- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Development of a Mouse Model of Hematopoietic Loss of Y Chromosome
Published: Vol 13, Iss 15, Aug 5, 2023 DOI: 10.21769/BioProtoc.4729 Views: 2090
Reviewed by: Gal HaimovichChris TibbittRAVI THAKUR

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Puromycin Proximity Ligation Assay (Puro-PLA) to Assess Local Translation in Axons From Human Neurons
Raffaella De Pace [...] Saikat Ghosh
Mar 5, 2025 3283 Views
Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
Yuki Yoshino [...] Natsuko Chiba
Apr 5, 2025 2427 Views
Efficient Fluorescent Labeling of Human Trophoblast Stem Cells via a CRISPR/Cas9-Mediated Knock-In Approach in a Safe Harbor Locus
Hengshan Zhang [...] Danny J. Schust
Jan 5, 2026 285 Views
Abstract
This protocol describes the generation of chimeric mice in which the Y chromosome is deleted from a proportion of blood cells. This model recapitulates the phenomenon of hematopoietic mosaic loss of Y chromosome (mLOY), which is frequently observed in the blood of aged men. To construct mice with hematopoietic Y chromosome loss, lineage-negative cells are isolated from the bone marrow of ROSA26-Cas9 knock-in mice. These cells are transduced with a lentivirus vector encoding a guide RNA (gRNA) that targets multiple repeats of the Y chromosome centromere, effectively removing the Y chromosome. These cells are then transplanted into lethally irradiated wildtype C57BL6 mice. Control gRNAs are designed to target either no specific region or the fourth intron of Actin gene. Transduced cells are tracked by measuring the fraction of blood cells expressing the virally encoded reporter gene tRFP. This model represents a clinically relevant model of hematopoietic mosaic loss of Y chromosome, which can be used to study the impact of mLOY on various age-related diseases.
Graphical overview

Background
Mosaic loss of Y chromosome (mLOY) is the most prevalent post-zygotic mutation in humans (Forsberg et al., 2014). More than 40% of men exhibit appreciable mLOY in hematopoietic cells by the age of 70 years (Thompson et al., 2019). The Y chromosome contains a relatively small number of protein-coding genes, and it can be viewed as a gene desert. Thus, while the phenomenon of mLOY was identified more than half a century ago, it has been regarded as a benign age-related alternation or as a biomarker for aging. However, recent research has found that mLOY in blood cells elevates the risk of all-cause mortality and a variety of human disorders, such as solid cancers, Alzheimer’s disease, and cardiovascular disease (Forsberg et al., 2014; Dumanski et al., 2016; Thompson et al., 2019; Sano et al., 2022). However, due to the descriptive nature of this epidemiological research, it cannot be determined whether mLOY in blood plays a causal role in the development of disease. To address this gap in knowledge, experimental studies can provide insights as to whether mLOY is causally linked to age-related illness.
The reconstitution of the hematopoietic system by the bone marrow transplantation (BMT) of gene-modified cells has been widely employed in mice to study the regulatory mechanisms that control the blood system. Recently, we employed CRISPR/Cas9-mediated gene editing to create mice with targeted mutations in genes to model the phenomenon of clonal hematopoiesis of indeterminate potential. In this method, lineage-negative cells isolated from the bone marrow of Cas9-transgenic mice are transduced with lentivirus vectors that encode gRNAs ex vivo. Hematopoietic lineage-negative cells are comprised of stem/progenitor cells that lack the expression of specific cell surface markers associated with various blood cell lineages. The lineage-negative cell pool has the ability to differentiate into multiple blood cell types. The resulting gene-edited cells are then transplanted into conditioned wildtype mice, achieving an editing efficiency of 90% within the transduced hematopoietic stem and progenitor cells (HSPC). More recently, we employed a similar CRISPR/Cas9 approach to ablate the entire Y chromosome within hematopoietic cells (Sano et al., 2022). In this application, gRNAs were designed to specifically target the repetitive sequences within the Y chromosome, allowing the creation of mice that lacked the Y chromosome in a portion of leukocytes.
Materials and reagents
BD Luer-LokTM disposable syringes, single use, 10 mL (BD, catalog number: 309604)
BD Luer-LokTM disposable syringes, single use, 50 mL (BD, catalog number: 309653)
BD PrecisionGlideTM needle, 18 G (BD, catalog number: 305195)
Insulin syringe, 29 G (EXLINT, catalog number: 26028)
CorningTM FalconTM 50 mL conical centrifuge tubes (Fisher Scientific, catalog number: 14-959-49A)
CorningTM FalconTM 15 mL conical centrifuge tubes (Fisher Scientific, catalog number: 14-959-53A)
Falcon® 100 mm TC-treated cell culture dish (Life Sciences, catalog number: 353003)
Falcon® 6-well clear flat-bottom TC-treated multi-well cell culture plate, with lid (Life Sciences, catalog number: 353046)
Corning® Costar® ultra-low attachment multiple well plate, 6 well (Millipore Sigma, catalog number: CLS3471)
Polypropylene centrifuge tubes (Beckman, catalog number: 326823)
FisherbrandTM sterile cell strainers, 70 μm (Fisher Scientific, catalog number: 22-363-548)
Mouse Millex-HV syringe filter unit
B6 mice (C57BL/6J, The Jackson Laboratory, stock number: 000664, bred in our laboratory or purchased)
Rosa26Cas9 knock-in mouse [B6(C)-Gt(ROSA)26Sorem1.1(CAG-cas9*,-EGFP)Rsky/J, The Jackson Laboratory, stock number: 028555, bred in our laboratory]
pLKO5.sgRNA.EFS.tRFP (Addgene, catalog number: 57823)
psPAX2 (Addgene, catalog number: 12260)
pMD2.G (Addgene, catalog number: 12259)
HEK 293T cell line (ATCC, catalog number: CRL-3216)
Collagen from calf skin (Millipore Sigma, catalog number: C9791)
PEI MAX (Polysciences, catalog number: 24765-1)
Phosphate-buffered saline (PBS), pH 7.4 (Thermo Fisher Scientific, catalog number: 10010023)
Dulbecco’s modified Eagle’s medium (DMEM), high glucose (Millipore Sigma, catalog number: D5796)
RPMI-1640 medium (Millipore Sigma, catalog number: R8758)
StemSpanTM SFEM (STEMCELL Technologies, catalog number: 09600)
Recombinant murine thrombopoietin (TPO) (Peprotech, catalog number: 250-03)
Recombinant murine stem cell factor (SCF) (Peprotech, catalog number: 315-14)
Polybrene infection/transfection reagent (Merck Sigma, catalog number: TR-1003-G)
Penicillin-streptomycin (10,000 U/mL) (Thermo Fisher Scientific, catalog number: 15140122)
Isoflurane liquid inhalation 99.9% (Henry Schein, catalog number: 1182098)
Lineage Cell Depletion kit (Miltenyi Biotec, catalog number: 130-090-858)
Lenti-X qRT-PCR Titration kit (Clontech, catalog number: 631235)
Circular Glass coverslips (12 mm) (Fisher, catalog number: 12-545-80P)
Rubber Cement (Amazon)
Microscope slides (Fisher, catalog number: 12-544-2)
Coplin Jars (glass) (Fisher, catalog number: 08816)
Coplin Jars (plastic) (Market Lab, catalog number: ML9801)
Coverslips, 22 × 50 mm (Fisher, catalog number: 12-518-105EP)
Diamond Tip Scribe (Fisher, catalog number: 08-675)
DNA probes (direct labeled) and validated (Vysis or Cytocell, XqF4/YqA1)
20× SSC (solution) (Fisher, catalog number: BP1325-1)
Purified water, D.I.U.F. (Fisher, catalog number: W2-4)
2× SSC (pH = 7.2) (Cell Line Genetics, in house)
70% ethanol solution (Cell Line Genetics, in house)
85% ethanol solution (Cell Line Genetics, in house)
100% ethanol, 200 Proof (Fisher, catalog number: BP2818-4)
1:20 VectaShield with DAPI (Cell Line Genetics, in house)
1% Formaldehyde (Cell Line Genetics, in house)
0.01 N HCl (Cell Line Genetics, in house)
PBD (phosphate buffered detergent) (Cell Line Genetics, in house)
Pepsin stock solution (Cell Line Genetics, in house)
Recombinant murine SCF (Peprotech, catalog number: 250-03)
Recombinant human IL-11 (Peprotech, catalog number: 200-11)
Recombinant human Flt3-Ligand (Peprotech, catalog number: 300-19)
Recombinant murine IL-3 (Peprotech, catalog number: 213-13)
IMDM (Gibco, catalog number: 12440053)
Doxycycline hyclate (Sigma-Aldrich, catalog number: D9891)
Colcemid Solution (Thermo Fisher, 15212012)
Cell culture media (see Recipes)
Wash buffer for cells (see Recipes)
MACS buffer (see Recipes)
Equipment
Forceps (Fine Science Tools)
Scissors (Fine Science Tools)
Eppendorf® centrifuge 5424R (Millipore Sigma, catalog number: 05-401-205)
Ultracentrifuge (Beckman, model: L8-70M)
Freezer, 4 °C
Deep freezer, -80 °C
Applied BiosystemsTM QuantStudioTM 6 Flex Real-Time PCR System (Thermo Fisher Scientific, catalog number: 4485699)
MACS® MultiStand (Miltenyi Biotec, catalog number: 130-042-303)
LS Columns (Miltenyi Biotec, catalog number: 130-042-401)
QuadroMACSTM Separator (Miltenyi Biotec, catalog number: 30-090-976)
RS-2000 X-ray irradiator (Rad Source Technologies, Inc.)
BD LSRFortessa flow cytometer (BD Biosciences)
Fluorescence microscope (Olympus, catalog number: BX-41)
Appropriate filters matched to excitation and emission spectra of fluorophores used (Chroma, model: Vysis Probes: Spectrum Orange/Spectrum Green, Cytocell Probes: Rhodamine/FITC)
TruTemp DNA micro heating system (Abbott Molecular, model: ThermoBrite)
Stir plate (Fisher, model: Isotemp)
pH meter (Fisher, catalog number: AE150)
Balance (OHaus, catalog number: EP613c)
Water bath (Fisher, model: Isotemp 205)
Quick Spin centrifuge (Fisher, model: Quick Spin)
Software
FlowJoTM software (BD Biosciences)
GraphPad Prism9 (https://www.graphpad.com/scientific-software/prism/)
BioRender (https://biorender.com/)
Procedure
Generation of lentivirus
Lentiviruses are produced by co-transfecting psPAX2, pMD2.G, and the lentivirus pLKO5.0.sg.EFS.tRPF into HEK 293T cells. Our standard procedure is to purify lentivirus particles from the cell culture supernatant of twelve 6-well plates (i.e., 72 wells in total). The volume of supernatant is approximately 216 mL (3 mL/well × 72 wells). The culture supernatant is collected 48 h after transfection and concentrated by ultracentrifugation. The lentiviral titer is measured using a commercially available qPCR-based assay. Optimized methods for high-titer lentivirus preparation and storage are discussed elsewhere (Cante-Barrett et al., 2016; Sano et al., 2019).
Seeding of HEK 293T cells
In advance, we usually prepare ten 10 cm dishes with 90% confluent HEK 293T cells.
Prepare an approximately 1:200 solution of commercially available 0.1% collagen from calf skin in PBS (i.e., final concentration of 0.0005%). We usually prepare 40 mL of 0.0005% collagen solution for one batch of lentivirus generation by adding 200 μL of 0.1% collagen solution to 40 mL of 1× PBS.
Add 500 μL of 0.0005% collagen solution to each well of 6-well plates and incubate at 37 °C in 5% CO2 for more than 30 min (coating wells with collagen). The collagen solution used for coating wells can be reused several times. The collected solution should be kept at 4 °C.
Seed HEK 293T cells at a density of 1 × 106 cells in 2 mL of complete DMEM per well (i.e., suspend the cells at a concentration of 0.5 × 106 cells/mL and use 2 mL of cell suspension per well) and incubate at 37 °C in 5% CO2 for 2 h until the cells attach firmly. More than 7.2 ×107 cells are required for twelve 6-well plates.
Transfection into HEK 293T cells
To prepare the mixture of three transfection plasmids, combine plasmids (0.9 μg of lentivirus vector, 0.6 μg of psPAX2, and 0.3 μg of pMD2.G for each well) and then achieve a total volume of 10 μL by adding deionized water. Adjust volumes accordingly depending on the number of wells (72 wells). The amount and ratio of each plasmid may need to be further optimized to suit an individual researcher’s needs.
Carefully add 50 μL of 1× PBS and 5 μL of the diluted PEI MAX (1.0 mg/mL) to the plasmid mixture for each well and incubate for 15 min at room temperature. Next, add 1 mL of complete DMEM to the mixture for each well.
Note: PEI MAX can be stored at 4 °C after reconstitution.
Aspirate the media from the 6-well plates, add 1 mL of the plasmid mixture prepared above, and incubate at 37 °C in 5% CO2 for 3 h.
Replace the media with 2 mL of fresh, complete DMEM and incubate at 37 °C in 5% CO2 for 24 h.
The following day, add 1 mL of fresh complete DMEM and incubate at 37 °C in 5% CO2 for an additional 24 h (total incubation time is 48 h).
Lentivirus purification
Transfer the culture supernatant to 50 mL Falcon tubes and centrifuge at 3,000 × g for 15 min to remove any free-floating cells.
Using a syringe, filter the supernatant through a 0.45 μm filter into new 50 mL Falcon tubes and transfer the filtrate to new polypropylene ultracentrifuge tubes.
Ultracentrifuge at 72,100× g at rmax for 3 h at 4 °C.
Carefully aspirate the supernatant after ultracentrifugation, leaving behind the white pellet. It is ideal to aspirate as much supernatant as possible to minimize residual liquid. However, be careful to not dry the pellets.
Add 200 μL of serum-free hematopoietic cell expansion medium to the pellet and incubate on ice for 1 h. Following this, resuspend pellets without aeration. Avoid bubble formation as much as possible. The volume of final solution is typically greater than 200 μL.
Acquire a 10 μL aliquot for measuring the viral titer and store the aliquot and all remaining viral suspension at -80 °C until needed. Snap freezing is not required.
Thaw a 10 μL aliquot (prepared in step A3f) and titrate the virus with a qPCR-based assay according to the manufacturer’s instructions.
Note: The procedures outlined above should be carried out in a biosafety class II cabinet. Unless otherwise specified, all procedures can be carried out at room temperature.
Isolation of lineage-negative cells from Cas9-knockin mice
Harvest of bone marrow cells (Sano et al., 2019)
Euthanize 8–10 male CRISPR/Cas9 knock-in mice (8–10 weeks of age) by 5% isoflurane followed by cervical dislocation; then, disinfect their skin with 70% ethanol.
Using dissecting scissors, make a small incision in the skin of the abdomen and peel the skin distally in both directions to expose the legs and arms.
Carefully separate the lower limbs from the hip bone by dislocating the hip joint. Cut along the femur head to remove the femur completely from the hip. Dislocate the knee and cut at the joint to separate the femur and tibia, while keeping the bone epiphysis intact. Dislocate the ankle joint and peel away the foot and extra muscle.
Using dissecting scissors, cut over the shoulder to detach the upper limbs. Dislocate the shoulder, then cut at the elbow joint to harvest the humerus bone.
Manually dislocate hip joints on both sides and harvest hip bones.
Use cellulose-fiber wipes to carefully remove muscles from the femurs, tibias, humeri, and hip bones. Take extra precaution to ensure that the bones do not break during this process.
Place the isolated bones into a 50 mL conical tube containing RPMI-1640 medium and place on ice.
Grasp the bone with blunt forceps and, using dissecting scissors, carefully cut both epiphyses (1–2 mm). Both ends of hip bones will be opened by this procedure.
Note: An insufficient cutting of bone will lead to an incomplete harvest of bone marrow, while overly aggressive cutting will result in cell loss.
Preparation of collecting tubes: make a small hole in the bottom of sterile 0.5 mL microcentrifuge tubes using an 18 G needle and place them in sterile 1.5 mL tubes containing 200 μL of ice-cold sterile PBS.
Place the cut bones inside the 0.5 mL microcentrifuge tubes and close the lids.
Place tubes in a centrifuge and spin at 10,000× g for 30 s at 4 °C (Figure 1).

Figure 1. Isolation of bone marrow cellsAfter centrifugation, all bone marrow cells should have passed through the hole in the 0.5 mL microcentrifuge tubes, and the cells should have been translocated to the bottom of the 1.5 mL tubes.
Note: Bones will become white and translucent if the bone cavity has been well flushed. If not, you can re-cut the bone ends and centrifuge again.
Discard the 0.5 mL microcentrifuge tubes that contain empty bones.
After the bone marrow has been collected in 1.5 mL tubes, transfer bone marrow to a 50 mL conical tube.
Add 10 mL of RPMI-1640 medium.
Make a single-cell suspension by drawing the bone marrow into a 10 mL syringe loaded with an 18 G needle. Do this 8–10 times.
Filter cell suspension through a 70 μm cell strainer into a 50 mL Falcon tube.
Centrifuge at 310× g for 10 min at 4 °C.
Aspirate the supernatant and resuspend the cell pellets in an appropriate volume of optimized separation buffer for the following lineage-negative cell isolation process.
Purification and lentivirus transduction of lineage-negative cells
Typically, lineage-negative cells account for 2%–5% of whole bone marrow nucleated cells. The purity is usually greater than 90% following isolation.
Isolate lineage-negative cells, using the Lineage Cell Depletion kit according to the manufacturer’s instructions.
Note: The purity of lineage-negative cells can be measured by flow cytometry. Lineages can be defined using specific makers such as CD3, CD90.2, CD19, B220, NK1.1, Ter119, Gr1 (Ly6G/Ly6C), and CD11b.
Resuspend the lineage-negative cells in 1 mL of serum-free hematopoietic cell expansion medium.
Seed the cells into an ultra-low attachment 6-well plate at a density of 1.5 × 106 cells/mL.
Add recombinant murine TPO and SCF into wells at final concentrations of 20 and 50 ng/mL, respectively.
Note: We routinely prepare small aliquots of recombinant proteins after reconstitution with water at 1,000× concentration (20 μg/mL for TPO and 50 μg/mL for SCF). Aliquots are stored at -80 °C and thawed as needed.
Pre-incubate cells at 37 °C in 5% CO2 for 2 h.
Gently add lentivirus at multiplicity of infection (MOI) = 100, 4 μg/mL polybrene infection/transfection reagent, and penicillin/streptomycin to the wells and incubate at 37 °C in 5% CO2 for 16–20 h.
Transplantation of transduced cells into lethally irradiated mice
Prepare recipient mice for transplantation by placing them in an eight-slice pie cage and exposing them to two doses of total body irradiation (550 Rad/dose, total dose = 1,100 Rad), with approximately 4 h between each irradiation session.
Note: The recipient mice should be male wild type.
Prepare the cells for injection by transferring all cells to a 15 mL conical tube. Add RPMI to 15 mL and centrifuge at 300× g for 10 min.
Suspend 5 × 105 lineage-negative cells in 200 μL of RPMI medium (5 × 105 cells/200 μL) and aspirate into an insulin syringe. Keep the cells at room temperature for 1–2 h until transplantation into lethally-irradiated recipient mice.
Note: Each mouse receives approximately 5 × 105 lineage-negative cells. These lineage-negative bone marrow cells were infected with a lentivirus vector at an MOI of 100, and then cultured for 16–20 h before collecting and washing as detailed previously in this section. All cells collected at this stage are injected into each recipient mouse.
Inject cells retro-orbitally into anesthetized lethally-irradiated recipient mice.
After the cells are injected, house the mice in sterilized cages and provide them with a soft diet and drinking water supplemented with antibiotics for 14 days.
Verification of Y chromosome deletion in hematopoietic cells
Fluorescent in situ hybridization (FISH)
FISH was performed in collaboration with Cell Line Genetics, Inc. and protocols utilized were provided by them.
Verify that the internal temperature of the 2× SSC pH 7.2 is 73 ± 1 °C and then add slides and incubate for 2 min.
Pepsin digest
i. Add 200 μL of Pepsin stock solution to the Coplin jar containing 40 mL of 0.01 N HCl in a 37 °C water bath and shake to mix well.
ii. Incubate slides for 13 min.
Wash the slides in 1× PBS for 5 min at room temperature.
Fix the slides in 1% formaldehyde for 5 min at room temperature.
Wash the slides in 1× PBS for 5 min at room temperature.
Dehydrate slides in 70%, 85%, and 100% ethanol for 2 min each and allow to air dry.
Notes:
i. Slides can be held at this point indefinitely. However, if left to stand for more than 2 h, repeat step f before probing.
ii. Perform the following steps under low light conditions.
Prepare the commercial probes for X chromosome (XqF4 region) and Y chromosome (YqA1 region).
Using a micro-pipettor, apply 3 μL of probe to the target area, cover with a circular coverslip, and seal with a bead of rubber cement.
Place the slides on the ThermoBrite instrument. Turn the instrument on and select the correct program to run (Program: 9, Denaturation: Temp 68, Time 2, Hybridization: Temp 37, Time 48 h).
Place slide in a moist hybridization chamber at 37 °C overnight (i.e., Tupperware-style container lined with a moist paper towel).
At the end of the day, add 40 mL of 0.4× SSC/0.3% NP-40 (pH = 7.2) into a plastic Coplin jar and place in a 74 °C water bath overnight. Make sure the lid is on tight to prevent evaporation.
Note: On the next day, perform the following steps under low light conditions.
Remove the rubber cement from the slides and slide the coverslips off gently with forceps.
Immediately transfer the slides to the Coplin jar of 0.4× SSC/0.3% NP-40 (pH = 7.2) stringency wash pre-warmed to 73 °C ± 0.5 °C for 2 min.
Drain the slides and wash in PBD in a Coplin jar for at least 30 s.
Drain the slide briefly and wipe the back of the slide.
Add 10–15 mL of 1:20 VectaShield with DAPI to each hybridization spot using the dropper, add the coverslip (22 mm × 50 mm), and blot the edges. Remove the excess DAPI stain by gently pressing on the slide sandwiched between a single layer of paper towels.
Store slides in the dark at -20 °C until ready to screen.
Karyotyping
We first immortalized lineage-negative cells by transducing them with lentivirus containing Hoxb8 (LV.T11.Hoxb8.Puro), before conducting karyotyping. In this system, the expression of Hoxb8 is induced by tetracycline, while the reverse transactivator M2 and the puromycin resistance genes are constitutively expressed in a bisistronic manner, driven by the human phosphoglycerate kinase promoter.
Karyotyping was performed in collaboration with KaryoLogic, Inc. Some information regarding the protocols used was provided by KaryoLogic, Inc. However, due to proprietary reasons, complete details of the protocols cannot be disclosed.
Immortalization of lineage-negative cells:
Prepare Hoxb8 lentivirus as described in section A.
Follow the procedure outlined in B.2 to isolate lineage-negative cells.
Culture the isolated lineage-negative cells in StemSpan SFEM medium supplemented with 50 ng/mL murine SCF (Stem Cell Factor), 100 ng/mL human IL-11 (Interleukin-11), 100 ng/mL human Flt3-ligand, and 20 ng/mL murine IL-3 (Interleukin-3).
Gently introduce HoxB8 lentivirus to the wells at a multiplicity of infection (MOI) = 100, with 4 μg/mL polybrene and 1% penicillin/streptomycin.
Incubate the cells at 37 °C in 5% CO2 for 16 h.
Collect the cells and resuspend them in IMDM supplemented with 10% heat-inactivated FBS, 1% penicillin/streptomycin, and cytokines (100 ng/mL murine SCF, 100 ng/mL human IL-11, 100 ng/mL human Flt3-Ligand, and 20 ng/mL murine IL-3) along with 2.4 μg/mL of doxycycline hyclate.
Note: It is recommended to prepare frozen aliquots in small amounts of cytokine solutions at a concentration of 1,000×.
Treat cells with puromycin at a concentration of 0.5 μg/mL for 48 h.
Sort the fraction of cells that are RFP-positive and culture them for an additional seven days before conducting karyotype analysis.
Karyotyping of immortalized lineage-negative cells:
Add Colcemid solution to the cell cultures to a final concentration of 0.5 μg/mL.
Note: In metaphase spreads, the cell is first treated with a chemical (e.g., colcemid) to arrest it at metaphase and then the chromosomes are spread out on a microscope slide and stained to allow for their visualization under a microscope.
Incubate the cells at 37 °C in 5% CO2 for 10 min.
Transfer the cells to centrifuge tubes. Centrifuge the cells at 500× g for 7 min.
Resuspend the cells in 0.075 M KCl hypotonic solution and incubate the cells at room temperature (approximately 22 °C) for 6 min. Centrifuge the cells at 500× g for 7 min.
Resuspend the cells in 3:1 methanol:acetic acid fixative and incubate at room temperature for 30 min. Centrifuge the cells at 500× g for 7 min.
Drop a single drop of cells in 0.5 mL of fixative onto each wet microscope slide to make the slides.
Bake the slides at 65 °C for 20 h.
Treat the slides with 0.1% trypsin-EDTA and stain them with Giemsa in Gurr’s buffer at pH 6.8.
Analyze metaphase spreads using a Leica DM2500 brightfield microscope at 1,000× and Leica Biosystems CytoVision software, version 7.4.
Data analysis
The Y chromosome ablation in hematopoietic cells from mLOY mice was confirmed by fluorescent in situ hybridization (FISH) analysis of X and Y chromosomes (Figure 2A: peripheral blood) and karyotype analysis (Figure 2B: bone marrow hematopoietic stem/progenitor cells). These data and the methods were derived from Figure 1 of the original article (Sano et al., 2022).
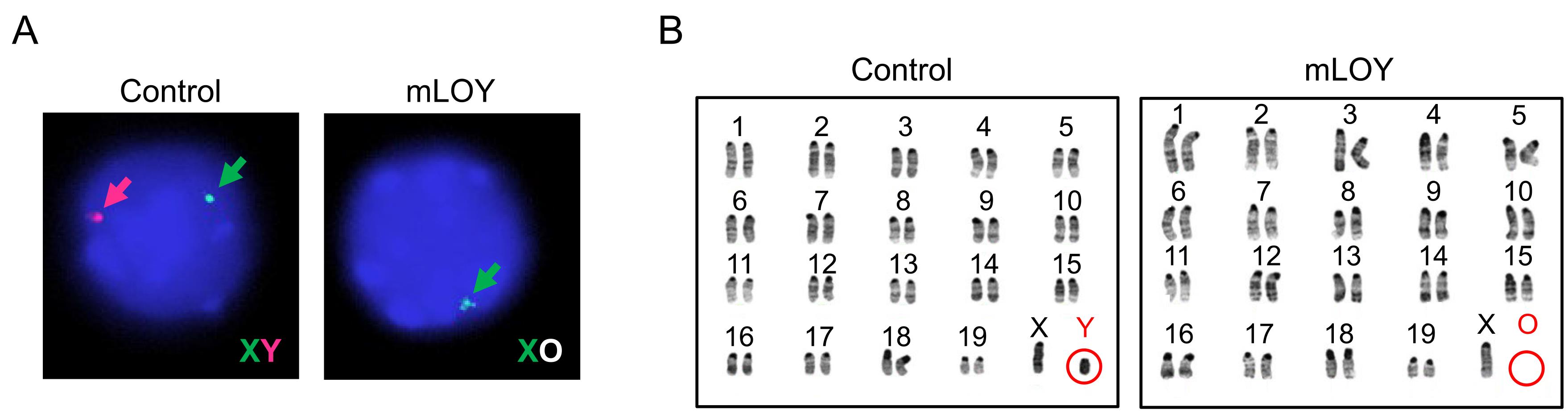
Figure 2. Depletion of the mouse Y chromosome with CRISPR/Cas9 system
In the original article, the control condition used a lentivirus encoding a gRNA designed not to target any region of the genome as a control (gRNA-NT). However, it was possible that cleavage of the genome itself may have an effect on the function of the transduced cells, regardless of the target genomic region. For example, studies have revealed that CRISPR/Cas9-mediated genome editing in normal cells can elicit a p53-induced DNA damage response (DDR), which induces the expression of downstream effectors, such as cell cycle inhibitor p21 (Haapaniemi et al., 2018; Janic et al., 2018). Thus, the expression of DDR-induced genes was assessed eight weeks after transplantation, using bone marrow lineage-negative cells from mice transplanted with gRNA that targets the centromere of the Y chromosome or gRNA-NT (Control). There was no difference between groups in the expression of p53, p21, and Bax (Figure 3).
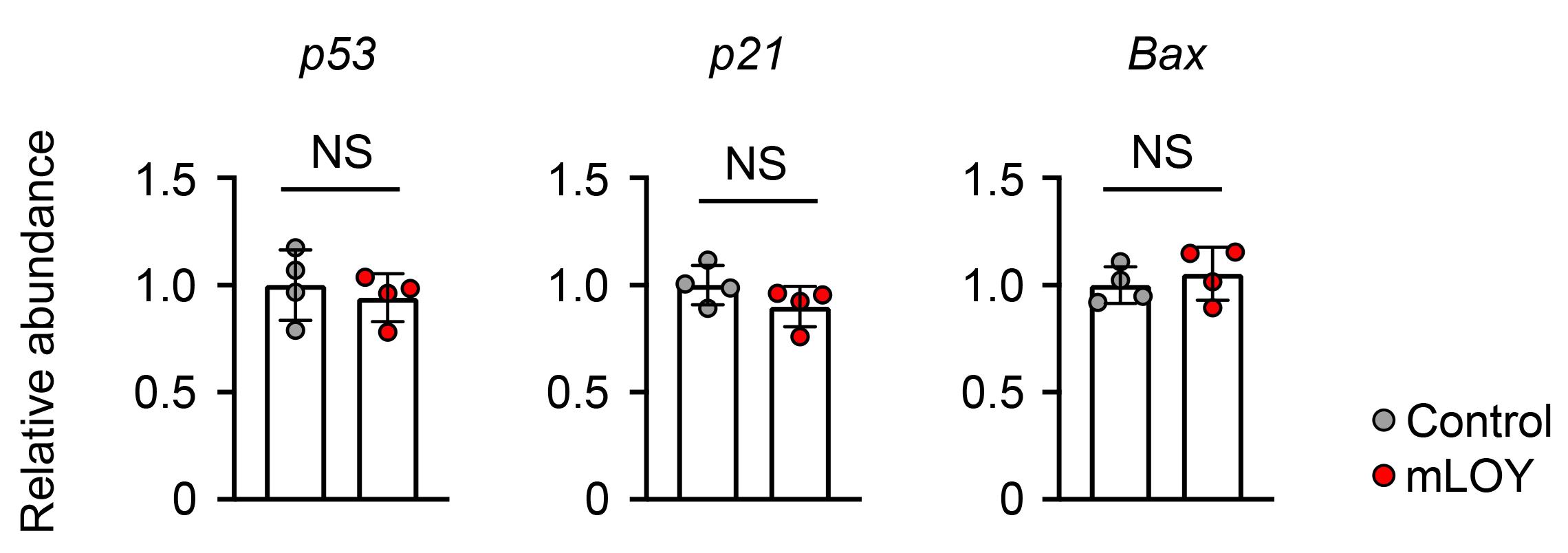
Figure 3. Expression of DNA damage response (DDR)-related genes in lineage-negative bone marrow cells harvested from mice transplanted cells treated with Y chromosome centromere-targeting gRNA or non-targeting gRNA
Next, to further rule out the possible confounding effects described above, we compared the action of gRNA-NT with that of a gRNA that targets the fourth intron of the alpha-actin gene (gRNA-Actin-Int4). In this experiment, we examined whether either version of control gRNA could influence heart function in response to stress in mice that had undergone BMT with transduced lineage-negative cells. For this purpose, mice undergoing BMT with hematopoietic cells treated with gRNA-NT or gRNA-Actin-Int4 were subjected to pressure overload by performing the transverse aortic constriction procedure. Compared to the data from the mLOY test condition (Sano et al., 2022), echocardiographic analyses revealed no differences in cardiac function in mice treated with gRNA-NT compared with those treated with gRNA-Actin-Int4 (Figure 4). These results indicate that CRISPR/Cas9-mediated genome cleavage in hematopoietic cells has no effect on long-term cellular function, at least in terms of cardiac function, possibly due to the disappearance of gRNA target sites following the initial editing of the genome.
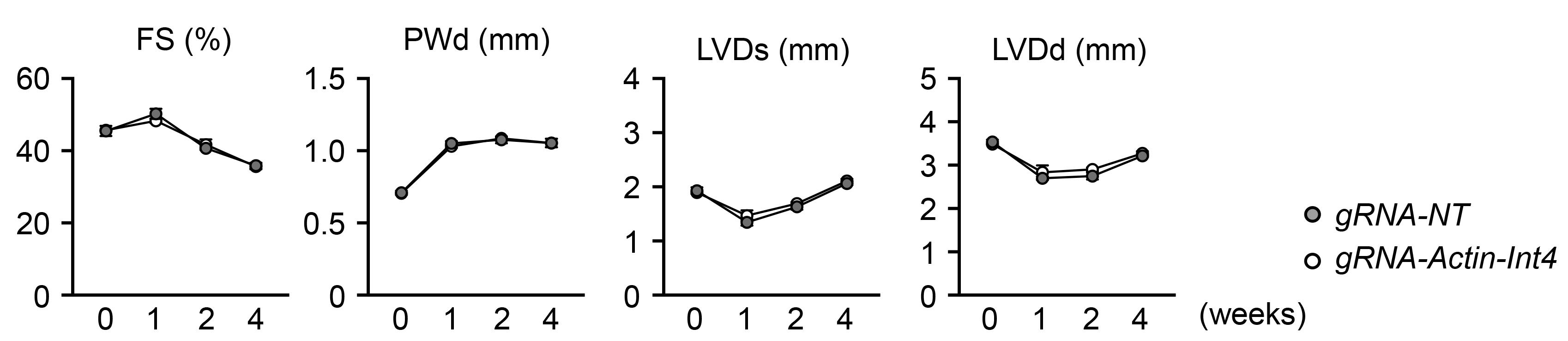
Figure 4. The impact of control gRNAs used in hematopoietic cells on the cardiac phenotype in response to pressure overload. FS; fractional shortening, PWTd; left ventricular posterior wall thickness at end-diastole, LVDs; left ventricular diameter at end-systole, LVDd; left ventricular diameter at end-diastole.
Recipes
Cell culture media
DMEM (high glucose) supplemented with 10% heat-inactivated FBS, 1% penicillin and streptomycin, warmed at 37 °C
Wash buffer for cells
PBS, warmed at 37 °C
MACS buffer
PBS supplemented with 0.5% BSA, 2 mM EDTA, chilled on ice
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants AG073249, AG072095, HL141256, HL139819, and HL142650 to K.W.; NIH grant HL152174 to S.S. and K.W.; and Grant-in-Aid for Research Activity Start-up 21K20879, Grant-in-Aid for Scientific Research C 22K08162, and grants from the Grant for Basic Research of the Japanese Circulation Society, the MSD Life Science Foundation, the Cardiovascular Research Fund, and the Japanese Heart Failure Society to S.S.
This protocol was derived from our previously published paper (Sano et al., 2022) with some modifications.
Competing interests
The authors declare no competing interests.
Ethical considerations
Animals were used in accordance with NIH guidelines and approved by the Institutional Animal Care and Use Committee at the University of Virginia.
References
- Cante-Barrett, K., Mendes, R. D., Smits, W. K., van Helsdingen-van Wijk, Y. M., Pieters, R. and Meijerink, J. P. (2016). Lentiviral gene transfer into human and murine hematopoietic stem cells: size matters. BMC Res Notes 9: 312.
- Dumanski, J. P., Lambert, J. C., Rasi, C., Giedraitis, V., Davies, H., Grenier-Boley, B., Lindgren, C. M., Campion, D., Dufouil, C., European Alzheimer’s Disease Initiative, I., et al. (2016). Mosaic Loss of Chromosome Y in Blood Is Associated with Alzheimer Disease.Am J Hum Genet 98(6): 1208-1219.
- Forsberg, L. A., Rasi, C., Malmqvist, N., Davies, H., Pasupulati, S., Pakalapati, G., Sandgren, J., Diaz de Stahl, T., Zaghlool, A., Giedraitis, V., et al. (2014). Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet 46(6): 624-628.
- Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. and Taipale, J. (2018). CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response.Nat Med 24(7): 927-930.
- Janic, A., Valente, L. J., Wakefield, M. J., Di Stefano, L., Milla, L., Wilcox, S., Yang, H., Tai, L., Vandenberg, C. J., Kueh, A. J., et al. (2018). DNA repair processes are critical mediators of p53-dependent tumor suppression.Nat Med 24(7): 947-953.
- Sano, S., Horitani, K., Ogawa, H., Halvardson, J., Chavkin, N. W., Wang, Y., Sano, M., Mattisson, J., Hata, A., Danielsson, M., et al. (2022). Hematopoietic loss of Y chromosome leads to cardiac fibrosis and heart failure mortality. Science 377(6603): 292-297.
- Sano, S., Wang, Y., Evans, M. A., Yura, Y., Sano, M., Ogawa, H., Horitani, K., Doviak, H. and Walsh, K. (2019). Lentiviral CRISPR/Cas9-Mediated Genome Editing for the Study of Hematopoietic Cells in Disease Models. J Vis Exp (152). doi: 10.3791/59977.
- Thompson, D. J., Genovese, G., Halvardson, J., Ulirsch, J. C., Wright, D. J., Terao, C., Davidsson, O. B., Day, F. R., Sulem, P., Jiang, Y., et al. (2019). Genetic predisposition to mosaic Y chromosome loss in blood. Nature 575(7784): 652-657.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sano, S. and Walsh, K. (2023). Development of a Mouse Model of Hematopoietic Loss of Y Chromosome. Bio-protocol 13(15): e4729. DOI: 10.21769/BioProtoc.4729.
- Sano, S., Horitani, K., Ogawa, H., Halvardson, J., Chavkin, N. W., Wang, Y., Sano, M., Mattisson, J., Hata, A., Danielsson, M., et al. (2022). Hematopoietic loss of Y chromosome leads to cardiac fibrosis and heart failure mortality. Science 377(6603): 292-297.
Category
Developmental Biology > Cell growth and fate > Ageing
Developmental Biology > Genome editing
Molecular Biology > DNA > Chromosome engineering
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








