- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Inoculation of Maize with Sugarcane Mosaic Virus Constructs and Application for RNA Interference in Fall Armyworms
Published: Vol 13, Iss 14, Jul 20, 2023 DOI: 10.21769/BioProtoc.4760 Views: 2200
Reviewed by: Zhibing LaiDemosthenis ChronisMalgorzata LichockaJuliane K Ishida

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
GUS Staining of Guard Cells to Identify Localised Guard Cell Gene Expression
Zhao Liu [...] Yu-Ling Chen
Jul 20, 2017 18062 Views
A Microplate-Based Expression Monitoring System for Arabidopsis NITRATE TRANSPORTER2.1 Using the Luciferase Reporter
Yoshiaki Ueda and Shuichi Yanagisawa
Dec 5, 2024 1768 Views
A Novel Gene Stacking Method in Plant Transformation Utilizing Split Selectable Markers
Guoliang Yuan [...] Xiaohan Yang
Feb 20, 2025 1965 Views
Abstract
Virus-mediated transient gene overexpression and gene expression silencing can be used to screen gene functions in plants. Sugarcane mosaic virus (SCMV) is a positive strand RNA virus in the Potyviridae family that has been modified to be used as vector to infect monocots, including maize (Zea mays), for transient gene overexpression and gene expression silencing. Relative to stable transformation, SCMV-mediated transient expression in maize has the advantages of being faster and less expensive. Here, we describe a protocol for cloning constructs into the plasmid vector pSCMV-CS3. After maize seedlings are transformed with pSCMV-CS3 constructs by particle bombardment, the virus replicates and spreads systemically in the plants. Subsequent infections of maize seedlings can be accomplished by rub inoculation with sap from SCMV-infested plants. As an example of a practical application of the method, we also describe virus-induced gene silencing (VIGS) of fall armyworm (Spodoptera frugiperda) gene expression. Transgenic viruses are created by cloning a segment of the fall armyworm target gene into pSCMV-CS3 prior to maize transformation. Caterpillars are fed on the virus-infected maize plants, which make dsRNA to silence the expression of the fall armyworm target gene after ingestion. This use of SCMV for plant-mediated VIGS in insects allows rapid screening of gene functions when caterpillars are feeding on their host plants.
Graphical overview
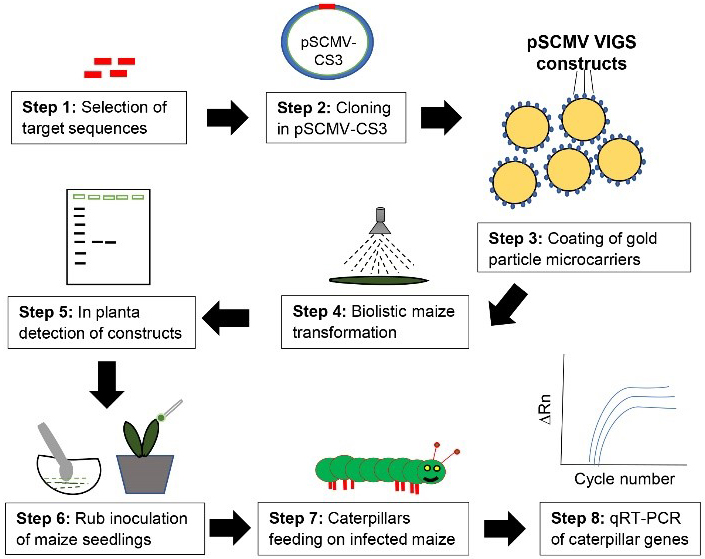
Background
Viruses can be engineered as vectors for overexpression of heterologous coding or non-coding RNA sequences in plants. Additionally, viruses carrying a fragment of a target gene can cause virus-induced gene silencing (VIGS) by activating the internal RNA interference (RNAi) machinery of their host plants (Pasin et al., 2019). The rapid replication of plant viruses makes VIGS an efficient approach for in vivo investigation of gene function in plants.
Several virus vectors have been used for VIGS in monocot plant species, including Chinese wheat mosaic virus (Yang et al., 2018), tobacco rattle virus (TRV) (Zhong et al., 2014), barley stripe mosaic virus (Buhrow et al., 2016), brome mosaic virus (Wang et al., 2021), bamboo mosaic virus, cymbidium mosaic virus (Hsieh et al., 2013), rice tungro bacilliform virus (Purkayastha et al., 2010), cucumber mosaic virus (Tzean et al., 2019), foxtail mosaic virus (Liu et al., 2016), maize rayado fino virus (Mlotshwa et al., 2020), and sugarcane mosaic virus (SCMV) (Mei et al., 2019; Chung et al., 2022). Several of these viruses, including SCMV, have been used for VIGS in maize (Mei et al., 2016; Wang et al., 2016; J. Zhang et al., 2017; Ding et al., 2018; Jarugula et al., 2018; Mlotshwa et al., 2020; Chung et al., 2022)
SCMV is a positive strand RNA virus of the Potyviridae family that infects many monocots including sugarcane, maize, wheat, and sorghum (Xiao et al., 1993). Previously, SCMV has been used as a vector for transient overexpression of non-maize genes (Mei et al., 2019), overexpression of maize endogenous genes (Chung et al., 2021), VIGS of maize endogenous genes (Chung et al., 2022), and plant-mediated VIGS of gene expression in corn leaf aphids (Rhopalosiphum maidis) feeding on maize (Chung and Jander, 2022). The plasmid vector pSCMV-CS3 (Mei et al., 2019; Chung et al., 2022) allows for both efficient gene overexpression and gene expression silencing by RNA interference. The entire SCMV virus is encoded between a cauliflower mosaic virus 35S promoter and a nopaline synthase terminator in the Escherichia coli plasmid pSCMV-CS3. A multiple cloning site, which allows cutting by the enzymes PspOMI, ApaI, PmeI, PstII, and SbfI, is inserted between the P1 and HC-Pro protein-coding regions of the virus. Since the virus is translated as a single polyprotein, prior to being cleaved by virus-encoded proteases, it is essential that there is no in-frame stop codon in the sequence that is cloned into pSCMV-CS3. A NIa protease cleavage site immediately downstream of the multiple cloning site catalyzes the release of the cloned protein from the endogenous viral proteins. Biolistic transformation of pSCMV-CS3 into plant cells causes the virus to be transcribed from the 35S promotor, thereby initiating a systemic viral infection.
The fall armyworm (Spodoptera frugiperda), a lepidopteran insect in the Noctuidae family, is one of the most important pests of maize, causing considerable damage to this important crop worldwide. It is a native North American species that has expanded its range to Africa and Asia in recent years (Rwomushana, 2019). The larval stage preferably consumes the tender shoots and leaves of maize plants (He et al., 2020). Currently available strategies such as use of chemical insecticides (Sisay et al., 2019) and transgenic maize producing Bt (Bacillus thuringiensis) toxin (Niu et al., 2016) are becoming less effective due to resistance that is developing in the pests (Banerjee et al., 2017; Zhang et al., 2020) However, RNAi has emerged as a promising approach for agricultural pest control (Zotti et al., 2018). RNAi of insect genes can be achieved by feeding them on host plants with either stable or transient expression of double-stranded RNA (dsRNA) targeting specific inset genes.
Plant-mediated RNAi has been used to silence expression of lepidopteran genes, including silencing of tobacco hornworm (Manduca sexta) genes by stable (Poreddy et al., 2017) or transient expression of dsRNA in tobacco using TRV (Kumar et al., 2012), and stable expression of dsRNA of cotton bollworm (Helicoverpa armigera) genes in Arabidopsis thaliana (Chen et al., 2019), Nicotiana benthamiana (Bally et al., 2020), cotton (Gossypium hirsutum) (Tian et al., 2015; Mao et al., 2007), and tomato (Lycopersicum esculentum) (Mamta et al., 2016). Most of these studies used expression of RNA in stable transgenic plants, an approach that is a laborious, relatively expensive, and low-throughput strategy for studying gene function in plant pests. By contrast, using viruses as vectors for transient expression of dsRNA is a robust method for generating a rapid and systemic RNAi signal in plants.
Here, we describe a general protocol for infecting maize with pSCMV-CS3 gene constructs, as well as the specific use of SCMV-mediated VIGS for silencing gene expression in fall armyworm caterpillars that are feeding on infected maize plants. When the maize RNAi pathway is activated by SCMV infection, viral dsRNA is cleaved into short interfering RNA (siRNA) of 21–24 nucleotides in length (Ding and Voinnet, 2007). Upon feeding on SCMV-infected maize, fall armyworm caterpillars ingest dsRNA and/or siRNA, which silences expression of the target gene.
Materials and reagents
Plants, insects, vectors, and cell lines
Zea mays inbred line P39 seeds (Maize COOP, http://maizecoop.cropsci.uiuc.edu)
Sweet corn variety Golden Bantam (Burpee Seeds, Warminster Township, PA, or www.amazon.com)
Fall armyworm (Spodoptera frugiperda) eggs (Benzon Research, Carlisle, PA, USA, www.benzonresearch.com)
pSCMV-CS3 (plasmid with a SCMV cloning site #3). This plasmid can be obtained with a material transfer agreement from Iowa State University; contact Dr. Steve Whitham (
swhitham@iastate.edu )Escherichia coli DH5α competent cells (Thermo Fisher, catalog number: 18265017)
pSCMV-GFP and pSCMV-GOI (Gene of Interest)
Oligonucleotides (Integrated DNA Technologies, USA)
Flanking multiple cloning site of pSCMV-CS3
SP7788: 5′-GCACAAATGGTTTCCAACG-3′
SP7789: 5′-ATGTTGCATGTCTTGCATG-3′
Reference maize gene primers for reverse-transcriptase PCR (RT-PCR)
Actin-F, 5′-GGTTTCGCTGGTGATGATGC-3′
Actin-R, 5′-CAATGCCATGCTCAATCGGG-3′
EF-1α-F, 5′-TGGGCCTACTGGTCTTACTACTGA-3′
EF-1α-R, 5′-ACATACCCACGCTTCAGATCCT-3′
Reference fall armyworm gene primers for quantitative reverse transcriptase-PCR (qPCR)
qrSF-RPL13-F, 5′-GCCTTAACCCTGCTTTTGCTAG-3′
qrSF-RPL13-R, 5′-GCTTCGCCCTTCAATACCTTC-3′
qrSF-EF1α-F, 5′-TGGGCGTCAACAAAATGGA-3′
qrSF-EF1α-R, 5′-TCTCCGTGCCAGCCAGAAAT-3′
Primers for VIGS target gene cloning and quantitative PCR (will vary based on the gene of interest)
Materials
Wizard® SV Gel and PCR clean-up system (Promega, catalog number: A9282)
Wizard® Plus SV Minipreps DNA purification system (Promega, catalog number: A1460)
Wizard® SV total RNA isolation system (Promega, catalog number: Z3100)
High-Capacity cDNA Reverse Transcription kits (Applied Biosystems, catalog number: 4374966)
GoTaq® Green Master Mix (Promega, catalog number: M7123)
PowerUpTM SYBRTM Green Master Mix (Applied Biosystems, catalog number: A25742)
Restriction enzymes: PspOMI, SbfI (NEB Biolabs, catalog number: R0653S and R3642)
T4 DNA ligase (Promega, catalog number: M1804)
100 bp DNA ladder (Promega, catalog number: G210A)
Nuclease-free water
Absolute ethanol (Sigma-Aldrich, catalog number: E7023)
Liquid nitrogen
Carborundum powder (Fisher, catalog number: C192-500)
Mortar and pestle
Pellet pestle (Fisher Scientific, catalog number: 12-141-363)
Biolistic Optimization kit [1.0 μm Gold Microcarriers, Macrocarrier disks, stopping screens, and 7,600 kPa (1,100 psi) rupture disks (Bio-Rad Laboratories, catalog number: 165-2279)]
0.1 M spermidine (Sigma-Aldrich, catalog number: 05292-1ML-F)
Pipettes and pipette tips (various)
Agarose low-EEO/multi-purpose/molecular biology grade (Fisher BioReagentsTM, catalog number BP160-100)
Microperforated plastic vented bread bags (30 cm × 60 cm) (www.amazon.com)
Plant trays without holes (28 cm wide × 54 cm long × 6 cm deep), plastic pots (9 cm square, 8 cm deep) (www.amazon.com)
qPCR plates (MicroAmpTM optical 384-well reaction plate with barcode (Applied Biosystems, catalog number: 43-098-49)
Cornell maize soil mix: [0.16 m3 Metro-Mix 360 (Scotts, Marysville, OH, USA), 0.45 kg finely ground lime, 0.45 kg Peters Unimix (Griffin Greenhouse Supplies, Auburn, NY, USA), 68 kg Turface MVP (Banfield-Baker Corp., Horseheads, NY, USA), 23 kg coarse quartz sand, and 0.018 m3 pasteurized field soil]
Fall armyworm artificial diet (Southland Products Inc, Lake Village, AR, USA, www.southlandproducts.net)
50 mg/mL kanamycin stock solution (Sigma-Aldrich, catalog number: K0254-20ML)
CaCl2·6H2O (Sigma-Aldrich, catalog number: 21108-500G)
Tryptone (Sigma-Aldrich, catalog number: T7293-1KG)
NaCl (Sigma-Aldrich, catalog number: S9888-1G)
Yeast extract (Sigma-Aldrich, catalog number: 70161-500G)
Agar (Sigma-Aldrich, catalog number: A1296-500G)
15 cm Petri dishes (Fisher Scientific, catalog number FB0875714)
0.22 μm syringe filters (Corning®, catalog number: CLS431219-50EA)
Monobasic potassium phosphate (KH2PO4) (Sigma-Aldrich, catalog number: P0662-500G)
Dibasic potassium phosphate, (K2HPO4) (Sigma-Aldrich, catalog number: P3786-500G)
Tris base (Sigma-Aldrich, catalog number: T1503-500G)
Acetic acid (Sigma-Aldrich, catalog number: A6283-1L)
EDTA (Sigma-Aldrich, catalog number: E4884-500G)
Glycerol (Sigma-Aldrich, catalog number: G7893-IL)
Buffers and solutions
2.5 M CaCl2 (see Recipes)
LB medium (see Recipes)
Inoculation buffer (see Recipes)
1× TAE buffer (see Recipes)
50% (v/v) glycerol (see Recipes)
70% ethanol (see Recipes)
LB agar plates (see Recipes)
Recipes
2.5 M CaCl2
Dissolve 11.0 g of CaCl2·6H2O in deionized water and make the volume up to 20 mL, filter sterilize with a 0.22 μm filter, and store at 4 °C.
LB medium
1% (w/v) tryptone
1% (w/v) NaCl
0.5% (w/v) yeast extract
pH 7.5
Inoculation buffer (50 mM potassium phosphate)
43.4 mL of 1 M KH2PO4
6.6 mL of 1 M K2HPO4
Bring volume to 1 L with deionized water
pH 6.0
1× Tris-Acetate-EDTA (TAE) buffer
242 g of Tris base
57.1 mL of glacial acetic acid
18.6 g of EDTA
Bring final volume to 1 L with deionized water
This is a 50 × TAE stock solution, dilute 1:50 to make 1× TAE buffer
50% (v/v) glycerol
50 mL of 100% glycerol in 50 mL of deionized water
70% ethanol
70 mL of absolute ethanol in 30 mL of deionized water
LB agar plates
LB medium (see Recipe 2)
1.5% w/v agar
Boil to dissolve the agar
Pour approximately 25 mL of heated LB agar into each Petri dish
Equipment
NanoDrop ND-1000 spectrophotometer (Thermo Fisher, USA)
PDS-1000/He biolistic particle delivery system with metal mesh (Bio-Rad laboratories, USA)
1600 MiniG® automated tissue homogenizer and cell lyser (SPEX® Sample Prep, USA)
QuantStudio 6 Flex real-time PCR system (Applied Biosystems, USA)
Ultrasonic water bath with 40 kHz frequency and 284 W heater power (FS140H, Fisher Scientific, USA)
Plate centrifuge (Benchmark Scientific, USA)
Vortex (American Scientific Products, USA)
Benchtop centrifuge (Thermo Scientific, USA)
Incubator with temperature set at 37 °C
Incubator with temperature set at 28 °C
Shaker with temperature set at 37 °C
Plant growth chambers
Procedure
Selection of target sequence and primer design
Select a sequence of size between 200 and 400 bp from the fall armyworm gene of interest, which should be a multiple of three base pairs, avoiding stop codons (in the antisense direction), and avoiding restriction sites of selected restriction enzymes. Check for off targets in the selected sequence against the fall armyworm and maize genomes using blastn at the National Center for Biotechnology Information (NCBI; https://blast.ncbi.nlm.nih.gov). Select a sequence with no off targets.
Design PCR primers to specifically amplify the selected sequence and clone the sequence into the pSCMV-CS3 vector (Figure 1 and Supplementary information) in the antisense direction using the SbfI and PspOMI restriction enzymes.
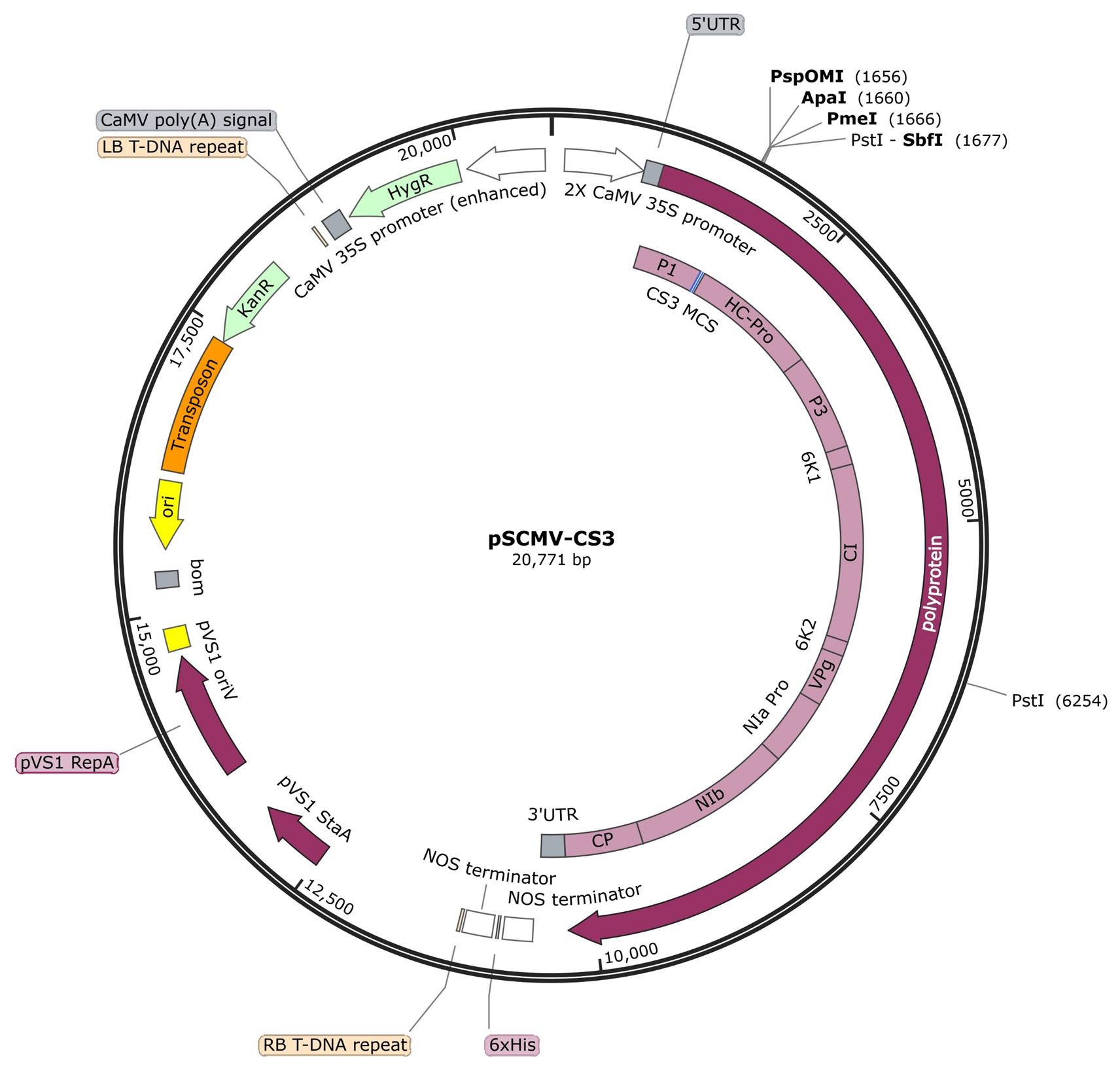
Figure 1. Plasmid map of pSCMV-CS3, showing the restriction sites used for cloning gene fragments into sugarcane mosaic virus (SCMV)Following the same parameters, clone a 240 bp fragment of the jellyfish green fluorescent protein (Gfp) gene into the pSCMV-CS3 vector to use as negative control for VIGS experiments.
Synthesis of SCMV-VIGS constructs
Hatch fall armyworm eggs on fall armyworm diet at 28 °C in an incubator. Use 7-day-old fall armyworm caterpillars for RNA isolation. Freeze caterpillars using liquid nitrogen. Grind the frozen larval tissue in a microfuge tube using a pellet pestle and isolate the total RNA using Wizard® SV Total RNA isolation system following the manufacturer’s instructions (Note 1).
Quantify the RNA and use 1 μg for reverse transcription using High-Capacity cDNA Reverse Transcription kits following the manufacturer’s instructions.
Using the cDNA as a template, amplify the fragment of target gene using 400 nM of each target-specific primer with GoTaq® Green Master Mix.
Analyze the specific amplification by 1.5% agarose electrophoresis using 1× TAE buffer, checking the correct size of the fragments by running a 100 bp DNA ladder in a parallel lane on the gel.
Purify the amplicon using Wizard® SV Gel and PCR clean-up system following the manufacturer’s instructions.
Double-digest the purified amplicons and pSCMV-CS3 vector with the selected restriction enzymes and, after analyzing the digestion by 1.5% agarose gel electrophoresis, purify fragments from gel using Wizard® SV Gel and PCR clean-up system following the manufacturer’s instructions.
Quantify the restricted purified products and ligate the target fragments in linearized pSCMV-CS3 using a 1:3 equimolar end ratio and T4 DNA ligase according to the manufacturer’s instructions.
Transform the ligation mixture into E. coli Top 10 competent cells (1:10 dilution) using a heat shock method and select the transformants after overnight growth at 37 °C on LB agar plates containing 50 μg/mL kanamycin.
Verify insertion of the fragment of interest in pSCMV-CS3 by screening a few colonies by PCR using the SP7788/SP7789 primers (flanking the multiple cloning site of the pSCMV-CS3 vector).
Verify the sequence of the inserted fragments by Sanger sequencing with the SP7788 primer, using plasmid DNA isolated using the Wizard® Plus SV Minipreps DNA purification system from positive colonies after overnight growth at 37 °C with shaking at 180 rpm in LB broth containing 50 μg/mL kanamycin.
Biolistic transformation of SCMV-VIGS constructs into maize and in planta detection of RNA expression
Prepare seedlings at the two-leaf stage by sowing five seeds of maize variety Golden Bantam in a pot containing Cornell maize mix. Allow seeds to germinate and grow for a week in a growth chamber with a 16:8 light/dark cycle at 23 °C with 60% humidity (Figure 2) (Notes 2 and 3). Prepare Gold microcarriers (1.0 μm) at the concentration of 100 mg/mL in 50% glycerol. For this, first vortex 50 mg of gold particles in 1 mL of 70% ethanol for 5 min at 2,000 rpm. Then, keep at room temperature for 15 min and collect the gold particles by centrifugation at 900× g for 20 s. Wash the gold particles three times with 1 mL of sterile water by following this sequence: vortex for 1 min at 2,000 rpm, incubation at room temperature for 1 min, and centrifugation for 10 s. Finally, resuspend particles in 500 μL of 50% glycerol (Note 4).

Figure 2. Maize variety Golden Bantam seedlings at the two-leaf stage for biolistic transformation. Five seedlings are planted in each pot.Place 30 μL of prepared gold microcarriers in a 1.5 mL microfuge tube and vortex for 30 s at 2,000 rpm, followed by sonication for 10 s in an ultrasonic water bath at room temperature with 40 kHz frequency.
Add 5 μg of the pSCMV-VIGS construct during sonication. After 10 s of further sonication, add 25 μL of 2.5 M CaCl2 and 10 μL of 0.1 M spermidine and vortex for 3 min at 2,000 rpm, followed by centrifugation at 2,700× g for 30 s to collect the pellet of gold microcarriers.
Discard supernatant and wash the gold particles with 150 μL of absolute ethanol followed by vortexing at 2,000 rpm for 10 s and centrifugation at 2,700× g for 10 s to collect the gold microcarriers.
Remove the supernatant and resuspend the DNA-coated gold particles in 50 μL of absolute ethanol (see Figure 3 for a flowchart of these steps).

Figure 3. Steps for coating of DNA on gold microcarriersPlace the microfuge tubes having DNA-coated gold microcarriers in an ultrasonic water bath, remove μL of microcarrier suspension, evenly spread it on macrocarrier, and air dry. Prepare four more macrocarriers with the remaining suspension of DNA-coated gold microcarriers.
Arrange the PDS-1000/He biolistic particle delivery system (gene gun) with a microcarrier disk, stopping screen, and 7,600 kPa rupture disk following the manufacturer’s instructions (https://www.bio-rad.com/webroot/web/pdf/lsr/literature/M1652249.pdf). Place one pot of maize seedlings in the instrument chamber and keep the leaves flat against the solid support with a metal mesh that is supplied along with the biolistic particle delivery system (Figure 4). After closing the door, turn on the vacuum and shoot the gold particles when the vacuum reaches 7,600 kPa.

Figure 4. Maize variety Golden Bantam seedlings in the chamber of the PDS-1000/He biolistic particle delivery systemWhen the helium pressure reaches approximately 7,600 kPa, a clear pop is heard. Release vacuum at this point, remove the pot, mist with water, and transfer the plants to a growth chamber.
Transfer the inoculated plants to individual pots the next day and grow them in a growth chamber under a 16:8 h photoperiod at 23 °C for three weeks, until the appearance of viral symptoms.
After three weeks, collect the tissue from the seventh leaf of each plant and immediately place it in liquid nitrogen.
Grind the leaf tissue using 1600 MiniG® automated tissue homogenizer and cell lyser and extract the total RNA using Wizard® SV Total RNA isolation system following the manufacturer’s instructions.
Quantify the RNA and use 1 μg of RNA for reverse transcription using High-Capacity cDNA Reverse Transcription kit following the manufacturer’s instructions.
Use SP7788 & SP7789 primers for PCR amplification and analyze the expression of target sequence in maize. Maize actin and EF-1α gene are amplified as reference genes using Actin-F, Actin-R & EF-1α-F, and EF-1α-R primer pairs. Analyze the amplicons by 1.5% agarose gel electrophoresis (Note 5).
Preparation of sap and rub inoculation of maize with SCMV-VIGS constructs
Separate the whole seventh leaf from the infected plant in which the expression of dsRNA has been confirmed, grind the tissue finely in a mortar and pestle in the presence of liquid nitrogen, wait for a few minutes until sap comes out of tissue, add inoculation buffer in 1:10 ratio (w/v), and grind again to prepare sap for inoculation (Figure 5A) (Note 6).
Grow maize inbred line P39 plants in individual pots in a growth chamber with a 16:8 h photoperiod at 23 °C for one week until the two-leaf stage (Note 7).
Spray the seedling leaves with deionized water and dust (light sprinkling) the leaves with 600 mesh carborundum.
Take a cotton swab, wet it with prepared maize sap, and use it to rub the top surface of both leaves (Figure 5B) (Note 8).
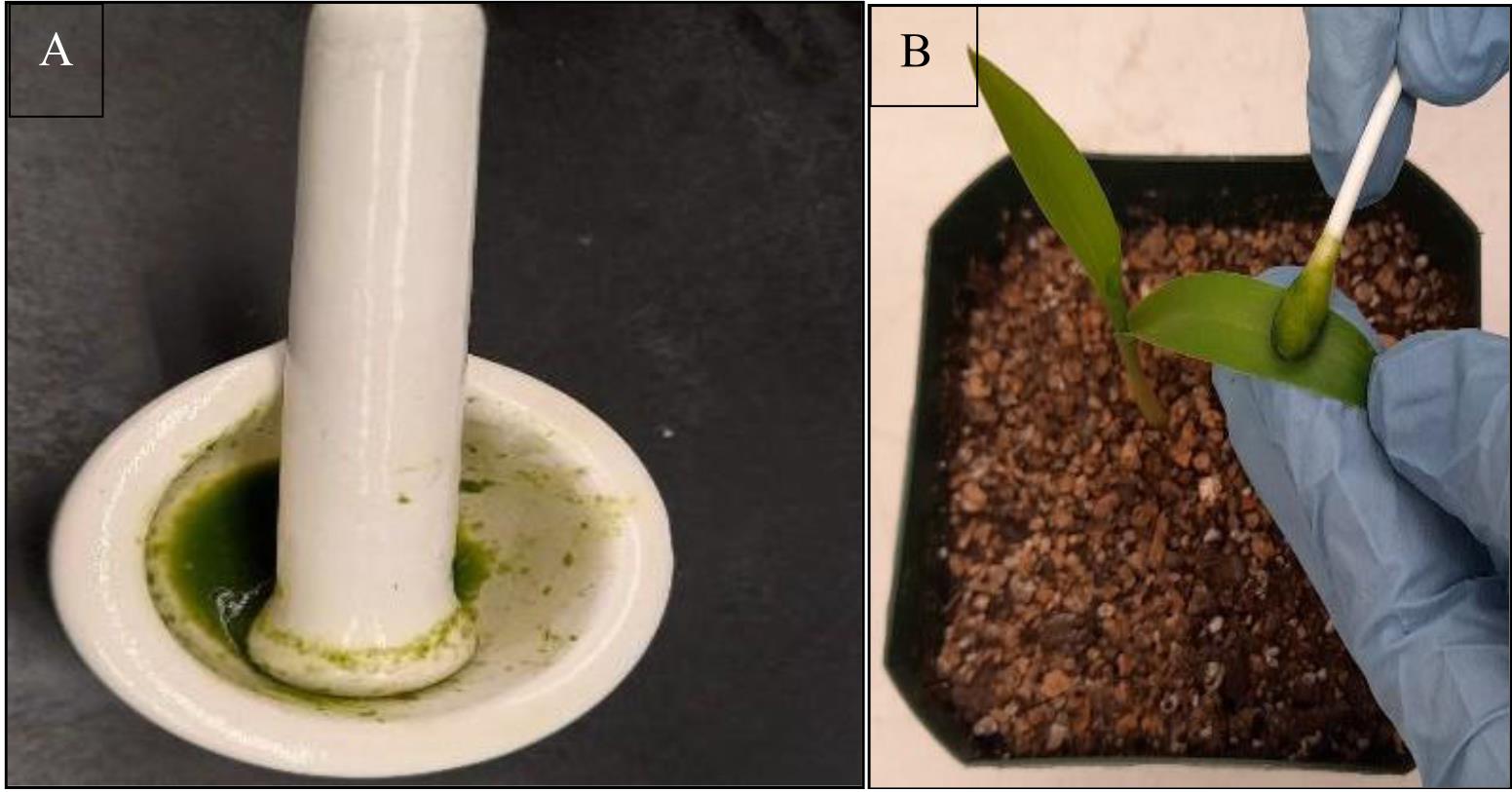
Figure 5. Sap preparation (A) and rub inoculation (B). Seven-day-old maize seedlings of inbred line P-39 at the two-leaf stage are rub inoculated with sap prepared from plants infected with pSCMV-VIGS constructs.Transfer the plants to the growth chamber and look for the appearance of viral infection symptoms after three weeks under a 16:8 h photoperiod at 23 °C.
The expression of viral RNA can be confirmed by PCR as described in section C, step 15, as plants without viral symptoms may also have active infections and expression of the gene of interest (Figure 6).
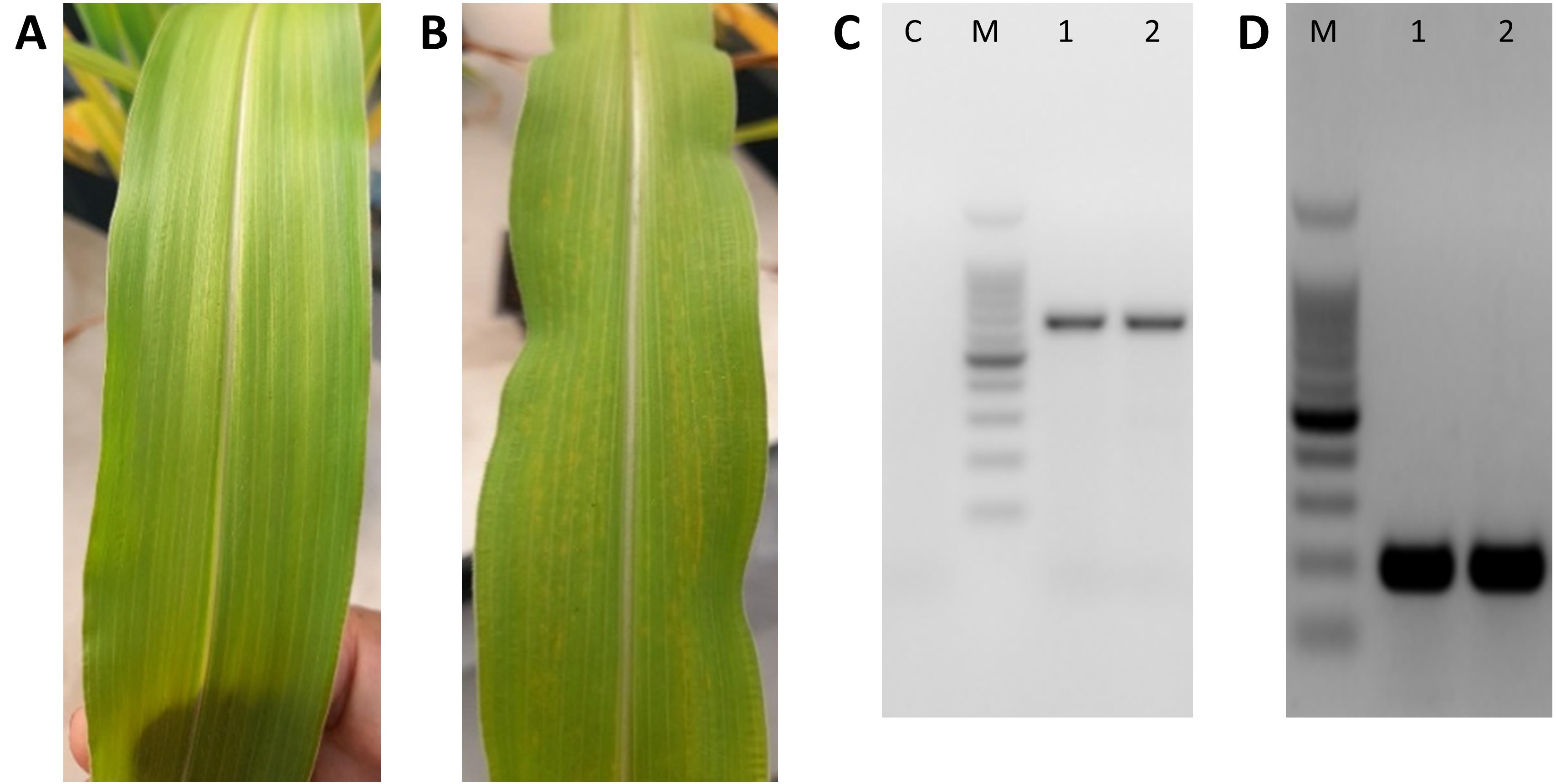
Figure 6. Maize leaves collected from sugarcane mosaic virus (SCMV)-induced gene silencing (VIGS)-infected plants and confirmation of viral infection by PCR. Leaves without (A) and with (B) mosaic symptoms can be positive for SCMV infection. (C) 1.5% agarose gel electrophoresis analysis of PCR for dsRNA from leaves with (lane 1) and without (lane 2) viral symptoms; negative control using water (lane C). (D) 1.5% agarose gel electrophoresis analysis of PCR for maize reference genes actin (lane 1) and EF-1α (lane 2) in the same virus-infected maize leaves.
Caterpillar bioassay for SCMV VIGS in maize and measurement of gene silencing in caterpillars by qRT-PCR
Three weeks after infecting the maize plants with pSCMV-GOI and pSCMV-GFP control constructs by rub inoculation, put five pre-weighed two-day-old caterpillars onto each plant. One caterpillar will be used for measurement of gene silencing after three days of feeding. Growth of the other caterpillars will be monitored after seven days of feeding. Cover the plants with the perforated plastic bags and tie the bags around the stems of the plants with wire twist ties to prevent caterpillar escape (Figure 7).

Figure 7. Bioassays with Spodoptera frugiperda caterpillars. (A) Experimental setup for caterpillar bioassay and gene expression analysis, starting three weeks post rub inoculation. Five two-day-old caterpillars are confined on each plant. The plants are covered with perforated bread bags, which are tied around the plant stem with wire twist ties. (B) Spodoptera exigua (fall armyworm) on a maize leaf.Transfer the plants back to the growth chamber under the same conditions (16:8 h photoperiod at 23 °C) and monitor the growth of caterpillars after seven days by collecting and weighing the surviving caterpillars.
For measurement of gene silencing, collect one caterpillar from each plant after three days of feeding on pSCMV-VIGS- and pSCMV-GFP-infected plants, place it in a 1.5 mL microfuge tube, and immediately place the tube in liquid nitrogen.
Grind collected caterpillars with a pellet pestle tissue homogenizer and extract the total RNA using Wizard® SV Total RNA isolation system following the manufacturer’s instructions.
Quantify the RNA using a Nanodrop and synthesize the cDNA with 1 μg of RNA using High-Capacity cDNA Reverse Transcription kit following the manufacturer’s instructions.
Perform quantitative PCR using a QuantStudio 6 Flex real-time PCR system, with a five-fold dilution of cDNA samples using qPCR primers and PowerUpTM SYBRTM Green Master Mix. The expression levels are normalized using the fall armyworm RPL13 and EF1α reference genes (with primers qrSF-RPL13-F, qrSF-RPL13-R, qrSF-EF1α-F, and qrSF-EF1α-R) as internal controls.
The gene expression levels are calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001) (Figure 8). Gene expression levels of caterpillars fed on pSCMV-GOI infected plants is compared to those on control plants (pSCMV-GFP infected plants).
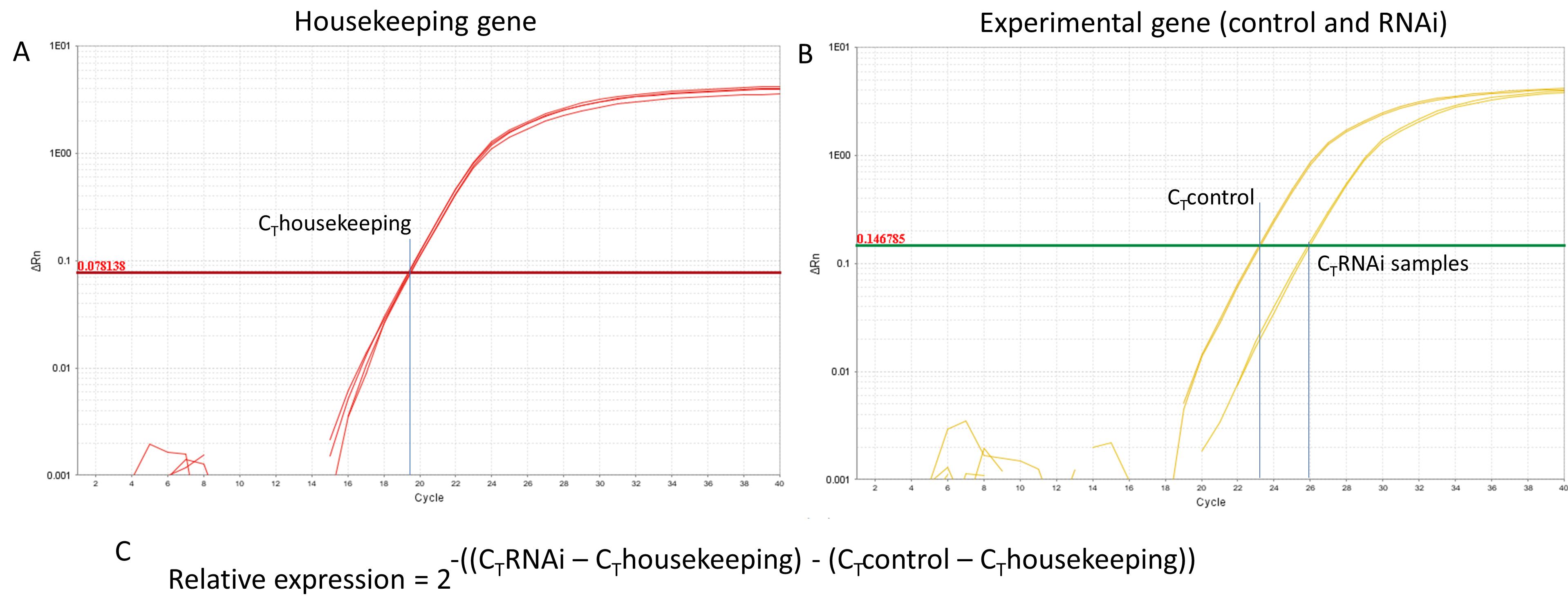
Figure 8. Sample quantitative PCR amplification plots from a QuantStudio 6 Flex real-time PCR system. (A) Amplification of a control housekeeping gene, showing a threshold cycle (CT) value of 19.5. (B) Amplification of a gene of interest from control and experimental (RNA interference) samples, showing CT values of 23.5 and 26, respectively. (C) Formula used to calculate the relative gene expression in the RNAi sample compared to that of the untreated control.
Notes
A similar approach can be used for isolating RNA from other insect species if their gene expression will be targeted by VIGS.
Use pots (9 cm square × 8 cm deep) to grow seedlings. Prepare five pots of seedling per construct transformation. In our hands, Golden Bantam works well for biolistic transformation of maize, but inbred line P39 can also be used.
The efficiency of transformation decreases after the two-leaf stage of seedling. Keep the seedlings in the dark for 24 h before biolistic transformation.
The prepared gold microcarriers can be stored at -20 °C and used for up to three months. Three milligrams of prepared microcarrier is sufficient for five bombardments per construct.
The amplified band is expected at 475 bp + the size of the target gene fragment. You can also confirm the sequence of the target fragment by Sanger sequencing.
Maize leaf sap can be stored indefinitely at -80 °C. Avoid frequent freeze thaw of the sap, as it will reduce the infection efficiency. You can also store the infected leaf tissue (0.25 g of leaf tissue per tube) at -80 °C after snap freezing in liquid nitrogen. This frozen tissue can be used to prepare fresh sap for inoculation of plants. Infection efficiency in both cases will be same.
Use of seedlings after the two-leaf stage results in low infection efficiency.
Change gloves and the cotton swab for sap from different pSCMV-VIGS constructs to avoid cross-contamination.
Acknowledgments
This work was supported by an International Postdoc Scholarship from the Punjab Higher Education Commission to I.G., NSF award 2019516 to G.J., and USDA award 2021-67014- 342237 to G.J.
Competing interests
The authors declare that they have no competing interests.
References
- Bally, J., Fishilevich, E., Doran, R. L., Lee, K., Campos, S. B., German, M. A., Narva, K. E. and Waterhouse, P. M. (2020). Plin‐amiR, a pre‐microRNA‐based technology for controlling herbivorous insect pests. Plant Biotechnol. J. 18(9): 1925-1932.
- Banerjee, R., Hasler, J., Meagher, R., Nagoshi, R., Hietala, L., Huang, F., Narva, K. and Jurat-Fuentes, J. L. (2017). Mechanism and DNA-based detection of field-evolved resistance to transgenic Bt corn in fall armyworm (Spodoptera frugiperda). Sci. Rep. 7(1): e1038/s41598-017-09866-y.
- Buhrow, L. M., Clark, S. M. and Loewen, M. C. (2016). Identification of an attenuated barley stripe mosaic virus for the virus-induced gene silencing of pathogenesis-related wheat genes. Plant Methods 12(1): e1186/s13007-016-0112-z.
- Chen, C. Y., Liu, Y. Q., Song, W. M., Chen, D. Y., Chen, F. Y., Chen, X. Y., Chen, Z. W., Ge, S. X., Wang, C. Z., Zhan, S., et al. (2019). An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. U.S.A 116(28): 14331-14338.
- Chung, S. H., Bigham, M., Lappe, R. R., Chan, B., Nagalakshmi, U., Whitham, S. A., Dinesh‐Kumar, S. P. and Jander, G. (2021). A sugarcane mosaic virus vector for rapid in planta screening of proteins that inhibit the growth of insect herbivores. Plant Biotechnol. J. 19(9): 1713-1724.
- Chung, S. H. and Jander, G. (2022). Inhibition of Rhopalosiphum maidis (Corn Leaf Aphid) Growth on Maize by Virus-Induced Gene Silencing with Sugarcane Mosaic Virus. In Vaschetto, L. M. (Ed.). RNAi Strategies for Pest Management: Methods and Protocols. Methods in Molecular Biology (pp. 139-153). Springer US: New York.
- Chung, S. H., Zhang, S., Song, H., Whitham, S. A. and Jander, G. (2022). Maize resistance to insect herbivory is enhanced by silencing expression of genes for jasmonate‐isoleucine degradation using sugarcane mosaic virus. Plant Direct. 6(6): e407.
- Ding, S. W. and Voinnet, O. (2007). Antiviral Immunity Directed by Small RNAs. Cell 130(3): 413-426.
- Ding, X. S., Mannas, S. W., Bishop, B. A., Rao, X., Lecoultre, M., Kwon, S. and Nelson, R. S. (2018). An Improved Brome mosaic virus Silencing Vector: Greater Insert Stability and More Extensive VIGS. Plant Physiol. 176(1): 496-510.
- He, H., Qin, X., Dong, F., Ye, J., Xu, C., Zhang, H., Liu, Z., Lv, X., Wu, Y., Jiang, X., et al. (2020). Synthesis, characterization of two matrine derivatives and their cytotoxic effect on Sf9 cell of Spodoptera frugiperda. Sci. Rep. 10(1): e1038/s41598-020-75053-1.
- Hsieh, M. H., Lu, H. C., Pan, Z. J., Yeh, H. H., Wang, S. S., Chen, W. H. and Chen, H. H. (2013). Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci. 201-202: 25-41.
- Jarugula, S., Willie, K. and Stewart, L. R. (2018). Barley stripe mosaic virus (BSMV) as a virus-induced gene silencing vector in maize seedlings. Virus Genes 54(4): 616-620.
- Kumar, P., Pandit, S. S. and Baldwin, I. T. (2012). Tobacco Rattle Virus Vector: A Rapid and Transient Means of Silencing Manduca sexta Genes by Plant Mediated RNA Interference. PLoS One 7(2): e31347.
- Liu, N., Xie, K., Jia, Q., Zhao, J., Chen, T., Li, H., Wei, X., Diao, X., Hong, Y., Liu, Y. (2016). Foxtail Mosaic Virus-Induced Gene Silencing in Monocot Plants. Plant Physiol. 171(3): 1801-1807.
- Livak, K. J. and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25(4): 402-408.
- Mamta, Reddy, K. R. K. and Rajam, M. V. (2016). Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol. Biol. 90(3): 281-292.
- Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., Huang, Y. P. and Chen, X. Y. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25(11): 1307-1313.
- Mei, Y., Liu, G., Zhang, C., Hill, J. H. and Whitham, S. A. (2019). A sugarcane mosaic virus vector for gene expression in maize. Plant Direct 3(8): e158.
- Mei, Y., Zhang, C., Kernodle, B. M., Hill, J. H. and Whitham, S. A. (2016). A Foxtail mosaic virus Vector for Virus-Induced Gene Silencing in Maize. Plant Physiol. 171(2): 760-772.
- Mlotshwa, S., Xu, J., Willie, K., Khatri, N., Marty, D. and Stewart, L. R. (2020). Engineering Maize rayado fino virus for virus‐induced gene silencing. Plant Direct 4(8): e224.
- Niu, Y., Head, G. P., Price, P. A. and Huang, F. (2016). Performance of Cry1A.105-selected fall armyworm (Lepidoptera: Noctuidae) on transgenic maize plants containing single or pyramided Bt genes. Crop. Prot. 88: 79-87.
- Pasin, F., Menzel, W. and Daròs, J. (2019). Harnessed viruses in the age of metagenomics and synthetic biology: an update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnol. J. 17(6): 1010-1026.
- Poreddy, S., Li, J. and Baldwin, I. T. (2017). Plant-mediated RNAi silences midgut-expressed genes in congeneric lepidopteran insects in nature. BMC Plant Biol. 17(1): e1186/s12870-017-1149-5.
- Purkayastha, A., Mathur, S., Verma, V., Sharma, S. and Dasgupta, I. (2010). Virus-induced gene silencing in rice using a vector derived from a DNA virus.Planta 232(6): 1531–1540.
- Rwomushana, I. (2019). Spodoptera frugiperda (fall armyworm). In: Invasive Species Compendium.CABI: Wallingford, UK.
- Sisay, B., Tefera, T., Wakgari, M., Ayalew, G. and Mendesil, E. (2019). The Efficacy of Selected Synthetic Insecticides and Botanicals against Fall Armyworm, Spodoptera frugiperda, in Maize. Insects 10(2): 45.
- Tian, G., Cheng, L., Qi, X., Ge, Z., Niu, C., Zhang, X. and Jin, S. (2015). Transgenic Cotton Plants Expressing Double-stranded RNAs Target HMG-CoA Reductase (HMGR) Gene Inhibits the Growth, Development and Survival of Cotton Bollworms. Inter. J. Biol. Sci. 11(11): 1296-1305.
- Tzean, Y., Lee, M. C., Jan, H. H., Chiu, Y. S., Tu, T. C., Hou, B. H., Chen, H. M., Chou, C. N. and Yeh, H. H. (2019). Cucumber mosaic virus-induced gene silencing in banana. Sci. Rep. 9(1): e1038/s41598-019-47962-3.
- Wang, R., Yang, X., Wang, N., Liu, X., Nelson, R. S., Li, W., Fan, Z. and Zhou, T. (2016). An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J. 86(1): 102-115.
- Wang, Y., Chai, C., Khatabi, B., Scheible, W. R., Udvardi, M. K., Saha, M. C., Kang, Y. and Nelson, R. S. (2021). An Efficient Brome mosaic virus-Based Gene Silencing Protocol for Hexaploid Wheat (Triticum aestivum L.). Front. Plant Sci. 12: e685187.
- Xiao, X. W., Frenkel, M. J., Teakle, D. S., Ward, C. W. and Shukla, D. D. (1993). Sequence diversity in the surface-exposed amino-terminal region of the coat proteins of seven strains of sugarcane mosaic virus correlates with their host range. Arch. Virol. 132: 399-408.
- Yang, J., Zhang, T. Y., Liao, Q. S., He, L., Li, J., Zhang, H. M., Chen, X., Li, J., Yang, J., Li, J. B. et al. (2018). Chinese Wheat Mosaic Virus-Induced Gene Silencing in Monocots and Dicots at Low Temperature. Front. Plant Sci. 9: e01627.
- Zhang, J., Yu, D., Zhang, Y., Liu, K., Xu, K., Zhang, F., Wang, J., Tan, G., Nie, X., Ji, Q. et al. (2017). Vacuum and Co-cultivation Agroinfiltration of (Germinated) Seeds Results in Tobacco Rattle Virus (TRV) Mediated Whole-Plant Virus-Induced Gene Silencing (VIGS) in Wheat and Maize. Front. Plant Sci. 8: e00393.
- Zhang, L., Liu, B., Zheng, W., Liu, C., Zhang, D., Zhao, S., Li, Z., Xu, P., Wilson, K., Withers, A. et al. (2020). Genetic structure and insecticide resistance characteristics of fall armyworm populations invading China. Mol. Ecol. Resour. 20(6): 1682-1696.
- Zhong, X., Yuan, X., Wu, Z., Khan, M. A., Chen, J., Li, X., Gong, B., Zhao, Y., Wu, J., Wu, C., et al. (2014). Virus-induced gene silencing for comparative functional studies in Gladiolus hybridus. Plant Cell Rep. 33(2): 301-312.
- Zotti, M., dos Santos, E. A., Cagliari, D., Christiaens, O., Taning, C. N. T. and Smagghe, G. (2018). RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest. Manage. Sci. 74(6): 1239-1250.
Supplementary information
The following supporting information can be downloaded here:
- Supplementary information
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Gull, I. and Jander, G. (2023). Inoculation of Maize with Sugarcane Mosaic Virus Constructs and Application for RNA Interference in Fall Armyworms. Bio-protocol 13(14): e4760. DOI: 10.21769/BioProtoc.4760.
Category
Plant Science > Plant molecular biology > DNA > Gene expression
Molecular Biology > DNA > DNA cloning
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








