- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Embryonic Cardiomyocytes and Cell Proliferation Assay Using Genetically Engineered Reporter Mouse Model
Published: Vol 13, Iss 17, Sep 5, 2023 DOI: 10.21769/BioProtoc.4802 Views: 1777
Reviewed by: Pilar Villacampa AlcubierreKyoung-Han KimMarina Sánchez PetidierChao WangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Primary Neuronal Culture and Transient Transfection
Shun-Cheng Tseng [...] Eric Hwang
Jan 20, 2025 2697 Views
Isolation and Characterization of Cervical Cancer-Associated Mesenchymal Stem Cells From Primary Tumors Using Explant Culture
Surbhi Singla [...] Shalmoli Bhattacharyya
Jun 20, 2025 2760 Views
Generation of Intestinal Epithelial Monolayers From Single-Cell Dissociated Organoids
Neta Felsenthal and Danijela Matic Vignjevic
Oct 5, 2025 2298 Views
Abstract
Congenital heart disease (CHD) is often associated with myogenic defects. During heart development, cardiomyocyte growth requires essential cues from extrinsic factors such as insulin-like growth factor 2 (IGF-2). To determine whether and how growth factors account for embryonic cardiomyocyte proliferation, isolation followed by culturing of embryonic cardiomyocytes can be utilized as a useful tool for heart developmental studies. Current protocols for isolating cardiomyocytes from the heart do not include a cardiomyocyte-specific reporter to distinguish cardiomyocytes from other cell types. To optimize visualization of cardiomyocyte proliferation, our protocol utilizes a Tnnt2-promoter-driven H2B-GFP knock-in mouse model (TNNT2H2B-GFP/+) for in vitro visualization of nuclear-tagged cardiomyocyte-specific fluorescence. A cardiomyocyte-specific genetic reporter paired with an effective proliferation assay improves the reproducibility of mechanistic studies by increasing the accuracy of cell identification, proliferated cell counting, and cardiomyocyte tracking.
Key features
• This protocol refines previous methods of cardiomyocyte isolation to specifically target embryonic cardiomyocytes.
• UsesH2B-GFP/+cardiomyocyte reporters as identified by Yan et al. (2016).
• Traces cell proliferation with Phospho-Histone 3 (p-H3) assay.
• Has applications in assessing the role of growth factors in cardiomyocyte proliferation.
Graphical overview
Background
Myocardial expansion is one of the most critical cardiac developmental events. Failure of this process gives rise to many forms of congenital heart disease (CHD), which remains the most common cause of infant death (Zaidi and Brueckner, 2017). During heart development, cardiomyocytes intimately communicate with adjacent nonmyocytes, which promote cardiomyocyte proliferation through paracrine signals (Jang et al., 2022).
To study whether extrinsic cues contribute to embryonic cardiomyocyte proliferation, isolated cardiomyocytes from embryonic hearts can be used to explore the underlying mechanisms of myocardial growth. Here, a newly introduced genetically engineered reporter mouse model (Tnnt2H2B-GFP/+) (Yan et al., 2016) was paired with a proliferation assay. Since previous protocols have outlined the rudimentary trypsin digestion method for embryonic cardiomyocyte isolation (Rodgers et al., 2009), isolated cells were primarily cardiomyocytes but also included cardiac fibroblasts and cardiac endothelial cells. Thus, cardiomyocyte staining would be needed for further tracking of cardiomyocyte-specific proliferation within isolated cells. To monitor cardiomyocyte proliferation over time without the need of further staining, we used a cardiomyocyte specific reporter, cardiac Troponin T (Tnnt2), to definitively confirm cell type for accuracy in subsequent cell counting. With the inclusion of a proliferation assay, this protocol provides a reliable approach for in vitro assessments of growth factor effects on myocardial development.
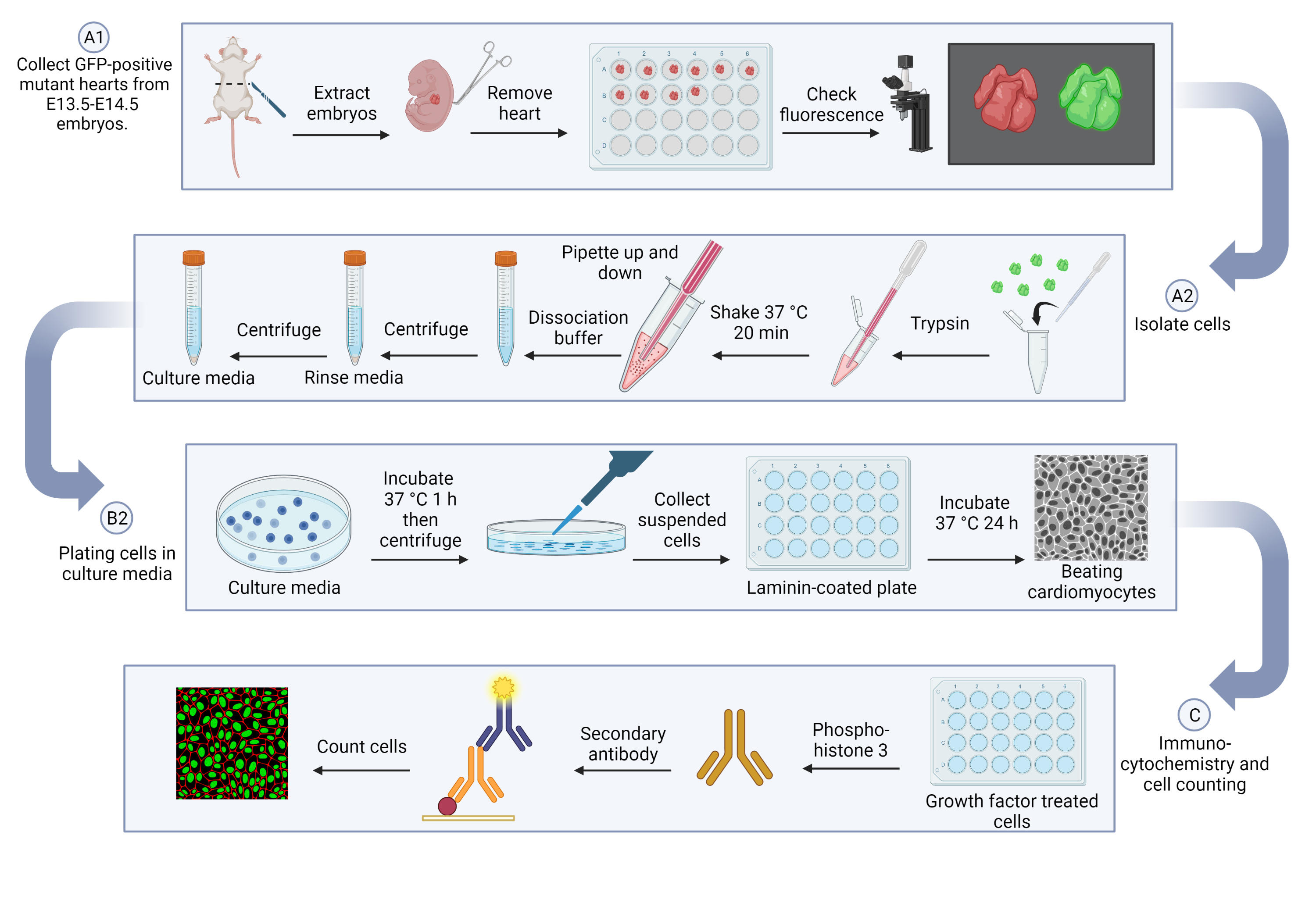
This protocol consists of three key steps: embryonic cardiomyocyte isolation, cardiomyocyte culture, and immunostaining of cell proliferation markers. First, the hearts will be collected and digested with trypsin, and then further separated with dissociation buffer. For cardiomyocyte culture, cells will be pre-plated in culture media to remove other cell types, such as fibroblasts and endothelial cells, and will then be moved into a laminin-coated dish to support myocardial cells. To measure cell proliferation, serum-free media will be added to starve the cells and prevent residual growth factor influences; cells will then be treated with insulin-like growth factor 2 (IGF-2) to stimulate cardiomyocyte proliferation. Finally, immunocytochemistry staining with Phospho-Histone 3 (p-H3) will be applied for counting the proliferating cells as well as total GFP-positive cardiomyocytes.
Taken together, the proliferation assay with the genetic reporter mouse model will allow us to dissect the general mechanisms of the mitotic properties of cells, which can be applied to developmental research.
Materials and reagents
Biological materials
p-H3 (rabbit) (Ser10) antibody (Cell Signaling, catalog number: 9701S), store at -20 °C
Donkey anti-rabbit IgG (H+L), Alexa Fluor 555 (Life, catalog number: A31572)
100 ng/mL IGF-2 recombinant protein (R&D Systems, catalog number: 792-MG-050)
TNNT2H2B-GFP/+mice (from Dr. Chen-Leng Cai, Background Strain: C57BL/6J)
Reagents
1× PBS pH 7.4 (Gibco, catalog number: 10010-023)
1 M HEPES (Gibco, catalog number: 15630-060), store at 4 °C
0.5 M EDTA pH 8.0 (Gibco, UltraPure, catalog number: 15575), store at 4 °C
1:250 Trypsin (Gibco, catalog number: 27250-018)
HBSS solution (Gibco, catalog number: 14175-095)
Horse serum (Gibco, catalog number: 16050-114), store at -80 °C
FBS (Gibco, catalog number: 10437-028), store at -80 °C
100× antibiotic-antimycotic (Gibco, catalog number: 15240-062)
1× DMEM [-] Ca2+(Gibco, catalog number: 21068-028), store at 4 °C
Laminin (Corning, catalog number: 354232), store at -80 °C
1× DPBS (Ca2+/Mg2+) (Corning, catalog number: 21-030-CV)
1× Opti-MEM (Gibco, catalog number: 31985-070), store at 4 °C
IC fixation buffer (Invitrogen, catalog number: 00822249), store at 4 °C
Triton X-100 (Fisher Biotech, catalog number: BP151-500)
Donkey serum (Fisher Brand, catalog number: 03-395-464)
0.1 μg/mL DAPI (1:1,000 working concentration) (Sigma, catalog number: D9542)
1× DMEM [+] Ca2+(Gibco, catalog number: 11965-092), store at 4 °C
Solutions
Trypsin solution (see Recipes)
Dissociation buffer (see Recipes)
Rinse media (see Recipes)
Culture media (see Recipes)
Recipes
Trypsin solution (in 1× HBSS)
Reagent Final concentration Quantity (25 mL) HEPES (1 M) 10 mM 250 μL EDTA (0.5 M) 0.54 mM 27 μL Trypsin 0.25% 0.0625 g HBSS solution (1×) n/a 25 mL Dissociation buffer (in 1× HBSS) at 4 °C
Reagent Final concentration Quantity (25 mL) Horse serum 10% 2.5 mL FBS 5% 1.25 mL HEPES (1 M)
HBSS Solution (1×)
n/a
n/a
250 μL
21.2 mL
Rinse media (in Ca2+free DMEM)
Reagent Final concentration Quantity (50 mL) Horse serum 10% 5 mL FBS 5% 2.5 mL Antibiotic-antimycotic
Ca2+-free DMEM
1%
n/a
0.5 mL
42 mL
Culture media (in Opti-MEM)
Reagent Final concentration Quantity (20 mL) Horse serum 10% 2 mL FBS 5% 1 mL Antibiotic-antimycotic
Opti-MEM
0.1%
n/a
20 μL
17 mL
Note: In isolating and plating the cardiomyocyte step, high quality and optimized concentration of FBS and horse serum are critical for cell viability and cell adherence.
Laboratory supplies
Surgical scissors (Roboz, catalog number: RS-5882)
Curved forceps (Stoelting, catalog number: 52102-54P)
7 mL transfer pipette (Globe Scientific, catalog number: 135038)
24-well culture plate (Denville, catalog number: T1024)
1.5 mL micro-centrifuge tube (Stellar Scientific, T3 Tube, catalog number: T17-300)
60 mm cell culture dish (Eppendorf, catalog number: 0030701111)
70 μm mesh cell strainer (Swish, catalog number: TC70-MT-3)
15 mL centrifuge tube (Denville, Thomas Scientific, catalog number: 1158R11)
Equipment
M205 FCA microscope (Leica)
Hybridization oven/shaker (Amersham Biosciences)
Centrifuge 5425 R (Eppendorf)
TSX60086A -80 °C freezer (Thermo Scientific)
Isotemp CO2incubator (Fisher brand)
Centrifuge 5702 (Eppendorf)
Handheld automated cell counter (Millipore, Scepter 3.0, catalog number: PHCC30000)
60 μm sensors (Millipore, Scepter 3.0, PHCC360050)
-20 °C freezer (Fisher Scientific, Isotemp)
4 °C fridge (Insignia)
BZ-X800 microscope (Keyence)
Standard orbital shaker, Model 5000 (VWR, catalog number: 89032-100)
Software and datasets
Procedure
Cardiomyocyte isolation
Collect GFP-positive mutant hearts from E13.5–E14.5 embryos (see General note 1).
Make the solutions for the experiment and prepare a sterile workspace. Ensure that the PBS is refrigerated ahead of time and kept on ice throughout the duration of the procedure.
Anesthetize dam at E13.5–E14.5 in accordance with IACUC guidelines.
Use a scalpel to make an abdominal incision and extract the embryos with curved forceps. Separate the embryo from the placenta and yolk sac with surgical scissors. Gently remove each heart (see General note 2) using curved forceps and use a transfer pipette to move it into a 24-well plate containing cold PBS. Each heart should be placed in an individually labeled well.
Check the hearts for fluorescence under a fluorescence-emitting microscope (Figure 1) and label the wells accordingly.
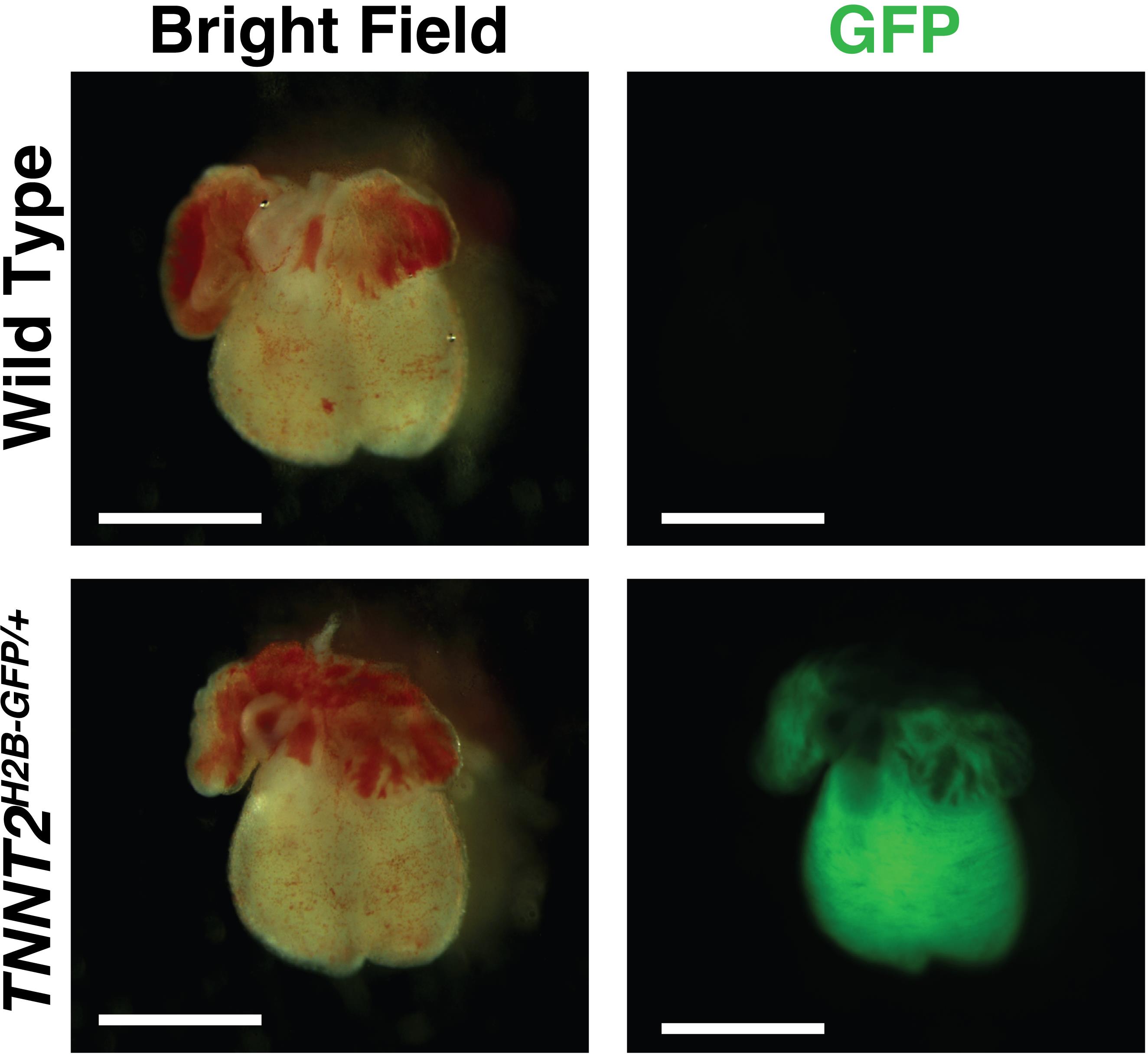
Figure 1. Embryonic day (E) 13.5 hearts from the same litter (TNNT2H2B-GFP/+) imaged in brightfield and GFP fluorescence at 10× magnification. Compared to wild type (above), the TNNT2H2B-GFP/+ mouse (below) has a positive indicator for the GFP-tagged TNNT2 gene. Scale bar, 1 mm.
Isolate cells (see General note 3).
Using a transfer pipette, move each GFP-positive sample from the 24-well plate to a 1.5 mL microcentrifuge tube. All GFP-positive samples from the litter will be pooled into the same tube and digested together. Then, pipette out excess PBS with a micropipette. Add 500 μL of trypsin solution (see Recipes) to the tube and gently pipette the solution up and down with the micropipette to mix.
Note: Each heart can be dissociated individually per embryonic age and experimental needs (see General note 4).
Use tape to secure the microcentrifuge tube to the platform of a hybridization shaker. Ensure that it is positioned such that the solution can move throughout the tube and mix properly. Shake at 70 strokes/min at 37 °C for 20 min.
Remove the tube from the shaker and gently pipette the liquid up and down with a 1 mL pipette 10–15 times until there are no observable tissue pieces.
Transfer the liquid from the 1.5 mL microcentrifuge tube to a 15 mL centrifuge tube using the same micropipette. Add 5 mL of cold (4 °C) dissociation buffer (see Recipes) to the 15 mL centrifuge tube. Pipette up and down with a 7 mL transfer pipette until the cells are disaggregated in the solution.
Centrifuge at 400×gfor 5 min at 4 °C.
In a sterile vent-hood with sanitized surfaces to avoid contamination, vacuum-pipette and discard the resulting supernatant from the tube.
Add 1 mL of rinse media in Ca2+free DMEM (see Recipes) with a micropipette.
Centrifuge at 400×gfor 5 min at 4 °C.
Vacuum-pipette and discard the resulting supernatant from the tube. Add 1 mL of culture media in Ca2+free DMEM (see Recipes) with a micropipette.
Use a new micropipette to disaggregate the pellet and centrifuge again (400×gfor 5 min at 4 °C). The resulting supernatant will be used for plating and should not be discarded.
Protocol continues in step B2.
Plating cells in culture media (see General note 3)
Laminin coating
Note: This step should be completed the day before plating.
To avoid damaging the laminin via excessive freezing and thawing, pipette out laminin (final concentration: 10 μg/mL) with a micropipette and transfer to a 1.5 mL microcentrifuge tube. Return the remaining laminin to a -80 °C freezer.
Dilute the laminin with 1× DPBS (Ca2+/Mg2+) (see General note 5) and add the solution to a 24-well culture plate (see General note 6). Ensure the entire surface is covered by the laminin coating solution.
Seal the plate with parafilm to prevent evaporation and contamination.
Incubate at 2–8 °C overnight. If a more rapid coating is required, incubate at 37 °C for 2 h.
Plating the cardiomyocytes
Using a 5 mL serological pipette attachment for an assisted pipette controller, add 4 mL of culture media to an uncoated 60 mm cell culture dish.
Through a 70 μm mesh cell strainer, add 1 mL of the supernatant from the microcentrifuge tube to the cell culture dish.
Check that the cells are present in the dish under fluorescence microscope (Figure 2).
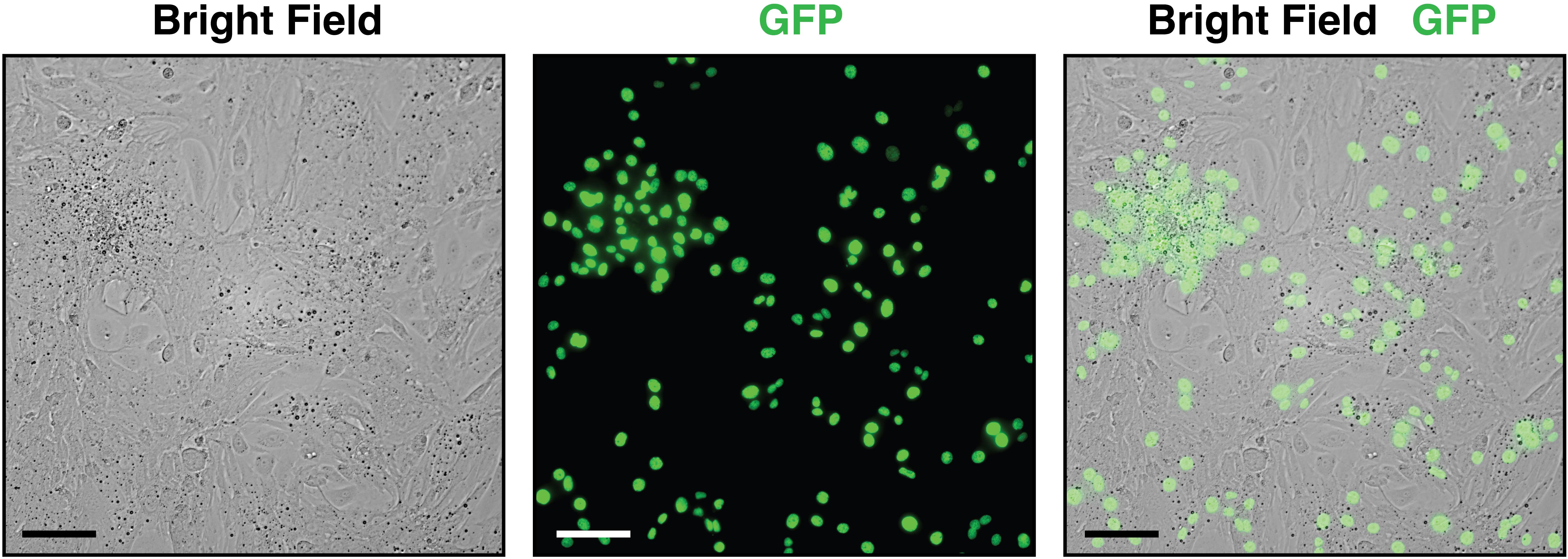
Figure 2. Visualization of cells collected from embryonic day (E) 13.5 hearts at 20× magnification plated in a 48-well cell culture plate with culture media. Brightfield imaging shows all the heart cells present in the dish, whereas GFP fluorescence identifies cardiomyocytes via the H2B-GFP tag. The combined brightfield/GFP image illustrates the ratio of non-cardiomyocyte to cardiomyocyte cells in the embryonic heart, necessitating the subsequent laminin-plating step for isolating cardiomyocytes. Scale bar, 100 μm.Pre-plate the dish for 1 h in a cell culture incubator at 37 °C in 5% CO2to allow nonmyocytes (fibroblasts and endothelial cells) to adhere to the dish.
Centrifuge the whole dish in the Eppendorf 5702 centrifuge with bucket rotors at 400×g for 5 min.
Collect the suspended cells carefully to avoid adherent cells, as they are primarily non-cardiomyocytes (e.g., fibroblasts and endothelial cells).
Load cells into a 60 μm sensor and run through the Scepter 3.0 handheld automated cell counter. The output will determine the size of wells needed for the laminin plating.
Use a vacuum pipette to remove excess laminin solution from the premade 24-well plate.
Transfer the cells to the premade laminin-coated plate (10 μg/cm2).
Incubate at 37 °C in 5% CO2.
After 24 h, cardiomyocytes can be seen beating under a microscope (Video 1) (see General note 7). Remove the solution and replace it with serum-free media (DMEM) for one hour to starve the cells.
Video 1. Embryonic day (E) 13.5TNNT2H2B-GFP/+ cardiomyocytes beating after 24 h in culture mediaWash the cells with PBS.
Treat the cells with 100 ng/mL recombinant IGF-2 protein growth factors.
Immunocytochemistry and cell counting (see General note 8)
Wash the cells with ice-cold PBS.
Fixate cells using 200 μL per well of IC fixation buffer for 10 min at room temperature.
Wash the cells three times with room temperature PBS for five minutes.
Permeabilize the sample by adding 50 µL (or enough to cover the cells) of PBS-T (PBS containing 0.1% Triton X-100) per well for 10 min at room temperature.
Wash the cells three times with room temperature PBS for five minutes.
Incubate cells with 50 µL (or enough to cover the cells) of 5% donkey serum in PBS per well for 1 h to block non-specific antibody binding.
Incubate cells with 50 µL (or enough to cover the cells) of 1:250 diluted primary antibody [p-H3 (rabbit) - donkey anti-rabbit] in 1% donkey serum in PBS per well overnight at 4 °C.
Decant the solution and wash the cells three times in PBS for five minutes.
Incubate cells with 50 µL (or enough to cover the cells) of 1:500 diluted secondary antibody [donkey anti-rabbit IgG (H+L), Alexa Fluor 555] in 1% donkey serum in PBS per well for 1 h at room temperature in the dark.
Decant the secondary antibody solution and wash the cells three times in PBS for five minutes in the dark.
Incubate cells in 50 µL (or enough to cover the cells) of 0.1 μg/mL 1:1,000 DAPI working concentration per well for 1 min.
Remove DAPI and add 200 µL of PBS to each well.
Image cell plate at 20× with Keyence microscope (Figure 3).
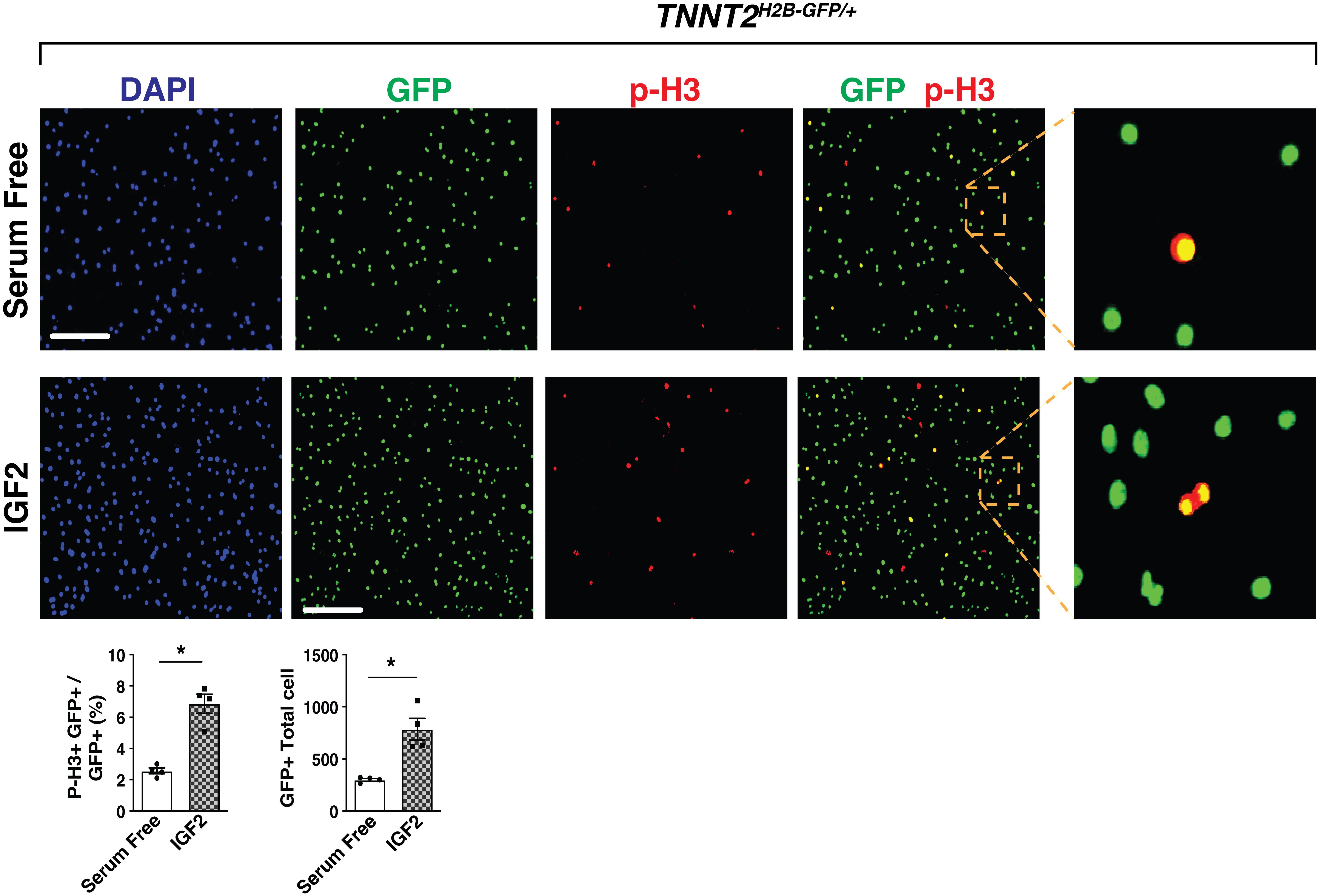
Figure 3. Fluorescence imaging at 20× magnification of p-H3 immunocytochemistry-stained embryonic day (E) 13.5 cardiomyocytes in serum-free media (above) and IGF-2 recombinant protein (below). When exposed to GFP fluorescence, H2B-GFP tagged cardiomyocytes are visible (column 1). Proliferating cells are identified with the p-H3 stain (column 2). The combined GFP/p-H3 image (columns 3 and 4) allows for quantification of GFP+p-H3+ vs. only GFP+ cardiomyocytes in serum free and IGF-2-treated E13.5 heart cells. Scale bar, 275 μm. Percentage of p-H3+ cardiomyocytes and total number of cardiomyocytes were quantitated. IGF2, n = 4 in each group. P-values were determined by the Mann-Whitney U test.Both the total number of cells (Green) and pH3+ cells (Red) are counted by ImageJ image processing software. For total cardiomyocyte number, we counted GFP-positive cardiomyocyte cells in ImageJ. For percentage of p-H3 within total cardiomyocytes, we counted each of p-H3-positive cells and GFP-positive cells.
General notes and troubleshooting
General notes
E13.5 is the ideal collection day for cardiomyocytes. If the hearts are collected prior to E12.5, there may not be enough cells, whereas if the hearts are collected after E14.5 there will be too many fibroblasts present in the samples.
As fluorescence cannot be observed in the whole embryo, hearts must first be removed to check fluorescence. For the purposes of this study, the whole heart was collected, but it is possible at this stage to separate the atria or ventricles if necessary. Additionally, this protocol can be used in wildtype (WT) hearts by staining cardiomyocyte markers such as cardiac Troponin T. GFP+ cell sorting by FACS can be performed before plating the cells.
All cell culture steps require sanitized gloved hands and surfaces in a sterile fume hood to prevent contamination.
As E13.5 hearts are approximately 1–2 mm in size, 500 μL of trypsin solution is enough to digest as many hearts as are collected from one litter.
DPBS with Ca2+and Mg2+should be used to dilute the laminin, since divalent cations are important for protein structure and function.
Based on the number of cells present in the pre-plated cell culture dish, the 24-well plate can be replaced with a 48-well plate (for 1 × 104cells) or a 96-well plate (for 4 × 104cells).
Twenty-four hours after plating the isolated cells, cardiomyocytes visibly beat and will reach confluency 48 h later. In this protocol, we treated the recombinant protein under serum-free conditions and did not passage the cells over seven days. However, in normal serum conditions, we can observe the beating cardiomyocytes 14 days after isolating the cells from the heart.
For each wash step, add approximately 200 μL per well or enough to cover the cells and remove after 5 min with a vacuum pipette; then, proceed to the next step with haste to prevent the cells from drying out. Steps for the immunocytochemistry procedure are performed on a standard orbital shaker.
Acknowledgments
We gratefully acknowledge Dr. Chen-Leng Cai (Indiana University, Indianapolis, Indiana, USA) for providingTNNT2H2B-GFP/+mice. This work was partially supported by American Heart Association-Career Development Award 23CDA1046244 (J.J.), the National Heart, Lung, and Blood Institute R01 grant HL153406 (D.L.) and American Heart Association-Transformational Project Award 20TPA35490132 (D.L.). Graphical overview created with BioRender.com.
Competing interests
There are no conflicts of interest or competing interests.
Ethical considerations
Tnnt2H2B-GFP/+mice were previously described. Animal protocol was approved by the Nationwide Children’s Institutional Animal Care and Use Committee (IACUC).
References
- Jang, J., Song, G., Pettit, S. M., Li, Q., Song, X., Cai, C. l., Kaushal, S. and Li, D. (2022). Epicardial HDAC3 Promotes Myocardial Growth Through a Novel MicroRNA Pathway. Circ. Res. 131(2): 151–164.
- Rodgers, L. S., Schnurr, D. C., Broka, D. and Camenisch, T. D. (2009). An improved protocol for the isolation and cultivation of embryonic mouse myocytes. Cytotechnology 59(2): 93–102.
- Yan, J., Zhang, L., Sultana, N., Oh, J. G., Wu, B., Hajjar, R. J., Zhou, B. and Cai, C. L. (2016). A series of robust genetic indicators for definitive identification of cardiomyocytes. J. Mol. Cell. Cardiol. 97: 278–285.
- Zaidi, S. and Brueckner, M. (2017). Genetics and Genomics of Congenital Heart Disease. Circ. Res. 120(6): 923–940.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Beall, M., Li, D. and Jang, J. (2023). Isolation of Embryonic Cardiomyocytes and Cell Proliferation Assay Using Genetically Engineered Reporter Mouse Model. Bio-protocol 13(17): e4802. DOI: 10.21769/BioProtoc.4802.
Category
Developmental Biology > Cell growth and fate > Proliferation
Cell Biology > Cell isolation and culture > Monolayer culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









