- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
(*contributed equally to this work) Published: Vol 13, Iss 13, Jul 5, 2023 DOI: 10.21769/BioProtoc.4763 Views: 2169
Reviewed by: Alba BlesaValentine V TrotterAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Producing GST-Cbx7 Fusion Proteins from Escherichia coli
Thao Ngoc Huynh and Xiaojun Ren
Jun 20, 2017 11523 Views
Combining Gel Retardation and Footprinting to Determine Protein-DNA Interactions of Specific and/or Less Stable Complexes
Meng-Lun Hsieh [...] Deborah M. Hinton
Dec 5, 2020 3581 Views
Assay for Assessing Mucin Binding to Bacteria and Bacterial Proteins
Lubov S. Grigoryeva [...] Nicholas P. Cianciotto
Mar 5, 2021 4763 Views
Abstract
Toxin–antitoxin (TA) systems are widespread bacterial immune systems that confer protection against various environmental stresses. TA systems have been classified into eight types (I–VIII) based on the nature and mechanism of action of the antitoxin. Type III TA systems consist of a noncoding RNA antitoxin and a protein toxin, forming a ribonucleoprotein (RNP) TA complex that plays crucial roles in phage defence in bacteria. Type III TA systems are present in the human gut microbiome and several pathogenic bacteria and, therefore, could be exploited for a novel antibacterial strategy. Due to the inherent toxicity of the toxin for E. coli, it is challenging to overexpress and purify free toxins from E. coli expression systems. Therefore, protein toxin is typically co-expressed and co-purified with antitoxin RNA as an RNP complex from E. coli for structural and biophysical studies. Here, we have optimized the co-expression and purification method for ToxIN type III TA complexes from E. coli that results in the purification of TA RNP complex and, often, free antitoxin RNA and free active toxin in quantities required for the biophysical and structural studies. This protocol can also be adapted to purify isotopically labelled (e.g., uniformly 15N- or 13C-labelled) free toxin proteins, free antitoxin RNAs, and TA RNPs, which can be studied using multidimensional nuclear magnetic resonance (NMR) spectroscopy methods.
Key features
• Detailed protocol for the large-scale purification of ToxIN type III toxin–antitoxin complexes from E. coli.
• The optimized protocol results in obtaining milligrams of TA RNP complex, free toxin, and free antitoxin RNA.
• Commercially available plasmid vectors and chemicals are used to complete the protocol in five days after obtaining the required DNA clones.
• The purified TA complex, toxin protein, and antitoxin RNA are used for biophysical experiments such as NMR, ITC, and X-ray crystallography.
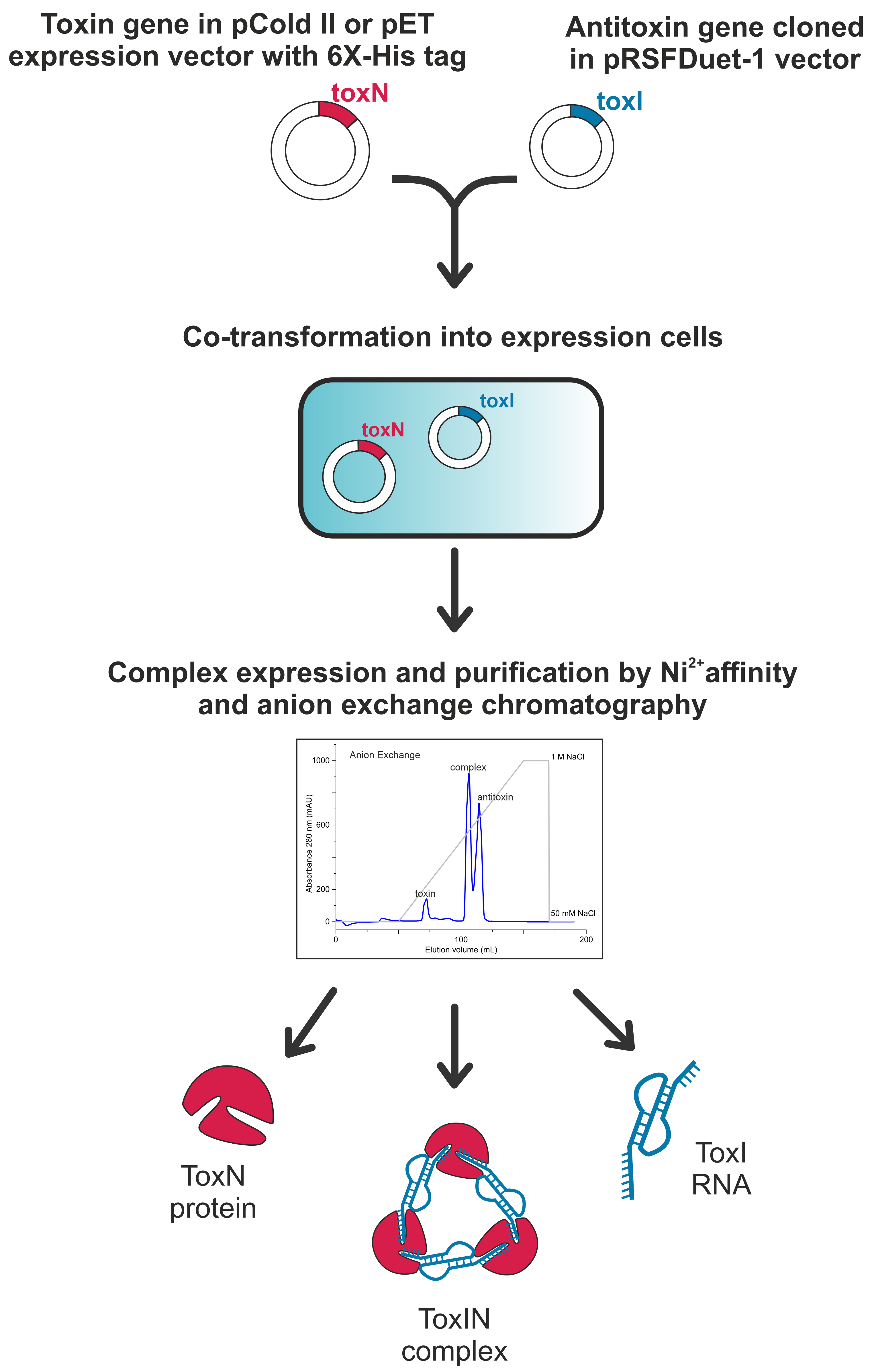
Graphical overview
Background
Type III toxin–antitoxin (TA) systems are genetic modules found in several bacteria, consisting of a protein toxin and a noncoding RNA antitoxin (Fineran et al., 2009). Antitoxin RNA repeats directly bind to the protein toxin, inhibiting its activity by forming a TA ribonucleoprotein (RNP) complex under homeostatic conditions. The primary functions of type III TA systems are attributed to the phage inhibition mechanism in bacteria (Song and Wood, 2020; LeRoux and Laub, 2022; Lin et al., 2023). Type III TA systems are widespread and present in several pathogenic bacteria, including Staphylococcus aureus, Yersinia pseudotuberculosis, and Fusobacterium nucleatum (Blower et al., 2012), and several bacteria in the gut microbiome. Thus, understanding the structure and assembly of type III TA systems could help in devising novel antibacterial strategies by targeting these systems to free the toxins in the pathogenic bacteria, resulting in bacterial growth arrest or death.
The type III TA systems are classified into three families: ToxIN, CptIN, and TenpIN, based on toxin sequence homology (Blower et al., 2012). Among these classes, the ToxIN family (here, protein toxin and antitoxin RNA are called ToxN and ToxI, respectively) is the most studied in terms of its structure, assembly, and functions (Blower et al., 2011; Short et al., 2012; Guegler and Laub, 2021; Manikandan et al., 2022). In a recent study, we classified E. coli ToxIN TA systems into five clusters based on the sequence analysis of the toxin proteins (Manikandan et al., 2022). The structural and functional analysis of a CptIN TA complex from Eubacterium rectale has also been reported (Rao et al., 2015).
It is often difficult to purify free toxins from E. coli due to their inherent toxicity to the bacteria. To circumvent this issue, toxin and antitoxin are often co-expressed and co-purified as non-toxic TA complexes. This is followed by denaturing the TA complex (e.g., in the presence of 6 M guanidinium hydrochloride or 8 M urea) to separate the toxin and antitoxin. The free denatured toxin can then be refolded back to its natural form. However, in vitro refolding of proteins is not always successful. Alternatively, in some cases, active-site toxin mutants have been generated, which can be expressed in E. coli and purified (Samson et al., 2013). While several experiments can be performed using the catalytic mutant protein, a wild-type active protein is often still required to understand the toxins’ mechanism of action.
Here, by adapting the methods published earlier (Blower et al., 2011; Short et al., 2012; Rao et al., 2015; Manikandan et al., 2022), we have optimized a protocol to co-express and co-purify the ToxIN complexes and their components from three clusters of ToxIN family from E. coli (Manikandan et al., 2022) for their biophysical and structural investigation [using X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy]. The method described here yields TA RNP complex, free protein toxin, and free antitoxin RNA of the ToxIN system with an approximate molar ratio of 3:2:2 (with an approximate yield of 7, 3, and 1.5 mg, respectively, from one litre of LB media). The availability of a robust purification method could potentially break the barrier between the discovery of new type III TA systems and their subsequent biophysical characterization to understand their mechanism of action.
Materials and reagents
Biological materials
E. coli BL21(DE3) competent cells (prepared in the lab following a standard protocol) (Sambrook and Russell, 2006)
E. coli DH5α competent cells (prepared in the lab following a standard protocol)
pColdTM II vector (Takara Bio, catalog number: 3362)
pET-21a(+) (Novagen, catalog number: 69740-3)
pETDuetTM-1 vector (Novagen, catalog number: 71146-3)
pRSFDuetTM-1 vector (Novagen, catalog number: 71341-3)
Reagents, chemicals, and kits
Acetic acid (glacial) (Sisco Research Laboratories, catalog number: 85801, CAS number: 64-19-7)
Acrylamide:Bis-acrylamide (19:1) for electrophoresis, 40% solution (Fischer BioReagentsTM, catalog number: BP1406-1, CAS number: 79-06-1), store at 4–8 °C
Agarose low EEO (Sisco Research Laboratories, catalog number: 36601, CAS number: 9012-36-6)
Ammonium persulfate (APS) (Sisco Research Laboratories, catalog number: 65553, CAS number: 7727-54-0)
Ampicillin (Sisco Research Laboratories, catalog number: 61314, CAS number: 69-52-3), store at 2–8 °C
Bradford reagent (Sigma, catalog number: B6916-500 mL), store at 2–8 °C
Bromophenol Blue indicator (Sisco Research Laboratories, catalog number: 11458, CAS number: 115-39-9)
Coomassie brilliant blue (Sisco Research Laboratories, catalog number: 93473, CAS number: 6104-58-1)
D/L-Dithiothreitol (DTT) (Sisco Research Laboratories, catalog number: 17315, CAS number: 3483-12-3), store at 0–4 °C
dNTPs (New England Biolabs, catalog number: N0447S), store at -20 °C
DpnI (New England Biolabs, catalog number: R0176), store at -20 °C
Ethanol (Changshu Hongsheng Fine Chemicals, analytical grade, UN No: 1170)
Ethylenediaminetetraacetic acid (EDTA) (Sisco Research Laboratories, catalog number: 50027, CAS number: 6381-92-6)
Formamide (Sisco Research Laboratories, catalog number: 71714 (062930), CAS number: 75-12-7)
Gel extraction kit (QIAquick®, Qiagen, catalog number: 28704)
Glycerol (Sisco Research Laboratories, catalog number: 62417, CAS number: 56-81-5)
Glycine (Sisco Research Laboratories, catalog number: 64072, CAS number: 56-40-6)
Hydrochloric acid (Sisco Research Laboratories, catalog number: 65955, CAS number: 7647-01-0)
Imidazole (Sisco Research Laboratories, catalog number: 61510-500G)
Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sisco Research Laboratories, catalog number: 67208, CAS number: 367-93-1), store at 0–4 °C
Kanamycin (Sisco Research Laboratories, catalog number: 99311, CAS number: 25389-94-0), store at 2–8 °C
Luria Bertani agar (LA), Miller (Himedia, catalog number: M1151-500G)
Luria broth (LB) (Himedia, catalog number: M575-500G)
Methanol for HPLC (SD Fine Chemical Limited, catalog number: 25217 L25)
NaCl (Sisco Research Laboratories, catalog number: 41721, CAS number: 7647-14-5)
NcoI (New England Biolabs, catalog number: R3193S), store at -20 °C
NdeI (New England Biolabs, catalog number: R0111S), store at -20 °C
PCR purification kit (Qiagen, QIAquick® PCR purification kit, catalog number: 28104)
Phusion High-Fidelity DNA polymerase (New England Biolabs, catalog number: M0530S), store at -20 °C
Plasmid extraction kit (QIAprep® spin Miniprep kit, Qiagen, catalog number: 27106)
Protease inhibitor cocktail (Roche, catalog number: 04693159001), store at 2–8 °C
Quick calf intestinal phosphatase (CIP) (New England Biolabs, catalog number: M0525S), store at -20 °C
Sodium dodecyl sulphate (SDS) (Sisco Research Laboratories, catalog number: 32096, CAS number: 151-21-3)
T4 DNA ligase (New England Biolabs, catalog number: M0202S), store at -20 °C
Tetramethylethylenediamine (TEMED) (Spectrochem private limited, catalog number: 012017), store at 2–8 °C
Toluidine Blue (Sisco Research Laboratories, catalog number: 22134, CAS number: 92-32-9)
Tris (Sisco Research Laboratories, catalog number: 71033-500G)
Urea (Sisco Research Laboratories, catalog number: 21113, CAS number: 57-13-6)
XhoI (New England Biolabs, catalog number: R0145S), store at -20 °C
β-Mercaptoethanol (Sisco Research Laboratories, catalog number: 83759, CAS number: 60-24-2)
Plastic and other materials
Centrifuge bottle 500 mL (Tarsons, catalog number: 544020)
Centrifuge tubes 15 mL (Tarsons, SpinwinTM Tube Conical bottom, catalog number: 520060)
Centrifuge tubes 50 mL (Tarsons, catalog number: 520061)
Membrane filters (0.22 μm) (Merck Life Science private limited, catalog number: GVWP04700)
Membrane filters (0.45 μm) (Merck Life Science private limited, catalog number: HVLP04700)
Microcentrifuge tubes 1.5 mL (Tarsons, catalog number: 500010)
Microcentrifuge tubes 2 mL (Tarsons, catalog number: 500020)
Oakridge centrifuge tubes (50 mL) (Thermo Scientific, catalog number: 3115-0050)
PCR tubes 0.2 mL flat cap (Tarsons, catalog number: 510051)
SnakeSkin dialysis bag 3.5 kDa cut off (dialysis bag) (Thermo Scientific, catalog number: 88242), store at 2–8 °C
Syringe filters (0.22 μm) (Sartorius Minisart®, catalog number: S6534-FMOSK)
Syringe filters (0.45 μm) (Sartorius Minisart®, catalog number: S6555-FMOSK)
Syringes 10 mL (Hindustan Syringes and Medical Devices Limited, Dispo Van, India)
Syringes 20 mL (Hindustan Syringes and Medical Devices Limited, Dispo Van, India)
PCR reaction mixture for all the amplifications (see Recipes)
Double digestion of vector and insert using restriction enzymes (see Recipes)
Ligation reaction of vector and insert (see Recipes)
Lysis buffer (see Recipes)
Wash buffer (see Recipes)
Elution buffer (see Recipes)
Final wash buffer (see Recipes)
Dialysis buffer (see Recipes)
Ion exchange chromatography buffers (see Recipes)
Size exclusion chromatography buffer (see Recipes)
Separating gel for 12% SDS-PAGE (see Recipes)
Stacking gel for SDS-PAGE (see Recipes)
5× loading dye for SDS-PAGE (see Recipes)
Staining solution for SDS-PAGE (see Recipes)
10× running buffer for SDS-PAGE (see Recipes)
SDS-PAGE destaining solution (see Recipes)
Urea-acrylamide gel (see Recipes)
Formamide dye (see Recipes)
SDS-loading dye (5×) (see Recipes)
Tris-borate-EDTA (TBE) buffer (see Recipes)
Recipes
PCR reaction mixture for all the amplifications
Reaction component Volume (μL) (per 20 μL reaction) 5× Phusion HF buffer 4 10 mM dNTPs 0.4 10 μM forward primer 1 10 μM reverse primer 1 Template DNA (100 ng/μL) 1 Autoclaved MilliQ H2O 12.4 Phusion® High-Fidelity DNA Polymerase 0.2 Total volume 20 Double digestion of vector and insert using restriction enzymes
Reaction component Volume (μL) (per 50 μL reaction) 10× Cutsmart buffer 5 Enzyme 1 1 Enzyme 2 1 Vector/insert DNA 5 μg/2 μg Autoclaved MilliQ H2O make up to 50 μL Total volume 50 Ligation reaction of vector and insert
Reaction component Volume (μL) (per 20 μL reaction) 10× ligase buffer 2 Digested purified vector (10 ng/μL)
Insert (10 ng/μL)
5
5
T4 DNA ligase
Autoclaved MilliQ H2O
1
7
Total volume 20 Lysis buffer
Reagent Final concentration Amount NaCl (4 M) 300 mM 37.5 mL Tris-HCl (1 M, pH 7.5)
Imidazole (5 M, pH 8.0)
Glycerol (100%)
β-mercaptoethanol (14.28 M)
50 mM
10 mM
10%
2 mM
25 mL
1 mL
50 mL
70 μL
MilliQ H2O n/a 386.43 mL Total n/a 500 mL Wash buffer
Reagent Final concentration Amount Lysis buffer n/a 49.9 mL Imidazole (5 M, pH 8.0) 20 mM 100 μL Total n/a 50 mL Elution buffer
Reagent Final concentration Amount Lysis buffer n/a 24 mL Imidazole (5 M, pH 8.0) 200 mM 1 mL Total n/a 25 mL Final wash buffer
Reagent Final concentration Amount Lysis buffer n/a 18 mL Imidazole (5 M, pH 8.0) 500 mM 2 mL Total n/a 20 mL Dialysis buffer
Reagent Final concentration Amount NaCl (4 M) 50 mM 25 mL Tris-HCl (1 M, pH 7.5) 50 mM 100 mL DTT 1 mM 308.48 mg MilliQ H2O n/a 1,875 mL Total n/a 2 L Ion-exchange chromatography buffers
Buffer A: same as dialysis buffer.
Buffer B:
Reagent Final concentration Amount NaCl (4 M) 1 M 125 mL Tris-HCl (1 M, pH 7.5) 50 mM 25 mL DTT 1 mM 77.12 mg MilliQ H2O n/a 350 mL Total n/a 500 mL Size-exclusion chromatography buffer
Reagent Final concentration Amount NaCl (4 M) 50 mM 6.25 mL Tris-HCl (1 M, pH 7.5) 20 mM 10 mL DTT 1 mM 77.12 mg MilliQ H2O n/a 483.75 mL Total n/a 500 mL Separating gel for 12% SDS-PAGE (prepare solution volume depending on the gel cast size)
Reagent Amount Acrylamide:Bis-acrylamide (19:1) (40% solution) 2.4 mL 1.5 M Tris, pH = 8.8 2 mL 10% SDS solution 80 μL 10% APS solution 80 μL TEMED 8 μL MilliQ H2O 3.432 mL Total 8 mL Stacking gel for SDS-PAGE (prepare solution volume depending on the gel cast size)
Reagent Amount Acrylamide:Bis-acrylamide (19:1) (40% solution) 0.75 mL 1.5 M Tris, pH = 6.8 1.25 mL 10% SDS solution 50 μL 10% APS solution 50 μL TEMED 5 μL MilliQ H2O 2.9 mL Total 5 mL 5× loading dye for SDS-PAGE
Reagent Amount β-Mercaptoethanol 5% Bromophenol Blue 0.02% Glycerol 30% SDS 10% Tris-Cl (pH = 6.8) 250 mM Staining solution for SDS-PAGE
Reagent Amount Coomassie brilliant blue 1 g Methanol 400 mL Acetic acid 100 mL MilliQ H2O 500 mL Total 1 L 10× running buffer for SDS-PAGE
Reagent Amount Tris base 30 g Glycine 144 g SDS 10 g MilliQ H2O Make up to 1L Total 1 L SDS-PAGE destaining solution
Reagent Amount Methanol 40 mL Acetic acid (glacial) 10 mL MilliQ H2O 50 mL Total 100 mL Urea-acrylamide gel (prepare solution volume depending on the gel cast size)
Reagent Concentration Acrylamide:Bis-acrylamide (19:1) 15% Urea 8 M TBE buffer 1× APS 1% TEMED 0.1% MilliQ H2O Make up the volume as per requirement Formamide dye
Prepared using a CSHL protocol for formamide gel-loading buffer (Cold Spring Harb Protoc, 2013)
SDS-loading dye (5×)
Prepared using a CSHL protocol (Cold Spring Harb Protoc, 2008)
Tris-borate-EDTA (TBE) buffer
Prepared according to a CSHL protocol (Cold Spring Harb Protoc, 2010)
Equipment
Amicon Ultra-15 centrifugal filter unit, 3.5 kDa cutoff (Millipore Sigma, catalog number: UFC9030)
Anion-exchange column (HiTrapTM, Q FF, 5 mL)
Centrifuge (Kubota, Model-6500, serial number: K60115-G000)
Centrifuge for 50 and 1 mL tubes (Eppendorf, model: 5804R)
Dry bath (Bionova, model: SLM-DB-120)
Electrophoresis equipment (BIOBEE® Tech)
Electrophoresis power supply (Bio-Rad, PowerPacTM Basic, 041BR178399)
Fast protein liquid chromatography (FPLC), with fraction collector (GE Healthcare, ÄKTA prime plus)
Laminar airflow (local make)
Mastercycler (Eppendorf Flexi lid Nexus, 6333, serial number: 6333DQ408627)
Mixer (Eppendorf Mixmate 22331 Hamburg, serial number: 5353DN316975)
NanoDrop (Thermo Scientific, NanoDrop 2000c spectrophotometer)
Ni2+-NTA column (GE Healthcare HisTrapTM HP, 5 mL)
Peristaltic pump (Bio-Rad, Econo gradient pump, serial number: 491BR 1897, catalog number: 731-9002)
pH meter (Thermo ScientificTM Orion StarTM A111)
Rotor (Kubota, 6 × 50 mL- AG-506R)
Rotor (Kubota, 6 × 500 mL- AG-5006A)
S200 column (HiLoadTM 16/600 superdexTM 200pg, ID-0059)
SDS-PAGE setup (Invitrogen, Minigel tank)
Shaker and incubator (BioTek, NB-205VQ)
Sonicator (LABMAN Scientific Instruments, model: pro650)
Spectrophotometer (Eppendorf Biophotometer D30, serial number: 6133D0400689)
Tabletop centrifuge (Eppendorf centrifuge 5418, FA-45-18-11 S/N:20881)
Vacuum pump (Tarsons, Rockyvac 400)
Water filtration source (Millipore, Sigma)
Weighing balance, milligram sensitivity (Sartorius, BSA 623S-CW)
Procedure
Molecular cloning of type III antitoxin RNA
Perform the DNA synthesis and cloning of the identified type III TA operon into a high copy number E. coli plasmid vector through a gene synthesis service provider. Sub-clone the antitoxin repeats and its natural promoter into pRSFDuet-1 vector using standard cloning protocols described below (Figure 1A and Supplementary Table 1).
Design forward and reverse primers to amplify the antitoxin region along with the natural promoter and terminator, with addition of restriction enzyme sites NcoI and XhoI at the 5′ and 3′ ends of the insert. PCR amplify the insert from the plasmid containing type III TA operon, using Phusion DNA polymerase (a PCR reaction setup is indicated in Recipe 1 and Table 1).
Table 1. Thermocycling conditions for the PCR reaction
Step Temperature (°C) Duration No. of cycles Initial denaturation 98 30 s 1 Denaturation 98 10 s 30 Annealing Ta 30 s Extension 72 30 s per kb Final extension 72 10 min 1 Hold 4 ∞ - Purify the PCR-amplified insert using the PCR purification kit following the manufacturer’s protocol and elute DNA using nuclease-free water.
Digest ~1–2 μg of the PCR purified insert using the restriction enzymes NcoI and XhoI by incubating at 37 °C for 2 h by setting up a reaction as described in Recipe 2.
Digest ~5 μg of the pRSFDuet-1 vector using the same restriction enzymes in a 50 μL reaction. After digesting the vector, incubate the reaction mixture at 80 °C for 20 min to heat-inactivate the restriction enzymes. Treat the mixture with 0.3 μL of calf intestinal phosphatase (CIP) at 37 °C for 1 h to prevent vector self-ligation.
Purify the digested insert and vector using 1% agarose gel electrophoresis. Visualize the digested insert and vector bands from the ethidium bromide–stained gel by UV irradiation at 254 nm. Excise the gel bands containing the vector and insert and isolate the respective DNA fragments using a gel extraction kit following manufacturer’s protocol; elute DNAs using nuclease-free water.
Use 100 ng of each digested vector and insert DNAs (~1:7 ratio) to set up a 20 μL ligation reaction as described in Recipe 3. Incubate the reaction mixture at room temperature for 1 h followed by incubation at 4 °C for 5–12 h.
Transform 10 μL of the ligation reaction mixture in E. coli DH5α competent cells and plate them in LB agar plate containing kanamycin (0.05 mg/mL) antibiotic. A control ligation reaction and transformation could be performed without the insert DNA in the reaction mixture. The control reaction should result in significantly fewer (~10-fold) colonies than the ligation with the insert.
Isolate plasmid DNA from the colonies obtained upon transformation using a plasmid isolation kit and confirm the positive clones by sequencing.
Molecular cloning of type III toxin protein
Design forward and reverse primers to clone the type III toxin into an E. coli expression vector under an inducible promoter with a hexahistidine tag. The vector must be compatible for co-transformation with the pRSFDuet-1 vector with a different antibiotic resistance and a different origin of replication (e.g., pCold II, pRSFDuet-1, pET21a(+), etc.) In our study, we cloned the type III toxin into a pCold II vector under a cold-shock inducible promoter with an N-terminal hexahistidine tag between the restriction enzyme sites NdeI and XbaI.
Obtain the ligated plasmid containing toxin insert in the vector of interest by following cloning steps A2–A7 of antitoxin cloning using the toxin insert and appropriate vector DNA.
Co-transform 10 μL of the ligation mixture along with 100 ng of the antitoxin DNA containing plasmid in E. coli DH5α competent cells and plate on an LB agar plate containing both kanamycin (0.05 mg/mL) and ampicillin (0.1 mg/mL) antibiotics.
Isolate the plasmid DNA mixture (toxin and antitoxin) from the colonies obtained and confirm the positive toxin clones by sequencing using primers specific to the toxin cloning site. The obtained plasmid DNA mixture will be used further to transform E. coli cells to express and purify the type III TA complex. In our study, the plasmid DNA mixture contains antitoxin and toxin cloned in vectors, as shown in Supplementary Table 1.

Figure 1. A general strategy for cloning type III toxins in expression vectors.(A) Schematic of cloning of antitoxin ToxI RNA in an expression vector. The antitoxin DNA was cloned along with its natural constitutive promoter in pRSFDuet-1 vector. (B) Schematic of cloning of toxin ToxN protein in an expression vector. The toxin DNA was cloned with an N-terminal hexahistidine tag in an IPTG inducible vector such as pCold II. (C) No colonies were obtained upon transformation of plasmids containing only toxin. (D) Positive colonies were obtained upon co-transforming plasmids containing toxin and antitoxin in different compatible vectors.
Transformation and inoculation
Place a vial of E. coli BL21 DE3 competent cells from -80 °C stock on ice to thaw for 15 min.
Add ~100 ng of the plasmid mixture (toxin and antitoxin) and incubate on ice for 20 min.
Provide heat shock by placing the vial in a dry bath at 42 °C for 45 s and immediately transfer the vial on ice and incubate for 5 min.
Add 250 μL of LB medium to the vial in a laminar airflow cabinet and incubate for 1 h in a shaker incubator maintained at 37 °C at 180 rpm.
Spread plate by using 150 μL of this culture on an agar plate containing double antibiotic [corresponding to the plasmid combination chosen; here, we used ampicillin (0.1 mg/mL) and kanamycin (0.05 mg/mL)] and incubate for 12–14 h to obtain the transformed colonies. We did not observe bacterial colonies post-transformation of only toxin plasmid (Figure 1C). However, the co-transformation of toxin and antitoxin plasmids resulted in bacterial colonies selected using dual antibiotics (Figure 1D).
Inoculate 100 mL of LB medium with both antibiotics with a single colony from the transformed plate. Incubate overnight at 37 °C at 180 rpm in a shaker incubator.
Depending on the plasmid in which the toxin and antitoxin are cloned (Table 1), one of the following steps can be followed:
Toxin cloned in pCold II: inoculate 10 mL of the overnight primary culture (in LB media) into a larger secondary culture (1 L of LB media) and incubate until OD600 reaches ~0.8–1.0. Incubate the culture at 15 °C without shaking for 30 min and induce by adding IPTG to a final concentration of 1 mM. Incubate at 180 rpm for 24 h at 15 °C for complex expression. Harvest the cells by centrifugation at 6,800× g for 15 min.
Toxin cloned in pET-21a(+): inoculate 10 mL of the overnight primary culture (in LB media) into a larger secondary culture (1 L of LB media) and incubate until an OD600 ~0.6–0.8. Induce by adding IPTG to a final concentration of 1 mM. Incubate the culture at 180 rpm for 4–5 h at 37 °C for complex expression. Harvest the cells by centrifugation at 6,800× g for 15 min.
Stop point: The cell pellet can be stored at -20 °C for up to a month before processing.
Cell lysis and Ni-NTA affinity chromatography
Resuspend the cells in lysis buffer (see Recipes) and add a tablet of protease inhibitor cocktail.
Lyse the cells by sonication with a pulse of 3 s on and 6 s off and an amplitude of 32%.
Centrifuge the lysed cells at 18,328× g for 45 min at 4 °C in the Oakridge centrifuge tubes.
Equilibrate Ni2+-NTA column connected to a peristaltic pump with lysis buffer. Filter the supernatant using a 0.45 μm syringe filter and load it onto the column at a 1.5 mL/min flow rate. After loading, wash the column with 50 mL of wash buffer (see Recipes). Elute the complex using elution buffer.
Collect three fractions: E1, the first 2 mL; E2, the next 15 mL; and E3, the final 5 mL. Store at 4 °C.
Wash the column with the final wash buffer to remove any bound protein to the column. Pass 25 mL of Milli-Q water and store the Ni2+-NTA column in 20% ethanol.
PAGE analysis of toxin and antitoxin: to analyse the flowthrough, wash, and elution fractions on 12% SDS-PAGE for the presence of toxin protein and on 15% Urea-PAGE for the presence of RNA antitoxin. Visualize the protein in SDS-PAGE gels with Coomassie brilliant blue staining solution followed by destaining using destaining solution (see Recipes). The RNA can be visualized by staining the urea-PAGE gels with 0.25% toluidine blue solution followed by destaining using water.
Dialysis
Collect all the elution fractions that contain the TA components for dialysis (usually, E2 has the TA components).
Dialysis: wash an appropriate length (which can hold the elution fraction volume containing the TA component) of the 6 M dialysis bag with a cutoff of 3.5 kDa with MilliQ H2O and equilibrate with dialysis buffer.
Carefully add the elution fractions into the bag and dialyse at 4 °C for 4–5 h.
Change the dialysis buffer and continue to dialyse for 12–14 h.
Ion-exchange chromatography
Equilibrate the anion-exchange column with 25 mL of ion-exchange buffer A.
Filter the dialysed complex using a 0.45 μm syringe filter and load it onto the anion-exchange column at a 1 mL/min flow rate connected to ÄKTA prime plus FPLC system. After loading, wash the column with 20 mL of ion-exchange buffer A.
Elute the complex by increasing the gradient of NaCl from 50 to 1,000 mM (using buffer A and buffer B), over a volume of 100 mL. This should yield separate fractions of ToxN protein (~0%–20% buffer B), TA RNP complex (~45%–55% buffer B), and antitoxin ToxI RNA repeat (~55%–75% buffer B) (Figure 2A), which can be confirmed by PAGE analysis of protein and RNA components.
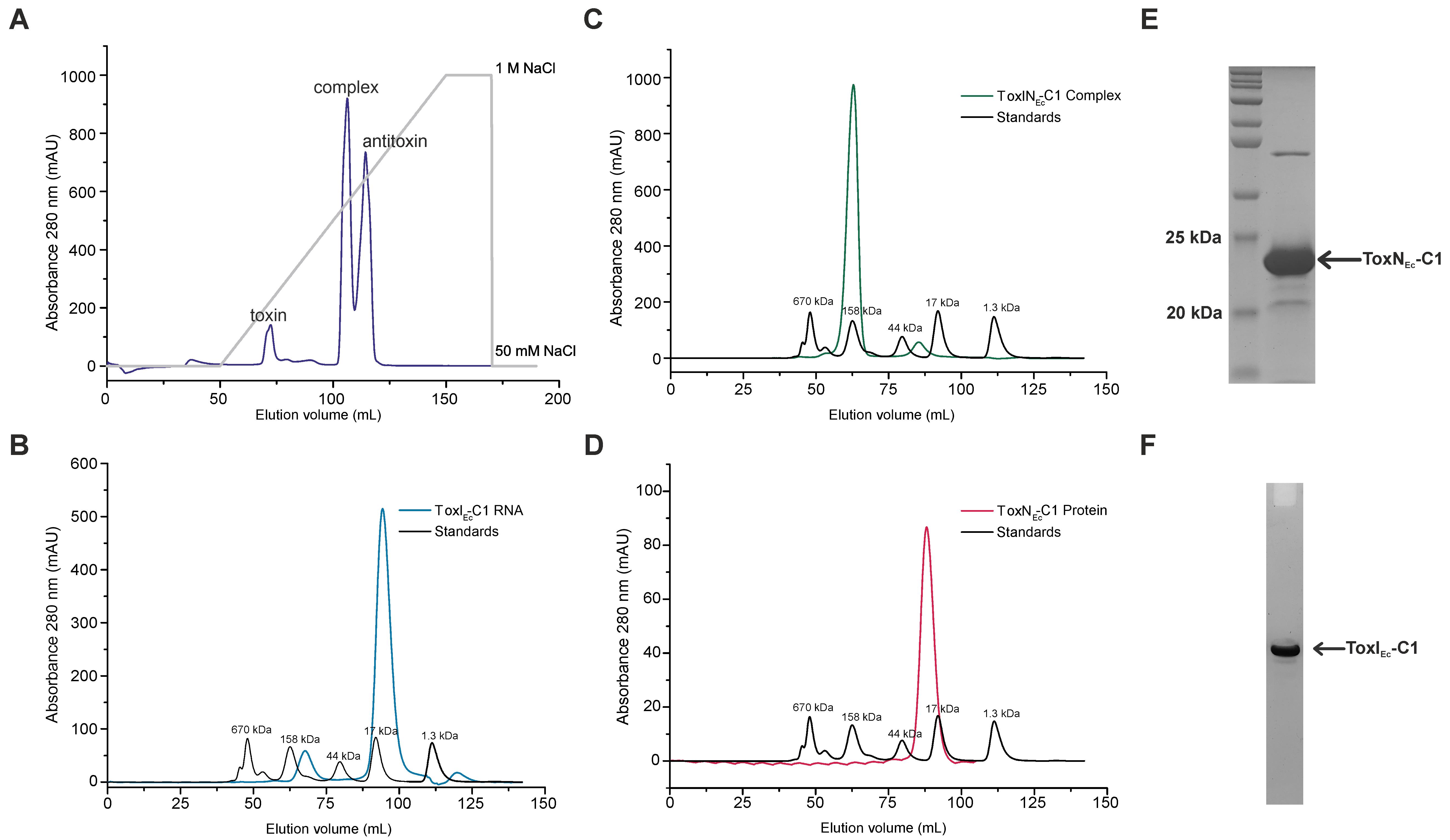
Figure 2. Expression and purification of type III toxin–antitoxin (TA) components from ToxINEc-C1. (A) Anion-exchange chromatography profile of ToxINEc-C1 complex shows the purification of individual toxin, antitoxin, and complex components of ToxINEc-C1. (B–D) Size exclusion chromatography (SEC) profiles of ToxIEc-C1 RNA (B), ToxNEc-C1 protein (C), and ToxINEc-C1 complex (D). The SEC profiles suggest the presence of the purified components in a single oligomeric state. (E) SDS-PAGE analysis of SEC-purified ToxINEc-C1 complex. Gel is stained using Coomassie stain for protein and shows the presence of ToxNEc-C1 protein. (F) Urea-PAGE analysis of SEC-purified ToxINEc-C1 complex. Gel stained using toluidine blue dye for RNA shows the presence of ToxIEc-C1 RNA repeat.
Size exclusion chromatography (SEC)
After ion-exchange chromatography, collect the fractions containing toxin, antitoxin, and complex separately.
Pool each set of fractions separately and concentrate using Amicon® Ultra-15 centrifugal filter (3.5 kDa) to a final volume of ~4 mL.
Filter the concentrated samples using a 0.45 μm syringe filter and inject into Sephacryl S-200 or Superdex S200 columns, pre-equilibrated with SEC buffer. Figure 2B–2D show the SEC elution profiles of antitoxin RNA (Figure 2B), TA RNP complex (Figure 2C), and toxin protein (Figure 2D).
Analyse the fractions on SDS-PAGE and urea-PAGE. Figure 2E and 2F show the SDS-PAGE and urea-PAGE analysis of SEC-purified ToxINEc-C1 complex for the presence of toxin protein and antitoxin RNA, respectively.
The fractions can be pooled together and concentrated appropriately depending on the experiment that needs to be performed, such as NMR spectroscopy, crystallization, or ITC. Using this protocol, we could also successfully purify two other ToxIN complexes and their components (from cluster 4 and 5 of E. coli) (Manikandan et al., 2022) (Supplementary Figure 1 and Supplementary Figure 2).
General notes and troubleshooting
The complete type III TA operon should contain i) the predicted natural promoter (including the -35 and -10 box regions), ii) the antitoxin RNA repeats, iii) the predicted transcription terminator, and iv) the toxin protein coding region (codon optimized for E. coli expression). In our study, the type III TA operon from E. coli ToxIN Cluster 1 (ToxINEc-C1) including natural promoter was synthesized and cloned into a pUC57 vector by GenScript (USA).
It is advisable to run the fractions obtained after every step of purification (Ni2+-NTA affinity chromatography, ion-exchange chromatography, and size exclusion chromatography) on the SDS-PAGE and Urea-PAGE to visualize the protein and RNA, respectively. This also ensures the visualization of impurities in the fractions eluted and whether another round of purification is required before proceeding to the biophysical experiments.
All the buffers used in the protocol were filtered with 0.22 μm sterile filters and used within one week of preparation.
For measuring the concentration of protein, any of the standard methods could be employed, such as absorbance at 280 nm or Bradford’s assay.
Absorbance at 260 nm was used for measuring the concentration of RNA.
In cases where it was difficult to measure the protein concentration accurately, 1:1 ratio of the RNA:protein was assumed to obtain an approximate concentration of the RNP TA complex.
Since the protocol involves the purification of RNA and RNA–protein complexes, it is important to be careful while handling the samples and to keep the columns away from RNases to obtain a good yield.
The toxin and antitoxin of ToxIN systems used in our study were cloned in separate compatible vectors. It is also possible to clone them in the same vector under different promoters to express and purify the components.
The toxin protein and TA complex fractions can be stored at 4 °C (up to a week) and the antitoxin RNA fractions can be stored at -20 °C (up to a month) or -80 °C (for more than a month).
Acknowledgments
The authors acknowledge the financial support received from the Department of Biotechnology (DBT), India (grant number BT/COE/34/SP15219/2015) and IISc-DBT partnership program. M.S. acknowledges the Indo-Poland grant from the Department of Science and Technology (DST), India (DST/INT/POL/P-47/2020). Authors acknowledge the DST and DBT, India, for the NMR and ITC facilities at the Indian Institute of Science, Bangalore. The authors acknowledge funding for infrastructural support from the following programs of the Government of India: DST-FIST, UGC-CAS, and the DBT-IISc partnership program. P.M. acknowledges the research fellowship from CSIR, India. K.N. acknowledges the Prime Minister’s Research Fellowship from the Ministry of Education, India. M.S. is a recipient of STAR award (award number STR/2021/000015) from the Science and Engineering Research Board (SERB), DST, India. This method is validated in the original research article (Manikandan et al., 2022).
Competing interests
There is no competing interest.
References
- Blower, T. R., Pei, X. Y., Short, F. L., Fineran, P. C., Humphreys, D. P., Luisi, B. F. and Salmond, G. P. C. (2011). A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 18(2): 185–190.
- Blower, T. R., Short, F. L., Rao, F., Mizuguchi, K., Pei, X. Y., Fineran, P. C., Luisi, B. F. and Salmond, G. P. C. (2012). Identification and classification of bacterial Type III toxin–antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 40(13): 6158–6173.
- Fineran, P. C., Blower, T. R., Foulds, I. J., Humphreys, D. P., Lilley, K. S. and Salmond, G. P. C. (2009). The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 106(3): 894–899.
- Cold Spring Harb Protoc. (2008). SDS loading dye. doi: 10.1101/pdb.rec11577
- Cold Spring Harb Protoc. (2010). TBE electrophoresis buffer (10X). doi:10.1101/pdb.rec12231
- Cold Spring Harb Protoc. (2013). Formamide Gel-Loading Buffer. doi:10.1101/pdb.rec073510
- Guegler, C. K. and Laub, M. T. (2021). Shutoff of host transcription triggers a toxin-antitoxin system to cleave phage RNA and abort infection. Mol. Cell 81(11): 2361–2373.e9.
- LeRoux, M. and Laub, M. T. (2022). Toxin-Antitoxin Systems as Phage Defense Elements. Annu. Rev. Microbiol. 76(1): 21–43.
- Lin, J., Guo, Y., Yao, J., Tang, K. and Wang, X. (2023). Applications of toxin-antitoxin systems in synthetic biology. Eng. Microbiol. 3(2): 100069.
- Manikandan, P., Sandhya, S., Nadig, K., Paul, S., Srinivasan, N., Rothweiler, U. and Singh, M. (2022). Identification, functional characterization, assembly and structure of ToxIN type III toxin–antitoxin complex from E. coli. Nucleic Acids Res. 50(3): 1687–1700.
- Rao, F., Short, F. L., Voss, J. E., Blower, T. R., Orme, A. L., Whittaker, T. E., Luisi, B. F. and Salmond, G. P. C. (2015). Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein–RNA toxin–antitoxin system. Nucleic Acids Res. 43(19): 9529–9540.
- Sambrook, J. and Russell, D. W. (2006). Preparation and Transformation of Competent E. coli Using Calcium Chloride. Cold Spring Harb. Protoc. 2006(1): pdb.prot3932.
- Samson, J. E., Spinelli, S., Cambillau, C. and Moineau, S. (2013). Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol. Microbiol. 87(4): 756–768.
- Short, F. L., Pei, X. Y., Blower, T. R., Ong, S. L., Fineran, P. C., Luisi, B. F. and Salmond, G. P. C. (2012). Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc. Natl. Acad. Sci. U.S.A. 110(3): E241-249.
- Song, S. and Wood, T. K. (2020). A Primary Physiological Role of Toxin/Antitoxin Systems Is Phage Inhibition. Front. Microbiol. 11: e01895.
Supplementary information
The following supporting information can be downloaded here:
- Supplementary Figure 1. Expression and purification of type III TA components from ToxINEc-Cluster 4.
- Supplementary Figure 2. Expression and purification of type III TA components from ToxINEc-Cluster 5.
- Supplementary Table 1. Table showing the details of source E. coli strains of the ToxIN systems studied and plasmid vectors used to express and purify different ToxIN complexes.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
How to cite
Manikandan, P., Nadig, K. and Singh, M. (2023). Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies. Bio-protocol 13(13): e4763. DOI: 10.21769/BioProtoc.4763.
Category
Microbiology > Microbial biochemistry > Protein > Interaction
Biochemistry > RNA > RNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








