- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Engineering Agrobacterium tumefaciens with a Type III Secretion System to Express Type III Effectors
Published: Vol 13, Iss 15, Aug 5, 2023 DOI: 10.21769/BioProtoc.4726 Views: 1378
Reviewed by: Wenrong HeYao XiaoYe XuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Simple and Fail-safe Method to Transform Miniprep Escherichia coli Strain K12 Plasmid DNA Into Viable Agrobacterium tumefaciens EHA105 Cells for Plant Genetic Transformation
Beenzu Siamalube [...] Steven Runo
Jan 5, 2025 2217 Views
Protocol for Inoculation of PGPR Staphylococcus sciuri to Seeds and Seedlings of Rice and Tomato Plants for Increased Root and Shoot Growth
Girija Somna [...] Dinakar Challabathula
Mar 20, 2025 2041 Views
In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1551 Views
Abstract
Plants elicit defense responses when exposed to pathogens, which partly contribute to the resistance of plants to Agrobacterium tumefaciens–mediated transformation. Some pathogenic bacteria have sophisticated mechanisms to counteract these defense responses by injecting Type III effectors (T3Es) through the Type III secretion system (T3SS). By engineering A. tumefaciens to express T3SS to deliver T3Es, we suppressed plant defense and enhanced plant genetic transformation. Here, we describe the optimized protocols for mobilization of T3SS-expressing plasmid to engineer A. tumefaciens to deliver proteins through T3SS and fractionation of cultures to study proteins from pellet and supernatants to determine protein secretion from engineered A. tumefaciens.
Keywords: Type III secretion systemBackground
The Type III secretion system (T3SS) is a specialized complex structure that translocates Type III effectors (T3Es) into the plant cell, where they interact with the host immune system. T3SS is present in many pathogenic Gram-negative bacteria (Cornelis, 2006). Heterologous expression of the genes encoding the type III secretion apparatus is functional in Pseudomonas fluorescens (Huang et al., 1988) and Escherichia coli (Ham et al., 1998). Gene cluster coding structural and regulatory components of T3SS from P. syringae have been cloned and are available in a plasmid called pLN18 (Jamir et al., 2004). We have transferred this plasmid into Agrobacterium tumefaciens to express T3SS and deliver T3Es to suppress plant defense and enhance plant genetic transformation (Raman et al., 2022). Because of the large size, we used triparental mating to mobilize pLN18 to A. tumefaciens in the presence of helper plasmid (pRK2013; containing tra genes) containing E. coli, using the protocol by Wise et al. (2006) with some modifications. In addition, we optimized culture growth and fractionation of the liquid culture into pellet and supernatant fractions to avoid cell lysis that leads to false positive results.
Materials and reagents
1.5 mL microcentrifuge tubes (USA Scientific)
50 mL Falcon tubes (Corning, catalog number: 352070)
0.45 μm Durapore PVDF membrane Millipore filter (Steriflip-HV Sterile Centrifuge Tube Top Filter Unit) (Millipore Sigma, catalog number: SE1M003M00)
Amicon Ultra-15 centrifugal filters (Millipore Sigma, catalog number: UFC901024)
Amicon Ultra-0.5 centrifugal filters (Millipore Sigma, catalog number: UFC501024)
A. tumefaciens GV2260 (Rifampicin resistance)
E. coli HB101 (pRK2013) (Kanamycin resistance)
Donor E. coli containing pLN18 (Kanamycin and Tetracycline resistance)
Luria Bertani (LB) medium (MP Biomedicals, catalog number: 3002011)
Luria Bertani (LB) medium with agar (MP Biomedicals, catalog number: 3002201)
PhiLOV-specific antibody (gift from Dr. John M. Christie, University of Glasgow)
Bacto peptone (BD, catalog number: 211677)
Yeast extract (BD, catalog number: 212750)
Bacto agar (BD, catalog number: 214010)
NaCl (J.T. Baker, catalog number: 3624-19)
K2HPO4 (J.T. Baker, catalog number: 4012-01)
NaH2PO4 (Fisher Scientific, catalog number: S369-500)
NH4Cl (Fisher Scientific, catalog number: A661)
KCl (Fisher Scientific, catalog number: P217-500)
MgSO4·7H2O (Sigma-Aldrich, catalog number: M1880)
CaCl2 (J.T. Baker, catalog number: 1313-01)
FeSO4·7H2O (Sigma-Aldrich, catalog number: F8263)
Sucrose (Invitrogen, catalog number: 15503-022)
Rifampicin (Sigma-Aldrich, catalog number: R3501-1G)
Kanamycin sulfate (Sigma-Aldrich, catalog number: 70560-51-9)
Tetracycline (Sigma-Aldrich, catalog number: T3258)
Yeast extract peptone (YEP) medium (1 L) (see Recipes)
20× AB buffer (1 L) (see Recipes)
20× AB salts (1 L) (see Recipes)
Water agar (see Recipes)
25% sucrose (see Recipes)
Agrobacterium minimal (AB)-sucrose agar medium or AB medium (see Recipes)
0.8% NaCl solution medium (see Recipes)
Hrp-derepressing (HDM) medium (see Recipes)
Equipment
28 °C incubator with shaker (New Brunswick Scientific)
37 °C incubator with shaker (New Brunswick Scientific)
Spectrophotometer (Bio-Rad, catalog number: 170-2525)
Eppendorf tabletop centrifuge (Eppendorf, catalog number: 5424)
Eppendorf tabletop refrigerated centrifuge (Eppendorf, catalog number: 5424R)
Eppendorf centrifuge with swing-bucket rotor (Eppendorf, catalog number: 5810R)
Procedure
Mobilization of T3SS to A. tumefaciens through triparental mating
Triparental mating is used to mobilize a large plasmid from E. coli to A. tumefaciens by using another E. coli carrying helper plasmid—in this case, HB101 (pRK2013).
Streak out the recipient A. tumefaciens strain (e.g., GV2260) from the -80 °C freezer onto YEP agar plates containing antibiotic [rifampicin (Rif); 10 μg/mL] and allow it to grow in a 28 °C incubator for two days. Streak out donor (pLN18, TetR) and helper (pRK2013, KmR) plasmid-containing E. coli strains onto LB agar plates containing appropriate antibiotics [kanamycin (Km), 50 μg/mL; and tetracycline (Tet), 5 μg/mL] and allow them to grow in a 37 °C incubator for a day.
The day before triparental mating, culture a single colony of A. tumefaciens in 5 mL of YEP medium supplemented with the appropriate antibiotics [rifampicin (10 μg/mL)] and leave overnight with shaking at 250 rpm and 28 °C. Culture a single colony of E. coli strains carrying donor and helper plasmids in 5 mL of LB liquid medium supplemented with the appropriate antibiotics, shaking at 250 rpm and 37 °C overnight.
On the day of mating, from the overnight cultures, inoculate 300 μL of A. tumefaciens to 3 mL of YEP liquid medium with rifampicin (10 μg/mL) and grow for 3–5 h at 250 rpm and 28 °C; inoculate 30 μL of E. coli to 3 mL of LB liquid medium with appropriate antibiotics and grow for 3–5 h at 250 rpm and 37 °C. Grow all the cultures until A600 reaches 0.3–0.5.
Triparental mating (Figure 1 and Figure 2)
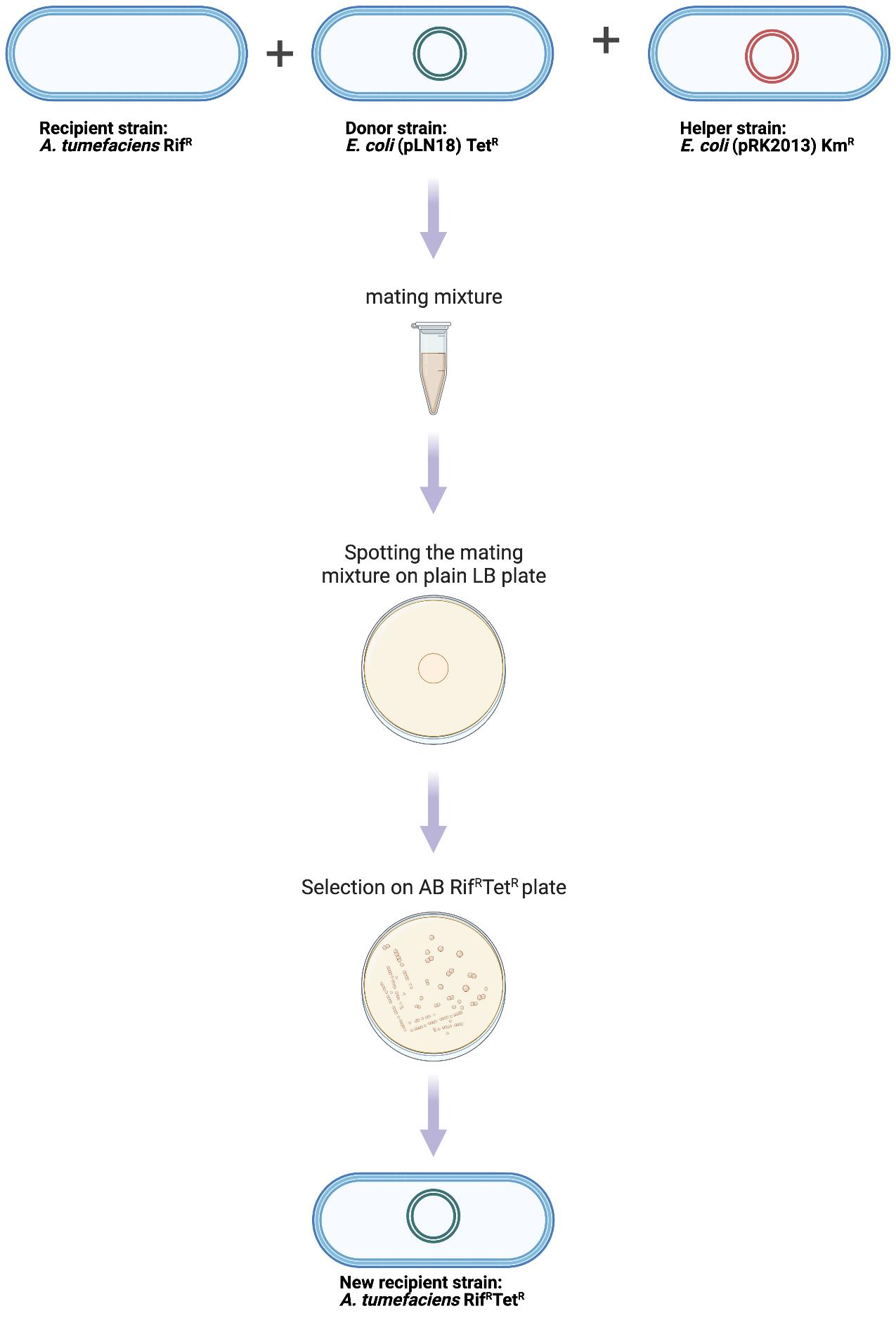
Figure 1. Experimental procedure for mobilizing pLN18 to A. tumefaciens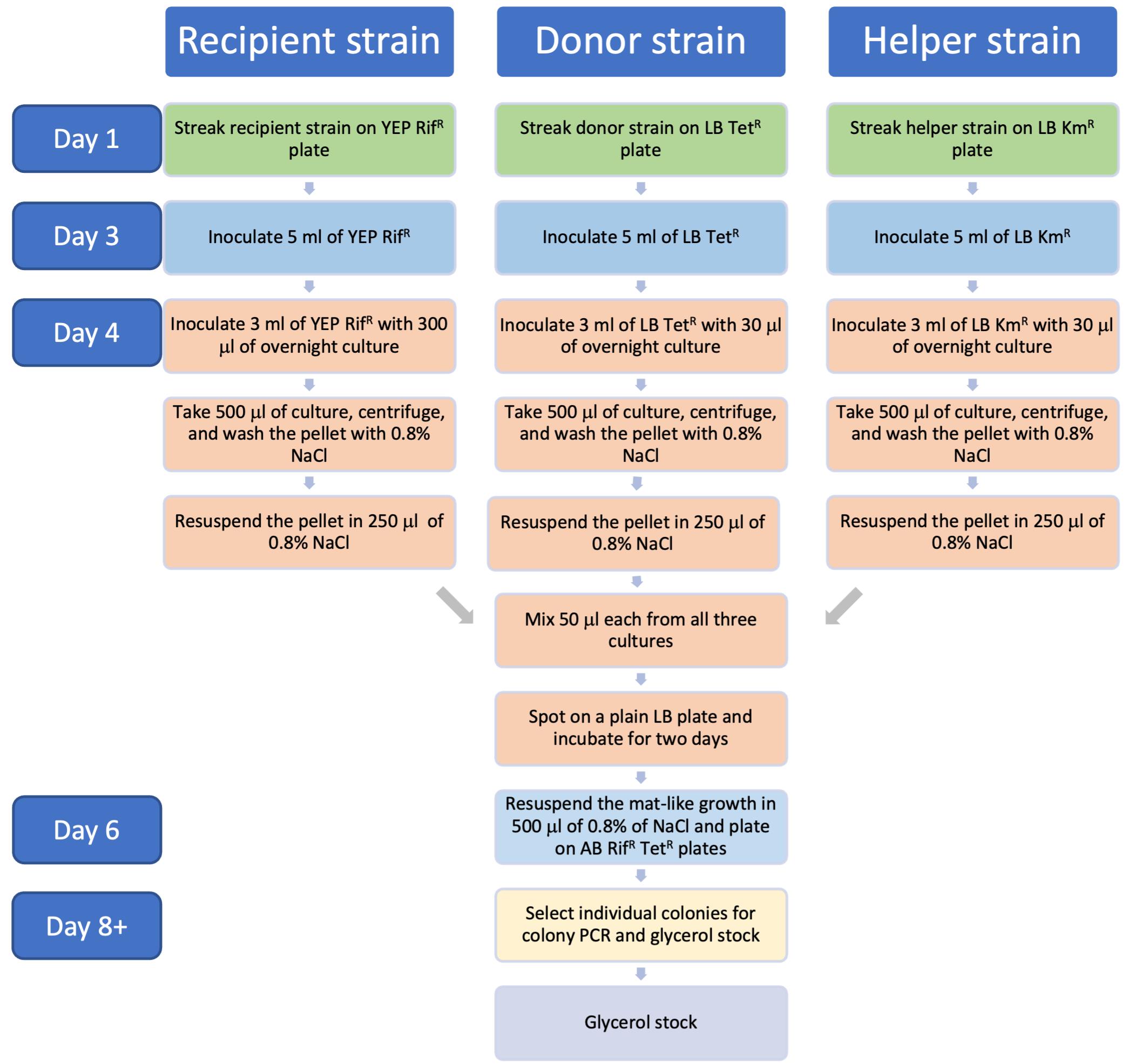
Figure 2. Flowchart describing timeline for the triparental mating protocolAliquot 500 μL each for all cultures in a 1.5 mL microcentrifuge tube separately and spin at 9,391× g for 2 min at room temperature (RT). Discard the supernatants. Wash once by resuspending the pellets in 500 μL of 0.8% NaCl and spin again.
Resuspend the pellet in 250 μL of 0.8% NaCl.
Mix 50 μL each of the three cultures in a microcentrifuge tube and spot it on a plain LB plate; allow it to dry for a few minutes and incubate the plate at 28 °C for two days.
Scrape off the mat-like growth (Figure 3) and resuspend in 500 μL of 0.8% NaCl in a 1.5 mL microcentrifuge tube. Spread 10 and 50 μL and the rest of the suspension in three separate AB (Gelvin, 2006) plates with Rif and Tet and incubate the plates at 28 °C for 2–4 days.
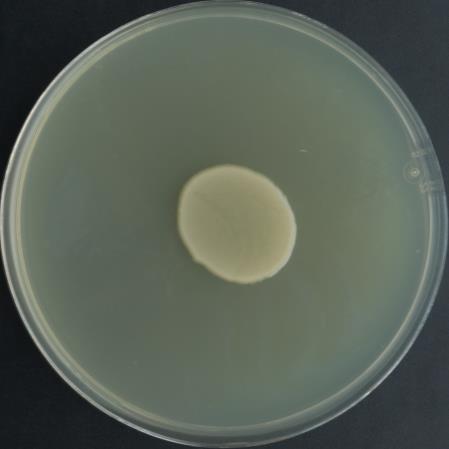
Figure 3. Representative image showing mat-like growth on plain LB plateIsolate single colonies for glycerol stocks and colony PCR.
Assays using A. tumefaciens expressing T3SS and T3Es
Either electroporation or the freeze-thaw method is used to transform GV2260 (pLN18) with a broad host range plasmid containing T3E (e.g., pBBR1MCS5-AvrPto-PhiLOV). Western blot and microscopy can be used to demonstrate effector secretion and delivery, respectively. In this protocol, we use western blot hybridization to show that T3Es are expressed and delivered out of the cell.
A. tumefaciens strains expressing T3SS and T3E are streaked onto YEP agar plates along with appropriate controls and incubated at 28 °C for two days. GV2260, GV2260 (pLN18), and GV2260 (pBBR1MCS5-AvrPto-PhiLOV) are used as controls.
Inoculate two colonies into 20 mL of HDM medium (Huynh et al., 1989) in 50 mL screw-cap tubes and incubate at 28 °C for 16 h shaking at 220 rpm.
Aliquot a small amount of culture (A600 = 0.25) in a 1.5 mL microfuge tube and centrifuge in a tabletop centrifuge at 9,391× g at RT for 2 min. Use the resulting pellet for pellet fraction analysis.
Centrifuge remaining cultures at 3,220× g for 15 min at 21 °C.
Using 25 mL pipettes, carefully remove the top 15 mL of the supernatant solutions without disturbing the pellet, by lowering the pipette slowly into the supernatant.
Transfer the supernatant fraction to 50 mL tubes and filter using a 0.45 μm Durapore PVDF membrane Millipore filter with lab vacuum connection.
Transfer 13 mL of the filtrate to Amicon Ultra-15 centrifugal filters and concentrate using a swinging bucket rotor at 3,220× g for 30 min at 4 °C.
Add the remaining 12 mL of the filtrate to the same Amicon Ultra-15 centrifugal filter and concentrate at 4 °C for another 30 min at 3,220× g.
Recover the concentrated filtrate from the bottom of the filter device and again concentrate to ~30 μL using Amicon Ultra-0.5 centrifugal filters at 14,000× g for 40–60 min at 4 °C.
Recover the concentrated sample by keeping the filter device upside down in a new microcentrifuge tube and spin for 2 min at 1,000× g in a refrigerated centrifuge. Store the concentrated sample at -80 °C until analysis.
Proteins from the pellet and supernatant fractions were subjected to electrophoresis through an SDS-PAGE gel using standard protocols, and immunoblot analysis was carried out using PhiLOV-specific antibody (dilution 1:5,000) (Figure 4).
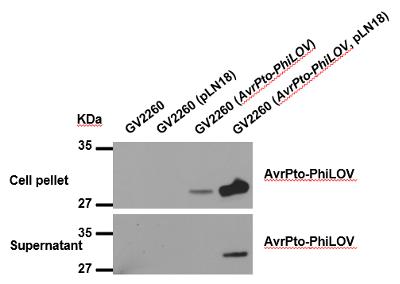
Figure 4. Western blot analysis of AvrPto expression and secretion in cell pellet and supernatant fractions
Recipes
YEP medium (1 L)
Bacto peptone 10 g
NaCl 5 g
Yeast extract 10 g
Bacto agar 15 g
Sterilize by autoclave at 121 °C for 15 min
20× AB buffer (1 L)
K2HPO4 60 g
NaH2PO4 20 g
Adjust the pH to 7.0
20× AB salts (1 L)
NH4Cl 20 g
KCl 3 g
MgSO47H2O 6 g
CaCl2 0.2 g
FeSO4·7H2O 50 mg
Water agar
Bacto agar 3.75 g in 220 mL of water
Autoclave 20× AB buffer, 20× AB salts, and water agar separately at 121 °C for 15 min
25% sucrose
12.5 g sucrose/50 mL water, mix well, and filter sterilize using 0.22 μm filter
AB-sucrose agar medium or AB medium
Add 20× AB buffer, 20× AB salts, and 25% sucrose when the water agar is warm
Water agar 220 mL
20× AB salts 12.5 mL
20× AB buffer 12.5 mL
25% sucrose 5 mL
0.8% NaCl solution medium (100 mL)
NaCl 0.8 g
Water 100 mL
Sterilize by autoclave at 121 °C for 15 min
HDM medium (1 L)
Fructose 1.801 g
(NH4)2SO4 1.004 g
MgCl2·6H2O 0.345 g
NaCl 0.099 g
1 M K2HPO4 6.6 mL
1 M KH2PO4 43.4 mL
Water 950 mL
pH 6.0 (No need to adjust the pH)
Sterilize by autoclave at 121 °C for 15 min
Acknowledgments
This work was supported by the National Science Foundation (grant # IOS-1725122 and IOS-2219792 to K.S.M.) and the Noble Research Institute, LLC. Media recipes described here are adapted from Gelvin (2006) and Huynh et al. (1989), and triparental mating procedure was modified from Wise et al. (2006). Protocols for fractionation of A. tumefaciens culture and western blot are from our published paper Raman et al. (2022). Figure 1 was generated with the help of Biorender (https://biorender.com/).
Competing interests
The authors declare no competing interests.
References
- Cornelis, G. R. (2006). The type III secretion injectisome. Nat rev Microbiol 4(11): 811-825.
- Gelvin, S. B. (2006). Agrobacterium virulence gene induction. Methods Mol Biol 343: 77-84.
- Ham, J. H., Bauer, D. W., Fouts, D. E. and Collmer, A. (1998). A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc Natl Acad Sci U S A 95(17): 10206-10211.
- Huang, H. C., Schuurink, R., Denny, T. P., Atkinson, M. M., Baker, C. J., Yucel, I., Hutcheson, S. W. and Collmer, A. (1988). Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J Bacteriol 170(10): 4748-4756.
- Huynh, T., Dahlbeck, D. and Staskawicz, B. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245(4924): 1374-1377.
- Jamir, Y., Guo, M., Oh, H. S., Petnicki-Ocwieja, T., Chen, S., Tang, X., Dickman, M. B., Collmer, A. and Alfano, J. R. (2004). Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J 37(4): 554-565.
- Raman, V., Rojas, C. M., Vasudevan, B., Dunning, K., Kolape, J., Oh, S., Yun, J., Yang, L., Li, G., Pant, B. D., et al. (2022). Agrobacterium expressing a type III secretion system delivers Pseudomonas effectors into plant cells to enhance transformation. Nat Commun 13(1): 2581.
- Wise, A. A., Liu, Z. and Binns, A. N. (2006). Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol Biol 343: 43-54.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Raman, V. and Mysore, K. S. (2023). Engineering Agrobacterium tumefaciens with a Type III Secretion System to Express Type III Effectors. Bio-protocol 13(15): e4726. DOI: 10.21769/BioProtoc.4726.
Category
Plant Science > Plant transformation > Agrobacterium
Microbiology > Microbe-host interactions > Bacterium
Biological Sciences > Biological techniques > Microbiology techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








