investigated further. However, currently there are no models to follow and interfere in pancreatic nerve–immune cell interactions over time in vivo.
Injection of neuromodulating agents such as neuropeptides, neurotransmitters, and receptor antagonists in mice has systemic effects, limiting isolation of microenvironmental changes. Separate culture and stimulation of cells in 2D cultures will, on the other hand, provide insufficient information. Microfluidics or organs on a chip are solutions used to measure hormone secretion and more complex cellular behavior in co-culture conditions, but the setup for these microfluidic models is usually challenging. Therefore, we developed a simple 3D co-culture model that allows imaging of interactions between sympathetic nerves, pancreatic islets, and macrophages.
In this protocol, we provide the instructions to construct and perform live cell imaging experiments in our co-culture model. We describe: 1) the design for the stamping device to make the imprint in the gel, 2) the isolation of sympathetic neurons, pancreatic islets, and macrophages, 3) co-culture conditions, and 4) how this can be used for live cell imaging. Finally, we discuss possibilities for a wider use of the model and suggestions for processing of the gel containing islets and cells after imaging for further data collection.
Using this new model, it is possible to study and interfere in neuro-immune interactions in the context of the pancreatic islet in a controlled environment. This environment can be manipulated by addition of neurotransmitters, peptides, or drugs. Instead of macrophages, it would be possible to seed other immune cells, such as T cells, dendritic cells, or monocytes. Due to the three seeding points for cells, it is also possible to seed different immune cells at the same time for example inflammatory and anti-inflammatory macrophages or differently activated Tcells. The versatility of this model makes it useful for studying pancreatic islet physiology and immune attack. In the future, it could also be adapted for the use of pancreatic islet organoids in combination with human immune cells. Altogether, this new co-culture model is useful to study nerve–immune cell–isletinteractions and will help to identify the functional relevance of neuro-immune interactions in the pancreas.
Materials and reagents
Biological materials
Two-day-old pups from a DsRed mouse litter [JAX strain 005441, Tg(CAG-DsRed*MST)1Nagy/J]
Mice expressing GFP [JAX strain 011106, Tg(CAG-GFP*)1Hadj/J]
Wildtype mice (Jackson Laboratory, C57BL/6 mice)
Reagents
Collagenase A, Type 4 (Roche, catalog number: 11088793001), use 10 mg/mL in media
12 mm poly-D-lysine(PLL)/laminin coated German Glass coverslip (Corning BioCoat, catalog number: 354087)
Nerve growth factor (NGF) (Peprotech, catalog number: 450-01)
Trypsin 0.25% EDTA, Mg2+ Ca2+-free (Corning Cellgro®, catalog number: 25-053-Cl)
Fetal bovine serum (FBS) (Sigma, catalog number: F7524-500ML)
Neurobasal media (Gibco, catalog number: 21103049)
B27 supplement (Gibco, catalog number: 17504044)
Penicillin-streptomycin (Pen/strep) 10,000 U/mL (Gibco, catalog number: 15140122)
UltraPure agarose (Invitrogen, catalog number: 1650010)
Low-gelling temperature agarose (Sigma, catalog number: A9045-5G)
RPMI 1640 medium (Thermo Fisher, catalog number: 21875-091)
D-(+)-Glucose (VWR, AnalaR NORMAPUR, catalog number: 101174Y)
Celltracker violet (Invitrogen, catalog number: C10094)
Macrophage colony stimulating factor (M-CSF) (Peprotech, catalog number: 3512-02)
L-glutamine (Life Technologies, catalog number: 25030-024)
Milli-Q (tapped from Purelab Ultra, model: USC214628)
Solutions
Neuron medium (see Recipes)
Islet medium (see Recipes)
Co-culture medium (see Recipes)
Agarose gel (see Recipes)
Low-gelling agarose gel (see Recipes)
Recipes
Neuron medium
Reagent Final concentration Quantity Neurobasal media - 492.8 mL B27 supplement 2% 200 μL Pen/strep 0.5% 2.5 mL L-glutamine 0.5 mM 5 mL Islet medium
Reagent Final concentration Quantity RPMI1640 435 mL FBS 10% 50 mL Pen/Strep 5% 5 mL L-Glutamine 0.5 mM 5 mL D-(+)-Glucose 11.1 mM [1 g in 5 mL] 5 mL Co-culture medium
Reagent Final concentration Quantity Islet medium (from recipe 2) - 50 mL M-CSF 50 ng/mL 50 μL NGF 10 ng/mL 50 μL *M-CSF and NGF are not stable; keep frozen as aliquots (add shortly before use)
Agarose gel
Reagent Final concentration Quantity UltraPure agarose 1.2% 0.48 g Autoclaved Milli-Q
RPMI1640 medium
-
-
10 mL
30 mL
Low-gelling agarose gel
Reagent Final concentration Quantity Low-gelling temperature agarose 1.2% 0.48 g Autoclaved Milli-Q
RPMI 1640 medium
-
-
10 mL
30 mL
Laboratory supplies
6 cm tissue culture dishes (VWR, Avantor, catalog number: 10861-588)
10 cm tissue culture dishes (Thermo Scientific, catalog number: 150464)
96-well U-bottom tissue culture plate (VWR, catalog number: 734-2328)
25 cm cell scraper (VWR, catalog number: 734-2602)
50 mL tubes (Falcon, catalog number: 352070)
50 mL Erlenmeyer (VWR, catalog number: 214-1130)
Parafilm M (VWR, catalog number: 291-1214)
Water bath (Grant, catalog number: TC120)
Equipment
Surgical stereo microscope (Leica, model: MZ10F)
Brightfield microscope (VWR, model: 020025)
Fluorescent microscope (Bio-Rad, ZOE Fluorescent Imager, model: 1450031)
Confocal microscope (Leica, model: TCS SP8, objective model: HC Fluotar L 25×/0.95 W)
FormTM Steri-CylceTM CO2 incubator (Thermo Fisher, model: 371)
Laminar flow hood (Kojair, model: KBS-125)
Microwave (Whirlpool, model VIP 20)
Heating device for imaging (LCI custom heating plate for 6 cm dishes)
3D printer (Asiga, model: MAX X 27)
Cell counting chamber (Neubauer, model: HIRS8100204)
Benchtop centrifuge (Beckman Coulter, model: Allegra X30R)
Software and datasets
ImageJ/Fiji for analyzing obtained imaging data, version number: 1.8.0 (free software)
Microscopy image analysis software, IMARIS, version number: 9.9 (requires a license)
Procedure
This protocol consists of three main parts: 1) the design of the stamp to create the imprint in the gel, 2) the protocol for culture in the device including cell isolation, gel preparation, building the agarose co-culture, and imaging, and 3) suggestions for interaction studies after imaging.
The design of the stamp to create the imprint in the agarose gel
Before the start of this protocol, make sure to 3D print the stamp needed to make the imprint in the agarose gel (Figure 1). The stamping device was 3D-printed in the material PlasGray (Asiga) with an Asiga Max X 27 3D printer (Asiga) with a layer height of 50 μM. The stamp includes holes to prevent air bubble formation during stamping in the agarose gel. The STL file is provided in the supplementary materials and on the SciLifeLab FigShare website DOI: 10.17044/scilifelab.24073062.
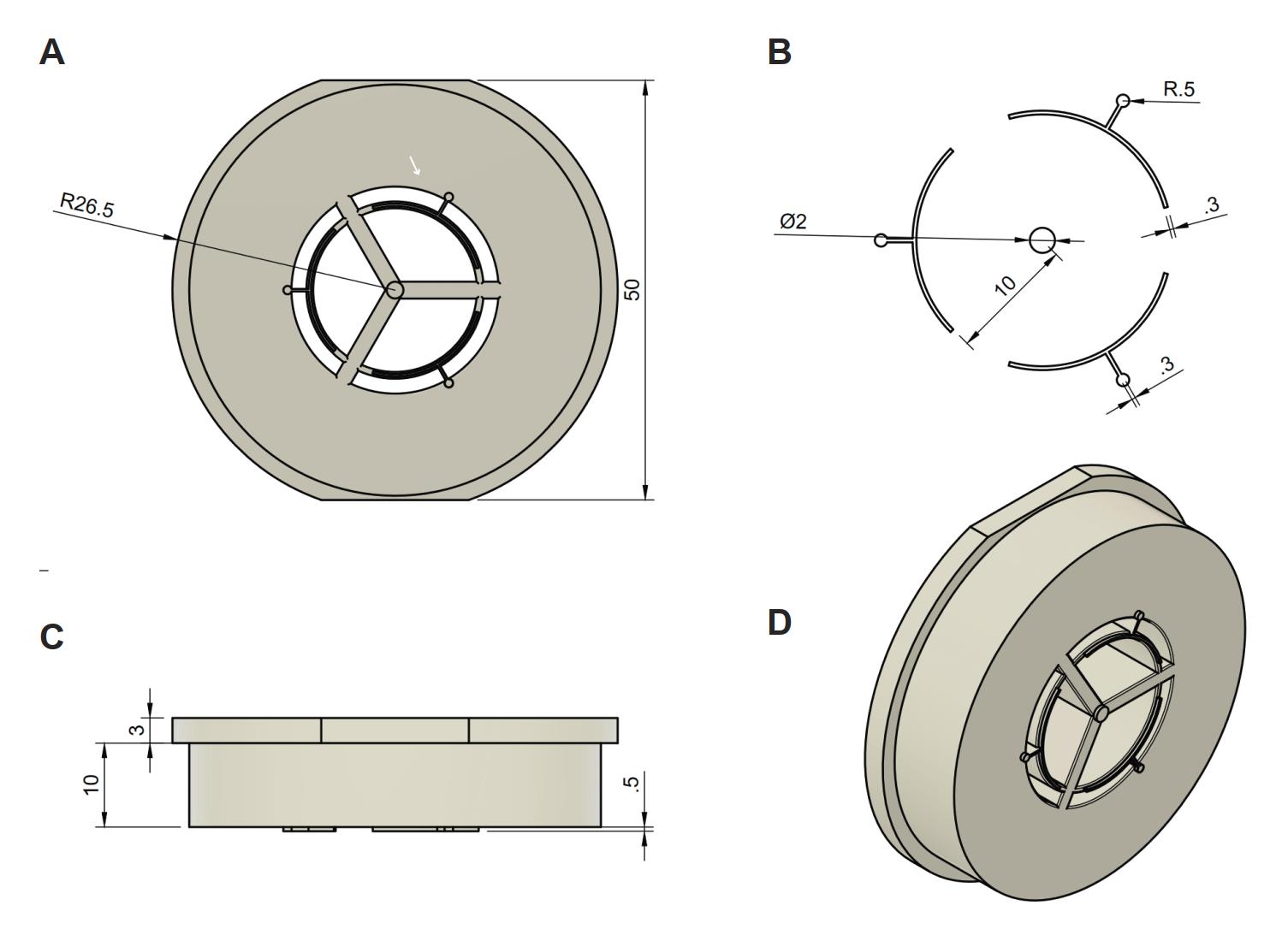
Figure 1. Stamp design. (A) Front view of stamp and measurements: radius (26.5 mm) and width (50 mm). (B) Part of the stamp that creates the imprint in the gel. Measurements for these parts: seeding point width (0.5 mm), capillary width (0.3 mm), islet seeding point width (2 mm), distance from islet seeding point until capillary circle (10 mm). (C) Side view from the device including measurements: height of the part that stays on the dish during stamping (3 mm), height of the stamp that fits in the dish (10 mm), and the depth of the stamp (0.5 mm). (D) Tilted view of the stamp.
Protocol for the co-culture including cell isolation, gel preparation, building the co-culture, and imaging
Isolation and culturing of sympathetic neurons
Isolate superior cervical ganglia from DsRed pups (maximum three days old) as described in Jackson’s protocol “Neuron Culture from Mouse Superior Cervical Ganglion” (Jackson and Tourtellotte, 2014), followed by neuron isolation from the ganglia as described below.
Note: Steps A2–A6 are adapted from Jackson and Tourtellotte (2014) .
Place superior cervical ganglia into a 50 mL tube with ~200 μL collagenase A (10 mg/mL) for a single ganglia (use 400 μL for four ganglia) for 30 min in a 37 °C water bath and agitate every 10 min.
Remove the collagenase solution by carefully pipetting around the ganglia. Replace with approximately 200 μL of 0.25% trypsin for 1–2 ganglia (use 500 μL for four ganglia). Place in 37 °C water bath for 15 min and agitate approximately every 5 min.
Note: The 50 mL tube can be swirled to bring ganglia together in the middle of the tube.
Add 1 mL of neurobasal media to inactivate trypsin and remove both media and trypsin by aspirating media around the ganglia.
Rinse ganglia three times in 1 mL of neuron medium (see Recipes).
Re-suspend ganglia in 200 μL of neuron medium and pipette up and down with barrier tips of 200 μL until the ganglia are no longer visible (~60–70 times).
Use a cell counter (counting chamber) to determine cell number. One ganglion yields approximately 35–40,000 cells.
Dilute neurons to 6,000 neurons/100 μL in neuron medium. Seed 3,000 neurons in droplets of 50 μL per 12 mm PLL/laminin-coated glass coverslip and place in incubator (37 °C, 5% CO2).
After one hour, check with a brightfield microscope if the cells are attached to the coverslip and carefully add an additional 450 μL of pre-warmed neuron medium supplemented with NGF (10 ng/mL) using the side wall of the plate (to not disturb the cells).
Note: In case the neurons are not attached after 2 h, the cells are probably not viable and should not be used.
Culture neurons in incubator (37 °C, 5% CO2) with medium changes every two days. To not disturb the cells, half of the neuron medium can be changed each time.
Note: Since only half of the medium is changed, the added NGF should be doubled (add 250 μL with 20 ng/mL NGF per well to reach a concentration of 10 ng/mL in the full 500 μL. For medium change, tilt the plate at an angle towards you to collect the medium in the bottom and add new medium along the wall of the plate to limit disturbance of the cells.
Neurons are ready to be used for co-culture after four days and can be kept in culture for 28 days (Figure 2A).
Make and stamp agarose gel
Warm up 40 mL of islet medium to 37 °C (see Recipes).
Weigh 0.48 g of agarose in a small Erlenmeyer flask (see also agarose gel recipe).
Note: We recommend using normal agarose to prevent the gel from melting during imaging.
Add 10 mL of autoclaved Milli-Q water to the agarose in a laminar flow hood.
Cover the Erlenmeyer flask with parafilm and heat it up in the microwave until agarose is completely dissolved.
Back in the hood, add 30 mL of pre-warmed 37 °C medium. Mix medium and dissolved agarose together by swirling and pipetting.
Pipet 4 mL of agarose-medium mix into a 6 cm dish, resulting in an approximately 5 mm thick layer.
Note: The pipette tip can be placed against the wall to prevent air bubble formation.
Wait until the gel starts to set (approximately 5–10 min).
Place the stamp on the set gel in the dish and apply light pressure for 10 s to make an imprint in the gel. Remove any remaining gel from the stamp with an ethanol-drenched tissue and quickly continue to the next one (Figure 2B).
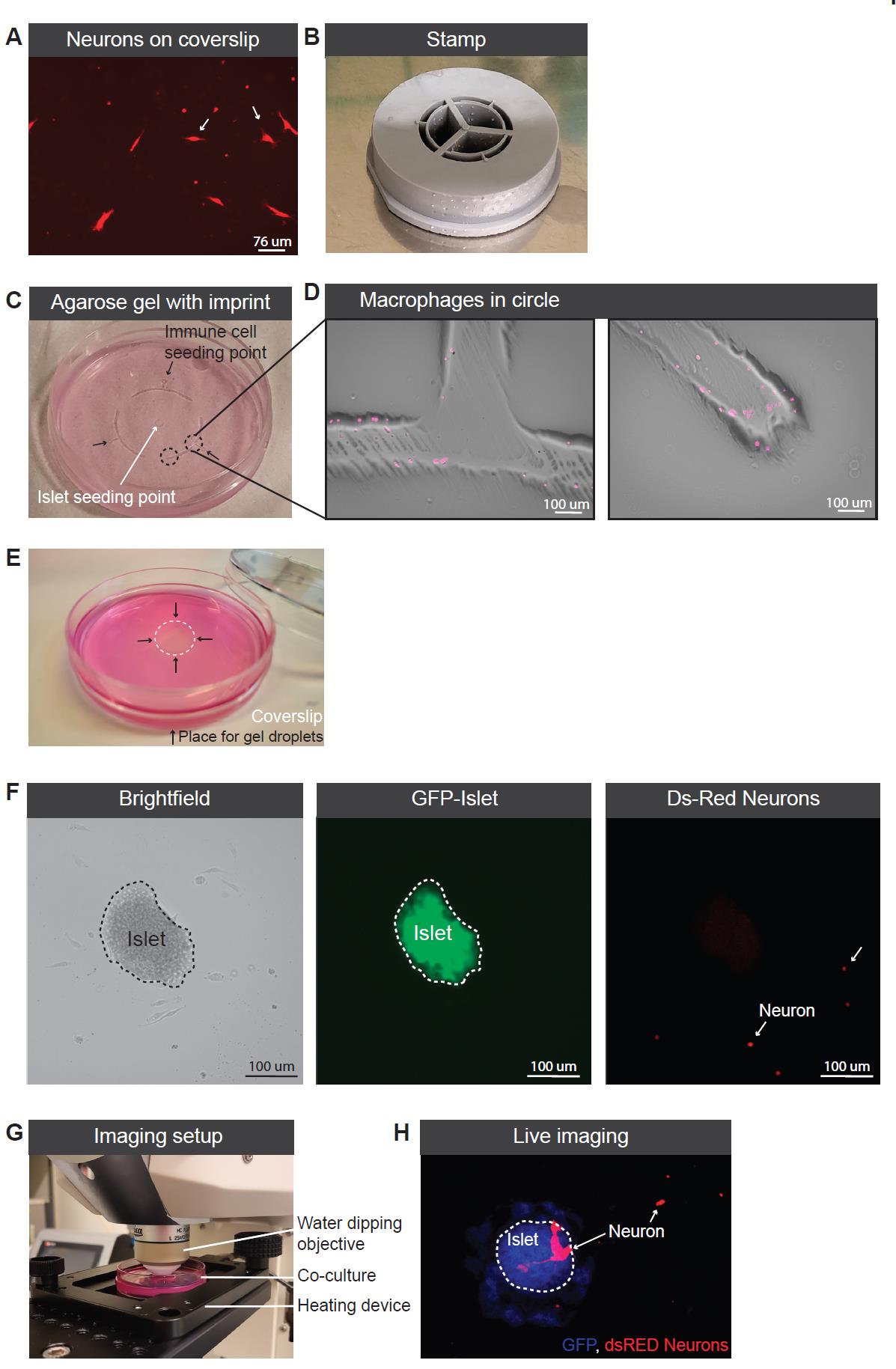
Figure 2. Co-culture setup. (A) Ds-Red neurons growing on coverslip. (B) Picture of the printed stamp. (C) Agarose gel with imprint providing seeding points for the pancreatic islets (white arrow) and the immune cells (indicated with three black arrows). Circled area is enlarged in Figure 2D. (D) Macrophages spread out from the seeding point into the half circled imprinted area. (E) Picture of coverslip and gel droplet placing to secure the location of the coverslip. (F) Picture of an islet in the co-culture model including brightfield to visualize all cells, GFP to visualize the islet, and Ds-Red to visualize the neurons. (G) Imaging setup of the co-culture. (H) Picture of the live imaging using the 488 laser to visualize the islet (indicated with white dotted line) and 512 to visualize the Ds-Red neurons.Let the gel set for an additional 10 min at room temperature without the lid to fully solidify (Figure 2C).
Continue to use the gel for the co-culture or put the lid back on and wrap the plate with parafilm before storage at 4 °C.
Collect pancreatic islets from a GFP mouse
Islets were isolated from a global GFP-expressing mouse as described elsewhere (Bohman et al., 2006).
After isolating using the gradient, hand pick the islets using a stereomicroscope.
Collect eight islets per well in a 96-well U-bottom plate in islet medium and keep in the incubator until ready for seeding (section E).
Isolate macrophages
For macrophages in the co-culture: isolate and grow bone marrow–derived macrophages as described previously in Haag and Murthy (2021).
Collect macrophages from a 10 cm dish using a cell scraper.
If not using macrophages expressing fluorescent protein: stain the macrophages with violet or far red dye cell tracer according to manufacturer’s instructions.
Count macrophages and dilute to 100,000 cells/μL in co-culture medium.
Seed GFP-islets and macrophages in the gel
Place the 6 cm dish containing the imprinted agarose gel (from section A) in a laminar flow hood.
Spin down the plate with islets (200 g, 2 min) and remove the medium in the hood. Resuspend islets in 10 μL of islet medium per well.
Let the islets sink to the lower part of the pipette tip and inject 5 μL of medium with islets into the middle circle of the gel.
Check if the placement was successful using a brightfield microscope.
Proceed with seeding 1 μL of macrophages per seeding point (three in total).
Note: Turn off lights in the hood and use a small light from the side for better visibility of the seeding points.
Check if macrophages are in the seeding points and have spread into the crescents by the capillary effect (Figure 2D).
Note: In case the macrophages do not spread into the crescents, it will not be possible to identify the exact starting location. However, it will still be possible to image interactions between neurons, islets, and macrophages.
Add 200 μL of liquid low melting 1.2% agarose gel on top to seal islets and macrophages in (see Recipes or section B).
Quickly add coverslip containing neurons on top of the new gel, so that it is located above the islets.
Put droplets of low-melting agarose gel on the sides of the glass slide to make sure the coverslip will stay in place (Figure 2E).
Check location of cells and islets in the gel using a brightfield and/or fluorescence microscope.
Add 4 mL of co-culture medium (see Recipes) and put the dish in the incubator.
Change co-culture medium every two days. For medium changes, the complete medium can be pipetted off and replaced with new co-culture medium.
At this stage, stimulants/inhibitors can be added to the medium at any time.
Cells in co-culture can be checked using brightfield or fluorescence microscopes (Figure 2F).
Start imaging after day six to provide the neurons with enough time to reach the islet (Figure 2F).
Imaging of co-culture with neurons, pancreatic islets, and macrophages
Check and mark location of islets and interesting spots, such as macrophages or islets.
Start up the microscope and heating device.
Place the co-culture in the heating device under the microscope (Figure 2G).
Use brightfield or GFP signal to find the islets.
Set the microscope to capture immune cells (violet), pancreatic islets (GFP), and neurons (DsRed).
Set Z-stack capturing the full islet. Immune cells and neurons might be slightly below or above the islet, so check their location and include them in the Z stack if wanted.
Start imaging. For our experiments, we imaged every 2 min during 30 min per islet with a Z slice size of 1–2 μm (depending on the size of the islet) (Figure 2H).
Note: Keep an eye on the fluid levels in the dish during imaging and add more medium if necessary to keep the connection between the objective and the fluid in the dish.
If using sterile imaging in a temperature-controlled chamber, the co-culture could be followed over time by keeping it in a tissue culture incubator and re-image on the required timepoints.
Suggestions for interaction studies after imaging
To study the interactions in the islet area further after imaging, the islets could be cut out of the gel using a stereomicroscope and scalpel. The gel containing islets can be fixed in 4% PFA overnight at 4 °C and used for additional fluorescent staining. Alternatively, it would even be possible to remove the agarose and use the isolated cells for flow cytometry, qPCR, or single-cell RNA sequencing.
Data analysis
For migration analysis, we would recommend making an overview (e.g., tile-scan) image to count all the macrophages that reach the middle. Additionally, the distance from the seeding point to the middle circle is known, making it possible to calculate the migrated distance of immune cells to the islets. If a picture is acquired every day from the start of the co-culture using a fluorescence microscope, the macrophage speed under various conditions can be followed. We would recommend tracking at least 30 macrophages per gel, but this number should be optimized by the end user depending on the goal of the experiment and variation in immune cell behavior.
Images and movies made with the microscope can be further analyzed using IMARIS or ImageJ. Using this software, it is possible to determine cell numbers and cell–cell interactions and follow cell tracks of macrophages, GFP+ cells that migrate out of the pancreatic islet, and DsRed nerves.
Acknowledgments
3D printing was performed at U-PRINT: Uppsala University’s 3D-printing facility at the Disciplinary Domain of Medicine and Pharmacy. We thank Adam Engberg for his advice for the design and printing of the stamping device. Funding: E.M. is funded by a Rubicon grant from the Netherlands Organization for Scientific Research (NWO). G.C. is supported by grants from the Swedish Research Council, the Swedish Society for Medical Research and the Göran Gustafsson foundation, and the Science for Life Laboratory (SciLifeLab). Kind support for the development of this model was also provided by the Family Ernfors Fund.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical considerations
All mouse procedures were approved by the local animal ethics committee for the Uppsala region.
References
- Bohman, S., Andersson, A. and King, A. (2006). No Differences in Efficacy Between Noncultured and Cultured Islets in Reducing Hyperglycemia in a Nonvascularized Islet Graft Model. Diabetes Technol. Ther. 8(5): 536–545.
- Christoffersson, G., Ratliff, S. S. and von Herrath, M. G. (2020). Interference with pancreatic sympathetic signaling halts the onset of diabetes in mice. Sci. Adv. 6(35): eabb2878.
- Guyot, M., Simon, T., Ceppo, F., Panzolini, C., Guyon, A., Lavergne, J., Murris, E., Daoudlarian, D., Brusini, R., Zarif, H., et al. (2019). Pancreatic nerve electrostimulation inhibits recent-onset autoimmune diabetes. Nat. Biotechnol. 37(12): 1446–1451.
- Haag, S. and Murthy, A. (2021). Murine Monocyte and Macrophage Culture. Bio Protoc 11(6): e3928.
- Jackson, M. and Tourtellotte, W. (2014). Neuron Culture from Mouse Superior Cervical Ganglion. Bio Protoc 4(2): e1035.
- Martinez-Sanchez, N., Sweeney, O., Sidarta-Oliveira, D., Caron, A., Stanley, S. A. and Domingos, A. I. (2022). The sympathetic nervous system in the 21st century: Neuroimmune interactions in metabolic homeostasis and obesity. Neuron 110(21): 3597–3626.
Supplementary information
The following supporting information can be downloaded here:
- STL file
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Muntjewerff, E. M., Josyula, V. S. and Christoffersson, G. (2023). Three-dimensional Co-culture Model for Live Imaging of Pancreatic Islets, Immune Cells, and Neurons in Agarose Gel. Bio-protocol 13(20): e4852. DOI: 10.21769/BioProtoc.4852.
Category
Cell Biology > Cell isolation and culture > 3D cell culture
Neuroscience > Cellular mechanisms > Cell isolation and culture
Immunology > Immune cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link







