- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Primary Mouse Invariant Natural Killer T (iNKT) Cell Purification and Transduction
Published: Vol 13, Iss 13, Jul 5, 2023 DOI: 10.21769/BioProtoc.4707 Views: 2169
Reviewed by: Luis Alberto Sánchez VargasXiaokang WuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Isolation of CD31+ Bone Marrow Endothelial Cells (BMECs) from Mice
Alhaji Osman Smith [...] Lingyu Zeng
Nov 20, 2021 4068 Views
A Rapid Protocol for Direct Isolation of Osteoclast Lineage Cells from Mouse Bone Marrow
Lei Dang [...] Jin Liu
Mar 5, 2022 4356 Views
Isolation and Culture of Primary Pericytes from Mouse
Tamara McErlain [...] Meera Murgai
Apr 20, 2025 2978 Views
Abstract
Invariant natural killer T (iNKT) cells are a non-conventional T-cell population expressing a conserved semi-invariant T-cell receptor (TCR) that reacts to lipid antigens, such as α-galactosyl ceramide (α-GalCer), presented by the monomorphic molecule CD1d. iNKT cells play a central role in tumor immunosurveillance and represent a powerful tool for anti-cancer treatment, notably because they can be efficiently redirected against hematological or solid malignancies by engineering with tumor-specific chimeric antigen receptors (CARs) or TCRs. However, iNKT cells are rare and require specific ex vivo pre-selection and substantial in vitro expansion to be exploited for adoptive cell therapy (ACT). This protocol describes a robust method to obtain a large number of mouse iNKT cells that can be effectually engineered by retroviral (RV) transduction. A major advantage of this protocol is that it requires neither particular instrumentation nor a high number of mice. iNKT cells are enriched from the spleens of iVα14-Jα18 transgenic mice; the rapid purification protocol yields a highly enriched iNKT cell population that is activated by anti-CD3/CD28 beads, which is more reproducible and less time consuming than using bone marrow–derived dendritic cells loaded with α-GalCer, without risks of expanding contaminant T cells. Forty-eight hours after activation, iNKT cells are transduced with the selected RV by spin inoculation. This protocol allows to obtain, in 15 days, millions of ready-to-use, highly pure, and stably transduced iNKT cells that might be exploited for in vitro assays and ACT experiments in preclinical studies.
Keywords: Primary iNKT cellsBackground
Invariant natural killer T (iNKT) cells are a non-conventional T-cell population expressing a conserved semi-invariant αβ T-cell receptor (TCR), which is formed in mice by an invariant Vα14-Jα18 chain paired with a limited set of diverse Vβ chains (Bendelac et al., 2007). iNKT cells react to CD1d-restricted self and non self-lipid antigens in stress conditions, and are strongly implicated in tumor immunosurveillance (Godfrey et al., 2018). iNKT cells are evolutionary conserved; their investigation benefits from the availability of mouse models and the unequivocal detection, in mice and humans, by antigen-loaded CD1d tetramers and monoclonal antibodies specific for the semi-invariant TCR.
Several studies have shown that iNKT cells are an active component of the tumor microenvironment (TME) and control tumor progression by restraining cancer-supporting myeloid populations, such as modulating myeloid-derived suppressor cells and killing tumor-associated macrophages (De Santo et al., 2008 and 2010; Song et al., 2009; Liu et al., 2012; Gorini et al., 2017; Janakiram et al., 2017; Cortesi et al., 2018). Retargeting iNKT cells against cancer cells, by transducing tumor-specific TCR genes, generates enhanced effectors able to concurrently kill malignant cells and modulate detrimental myeloid cells in TME (Delfanti et al., 2022). This enhanced anti-tumor activity is peculiar of TCR-engineered iNKT, in which both the endogenous invariant TCR and the exogenous tumor-specific TCR exert anti-tumor effects—unlike conventional T cells, in which the endogenous TCRs are irrelevant for the therapeutic action, or even detrimental for possible off-target effects (Wolf et al., 2018). Therefore, iNKT cells are an attractive adoptive cell therapy (ACT) platform for engineering with anti-tumor TCRs or CAR (Heczey et al, 2020; Rotolo et al, 2018; Landoni et al, 2020; Delfanti et al., 2022), providing an appealing alternative to conventional T cells for the treatment of solid and hematological malignancies.
Because they are rare, iNKT cell harnessing for ACT requires two obligatory steps, consisting of ex vivo pre-selection followed by substantial in vitro expansion. We previously set up an optimized protocol for splenic iNKT cell purification and expansion (Delfanti et al., 2021) that can be used as the starting point for iNKT cell engineering. iNKT cells are enriched from the spleen of iVα14-Jα18 transgenic mice (iVα14 tg) (Griewank et al., 2007), in which iNKT cells are 30 times more frequent than in wildtype mice, significantly reducing the number of mice needed for the procedure. The setup of the cell culture requires only one day, instead of one week required by another widespread published protocol in which iNKT cells are stimulated with bone marrow–derived dendritic cells pulsed with the strong iNKT cell agonist α-GalCer (Chiba et al., 2009). In our protocol, iNKT cells are activated by anti-CD3/CD28 beads, and this activation is also useful for the retroviral (RV) transduction, which is done 48 h after. This procedure allows the generation of millions of ready-to-use, highly pure, and stably transduced iNKT cells within 15 days, so that they can be easily employed for functional studies (Figure 1). Indeed, by this method, we could engineer iNKT cells to express a second anti-tumor TCR (OT-I TCR), while maintaining the expression of the endogenous one, and we proved their efficacy both in vitro and in vivo (Delfanti et al., 2022). This protocol is also suitable for iNKT cell engineering with other effector molecules such as CARs or cytokines. A reporter gene, such as GFP, can also be introduced in the RV sequence to track iNKT cell transduction by flow cytometry over time.
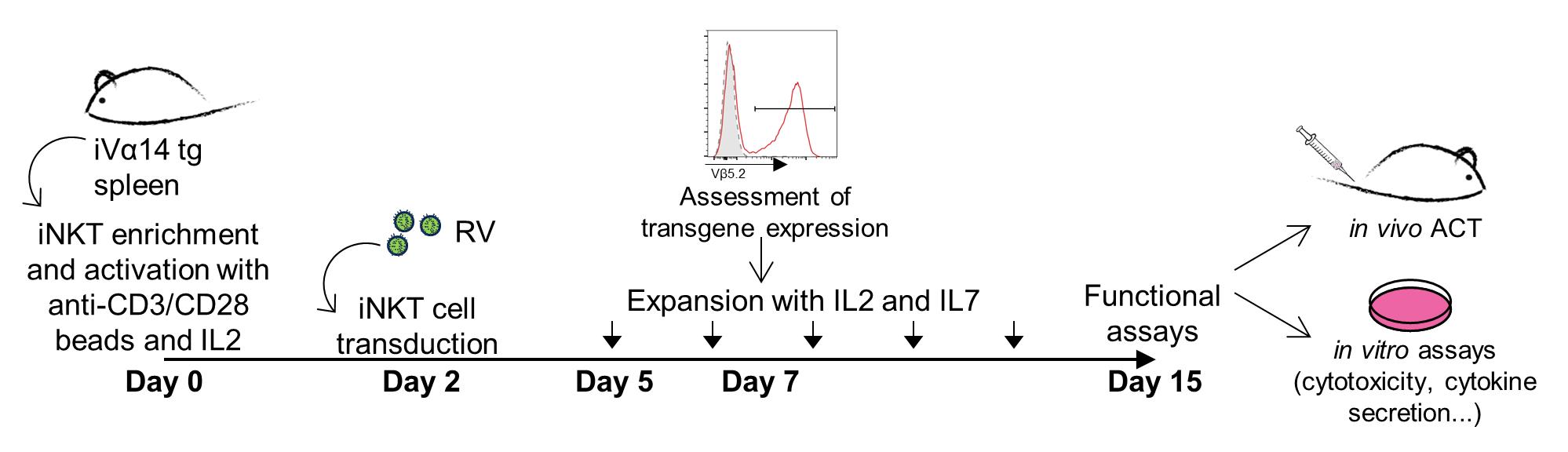
Figure 1. Timeline for engineered invariant natural killer T (iNKT) cell production. Schematic representation of the protocol passages that lead to the generation of engineered iNKT cells.
Materials and reagents
Millex-HV 0.45 μm filter (Merck Millipore, catalog number: SLHVM33RS)
Polypropylene centrifuge tubes (Beckman Coulter, catalog number: 326823)
Microplate 96-well shape (Greiner Bio One, catalog number: 650101)
75 cm2 flask (Corning, catalog number: 430720U)
100 mm × 20 mm tissue culture dish (Falcon, catalog number: 353003)
5 mL polystyrene round-bottom tube (Falcon, catalog number: 352052)
70 μm cell strainer (Falcon, catalog number: 352350)
15 mL polypropylene conical tube (Euroclone, catalog number: ET5015B)
50 mL polypropylene conical tube (Euroclone, catalog number: ET5050B)
iVα14-Jα18 tg mice, strain name: C57BL/6-Tg(Cd4-TcraDN32D3)1Aben/J, purchased from the Jackson Laboratory (USA)
IMDM, with HEPES and L-Gln (Lonza, catalog number: 12-277F)
Hyclone FetalClone I (Cytiva, catalog number: SH30080.03)
Penicillin/streptomycin solution (Gibco, catalog number: 15140-122)
Phoenix-eco packaging cell line (ECO) cells, purchased from ATCC
Chloroquine (Sigma-Aldrich, catalog number: C6628)
dH2O, tissue-culture grade (Sigma-Aldrich, catalog number: W-4502)
TE buffer (Invitrogen, catalog number: 8019005)
Vector DNA plasmid, purified using PureLink, HiPure Plasmid Filter Maxiprep kit (Invitrogen, catalog number: K210017)
Dulbecco’s phosphate buffered saline (PBS) (Euroclone, catalog number: ECB4004L)
Fetal bovine serum (FBS), ultra-low endotoxin (Euroclone, catalog number: ECS0186L)
Purified rat anti-mouse CD16/CD32 (mouse BD Fc block), clone: 2.4G2 (BD Pharmingen, catalog number: 553142)
CD19-FITC, clone: 6D5 (BioLegend, catalog number: 115506)
H2(IAb)-FITC, clone: AF6-120.1 (BioLegend, catalog number: 114406)
TCRβ-APC, clone: H57-597 (BioLegend, catalog number: 109212)
Vβ5.2 PE/Cy7, clone MR9-4 (BioLegend, catalog number: 139508)
Mouse PBS57-CD1d-tetramer, provided by NHI tetramer core facility
DAPI (Santa Cruz, catalog number: SC-3598)
Live/Dead Zombie Violet Fixable Viability kit (BioLegend, catalog number: 423114)
Bovine serum albumin (BSA) fraction V (Roche, catalog number: 10735094001)
EDTA stock solution (Cayman Chemical Company, catalog number: 600215)
Anti-FITC microbeads (Miltenyi Biotec, catalog number: 130-048-801)
Anti-PE microbeads (Miltenyi Biotec, catalog number: 130-048-701)
LD column (Miltenyi Biotec, catalog number: 130-042-901)
Pre-separation filter (Miltenyi Biotec, catalog number: 130-041-407)
LS column (Miltenyi Biotec, catalog number: 130-042-401)
MS column (Miltenyi Biotec, catalog number: 130-042-201)
Dynabeads mouse T-Activator CD3/CD28 (Gibco, catalog number: 11452D)
RPMI 1640-GlutaMAX (Gibco, catalog number: 61870-010)
Non-essential amino acids (Gibco, catalog number: 11140-035)
Sodium pyruvate (Gibco, catalog number: 11360-039)
β-mercaptoethanol (Gibco, catalog number: 31350010)
Polybrene hexadimethrine bromide (Sigma-Aldrich, catalog number: H9268)
(hr)IL-2 (Chiron Corp)
Recombinant mouse IL-7 (R&D System, catalog number: 407-ML-025)
Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: P6148)
CaCl2 (Sigma-Aldrich, catalog number: C9702)
NH4Cl (Sigma-Aldrich, catalog number: A0171)
KHCO3 (Sigma-Aldrich, catalog number: 237205)
N2EDTA (BDH, catalog number: 280254-D)
NaCl (Sigma-Aldrich, catalog number: S5886)
HEPES (Sigma-Aldrich, catalog number: H4034)
Na2HPO4 (Sigma-Aldrich, catalog number: S3264)
IMDM + 10% Hyclone (see Recipes)
TE 0.1-H2O (see Recipes)
2× HEPES buffered saline (HBS) (see Recipes)
ACK (ammonium-chloride-potassium) lysing buffer (see Recipes)
MACS separation buffer (see Recipes)
Complete RPMI (see Recipes)
Equipment
MACS® MultiStand (Miltenyi Biotec, catalog number: 130-042-303)
QuadroMACSTM Separator (Miltenyi Biotec, catalog number: 130-090-976)
DynaMagTM-15 Magnet (Invitrogen, catalog number: 12301D)
Polyallomer centrifuge tubes (Beckman Coulter, catalog number: 326823)
Eppendorf centrifuge (Eppendorf, model: 5810 R)
Ultracentrifuge (Beckman Coulter, model: Optima XPN-90)
Rotor (Beckman Coulter, model: SW32Ti)
S@FEGROW 188 incubator (Euroclone)
FACS Canto II (BD Biosciences)
Software
FlowJo_V10 (BD Biosciences)
Procedure
Retrovirus production
Note: Universal precautions must be taken while handling RV supernatants and RV-transduced samples, and all experiments must be carried out in at least class II biological safety cabinets and using appropriate protection equipment. RV must be handled and disposed of in accordance with your institutional biohazard control regulations.
Maintain Phoenix-ECO cells in 10 mL of IMDM + 10% Hyclone (see Recipe 1) in a T75 flask in a 37 °C incubator supplied with 5% CO2.
Seed 2 × 106 Phoenix-ECO cells into a 100 mm × 20 mm tissue culture dish with 9 mL of IMDM + 10% Hyclone.
Transfect Phoenix-ECO cells at ~70%–80% confluency, normally ~20–24 h after seeding.
Ten minutes before transfection, add 25 μL of 10 mM chloroquine to the cell and incubate at 37 °C.
Prepare the plasmid mix:
125 μL of 1 M CaCl2
20 μg of vector plasmid DNA (20 μL if DNA concentration is 1 μg/μL)
and the proper volume of TE 0.1-H2O (see Recipe 2) to reach a final volume of 500 μL
While vortexing the plasmid mix, add 500 μL of 2× HBS (see Recipe 3) (use the pipette gun and a 2 mL pipette).
Immediately add (with the same pipette) the precipitate to the Phoenix-ECO cells drop by drop.
Return cells to the incubator.
After 14–16 h, completely replace the medium and add 7 mL of fresh prewarmed IMDM + 10% Hyclone.
Twenty-four hours after step A9, collect culture media and replace with 7 mL of fresh prewarmed IMDM + 10% Hyclone.
Centrifuge the collected media at 300× g for 5 min at 4 °C and then pass through a 0.45 μm filter into a clean 50 mL tube. Store filtered media at 4 °C.
Twenty-four hours after step A11, repeats steps A10 and A11 and combine filtered media into 50 mL tubes.
Make 32 mL aliquots of retroviral supernatant in ultracentrifuge tubes (Polyallomer centrifuge tubes) carefully balancing the volumes (add medium or PBS if necessary).
Ultracentrifuge at 69,000× g for 2 h at 4 °C (rotor SW32Ti).
Discard supernatant and pay attention to dry all drops that may remain on the tube walls.
Add 70 μL of PBS per tube and wait 15 min at room temperature (RT).
Resuspend the samples at least 10 times setting the pipette volume at 60 μL, without creating bubbles, and pool all the samples in a 1.5 mL Eppendorf tube.
Parafilm the Eppendorf tube and rotate on a wheel for 30 min at 4 °C.
Make 20 μL aliquots and store at -80 °C. Under these conditions, RV may be stored up to two years after production.
Primary iNKT cell purification
Note: All the procedures must be performed under sterile conditions.
Spleen processing
Euthanize iVα14-Jα18 mice according to institutional policy, by inhalation of CO2.
Note: iVα14-Jα18 mice must be eight weeks old or older.
To dissect the spleen, place the animal on a clean dissection board and rinse with 70% reagent alcohol. Incise the abdominal cavity and collect the spleen using scissors and tweezers. The spleen is located to the left side of the abdomen, inferior to the stomach (Dowling et al., 2020).
Place a 70 μm cell strainer on a 50 mL tube and prepare the strainer by rinsing with 3 mL of PBS + 2% FBS (hereafter PBS-FBS).
Place the spleen on the strainer and homogenize the tissue via grinding in 10–20 mL final volume of PBS-FBS.
Centrifuge at 450× g for 5 min at 4 °C.
Remove the supernatant by inversion and resuspend the cell pellet with 1 mL of sterile ACK lysing buffer (see Recipe 4); for red blood cell lysis, incubate for 3 min at room temperature and block with 5 mL of PBS-FBS.
Centrifuge at 450× g for 5 min at 4 °C.
Remove the supernatant by inversion and resuspend the cell pellet in 3 mL of PBS-FBS. Remove fat residues by pipetting and determine cell number.
Note: Expected recovery from the spleen of one iVα14-Jα18 mouse is 2 × 106 iNKT cells; if more iNKT cells are needed, increase the number of mice accordingly. Cells coming from different mice can be pooled at this point prior to the determination of cell number.
T-cell enrichment
Note: For the enrichment steps, work fast, keep the cells cold, and use solutions pre-cooled at 4 °C overnight and then kept on ice.
Resuspend the total spleen cells in the appropriate volume of PBS-FBS (500 μL for 107 cells) + Fc blocker (5 μL × 107 cells) and incubate for 15 min at RT.
Wash with 1–2 mL of MACS separation buffer (MB) (see Recipe 5) per 107 total cells and centrifuge at 450× g for 10 min at 4 °C.
Remove the supernatant by inversion and stain the cell with CD19-FITC and H2(IAb)-FITC (use 5 μL × 107 cells in 100 μL of MB); mix well and incubate for 15 min in the dark at 4–8 °C.
Wash cells by adding 1–2 mL of MB per 107 cells and centrifuge at 450× g for 10 min at 4 °C.
Pipette off the supernatant completely and resuspend the cell pellet in 90 μL of MB per 107 total cells.
Add 10 μL of anti-FITC microbeads per 107 total cells. Mix well and incubate for 15 min in the dark at 4–8 °C.
Wash the cells by adding 1–2 mL of MB per 107 cells and centrifuge at 450× g for 10 min at 4 °C.
Pipette off the supernatant completely and resuspend up to 1.25 × 108 cells in 500 μL of MB.
Place a LD column in the magnetic field of MACS Separator to proceed with the depletion. To avoid clogging, apply a pre-separation filter on the LD column and rinse with 2 mL of MB. When the column reservoir is empty, apply the cell suspension onto the filter. This step depletes B and myeloid antigen-presenting cells from the spleen cell suspension.
Collect the unlabeled cells that pass through the column.
Wash three times with 1 mL of MB, only when the column reservoir is empty.
Collect the total effluent, which will be enriched in T cells, and count the cells.
iNKT cell enrichment
Centrifuge the T cell–enriched suspension at 450× g for 5 min at 4 °C and remove the supernatant by inversion.
Stain the cells with CD1d-tetramer-PE (mouse PBS57-CD1d-tetramer), according to the antibody titration, in 50 μL of MB per 106 cells. Mix well and incubate for 30 min in the dark on ice.
Wash the cells by adding 1–2 mL of MB per 107 cells and centrifuge at 450× g for 10 min at 4 °C.
Pipette off the supernatant completely and resuspend the cell pellet in 80 μL of MB per 107 total cells.
Add 20 μL of anti-PE microbeads per 107 total cells. Mix well and incubate for 15 min in the dark at 4–8 °C.
Wash cells by adding 1–2 mL of MB per 107 cells and centrifuge at 450× g for 10 min at 4 °C.
Pipette off the supernatant completely and resuspend up to 108 cells in 500 μL of MB; otherwise, if cells exceed 108, adjust the volume accordingly.
According to cell count, place a LS (up to 108) or MS (up to 107) column in the magnetic field of MACS Separator. Rinse the column with MB (3 mL for LS, 500 μL for MS).
Apply the cell suspension onto the column. Collect unlabeled cells that pass through.
Wash column three times by adding the appropriate volume of MB (3 × 3 mL for LS column, 3 × 500 μL for MS column), only when the column reservoir is empty. The total effluent is the negative fraction.
Remove column from the magnetic field and place it on a new collection tube.
Add MB onto the column (5 mL for LS column or 1 mL for MS column), push the provided plunger into the column, and flush out the positive fraction (enriched in iNKT cells).
To further increase the iNKT cell recovery, centrifuge the negative fraction at 450× g for 10 min at 4 °C and repeat steps h–l with a new LS or MS column.
Pool the positive fractions and determine cell count.
Check the purification steps by FACS analysis. Samples are spleen ex vivo, T cell–enriched fraction, iNKT-positive fraction, and iNKT-negative fraction. Stain the cells with CD19-FITC, IAb-FITC, CD1d-tetramer-PE, TCRβ-APC, and DAPI (Figure 2 and Figure 3).
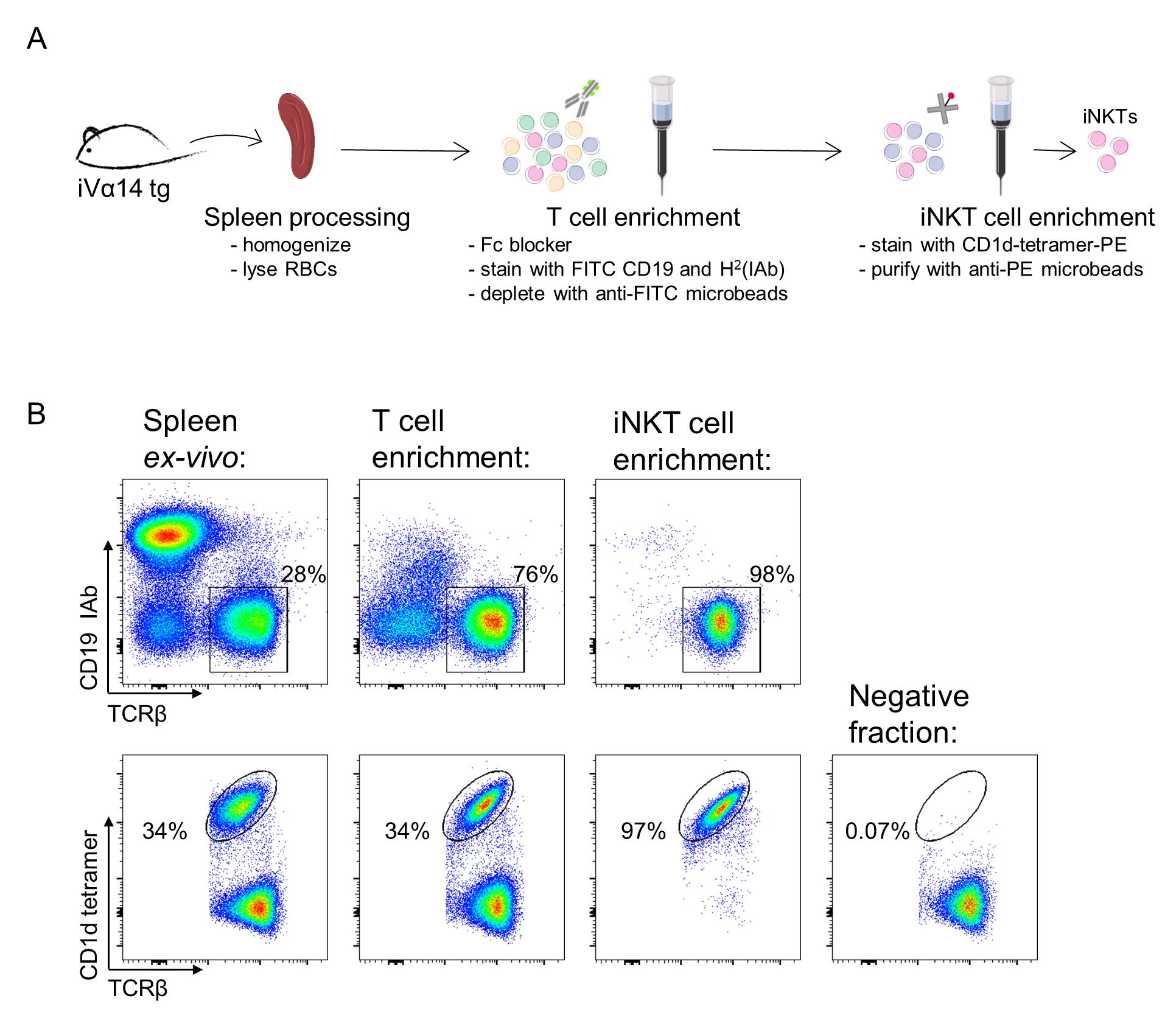
Figure 2. Primary invariant natural killer T (iNKT) cell purification. A. Schematic representation of the purification protocol. B. Flow cytometry analysis of each enrichment step. Upper plots show the percentage of T-cell frequencies, gated on viable lymphocytes (see data analysis section for details). Lower plot show the percentage of iNKT cells along each step, gated on viable CD19- IAb- TCRβ+ lymphocytes. Adapted from Delfanti et al. (2021).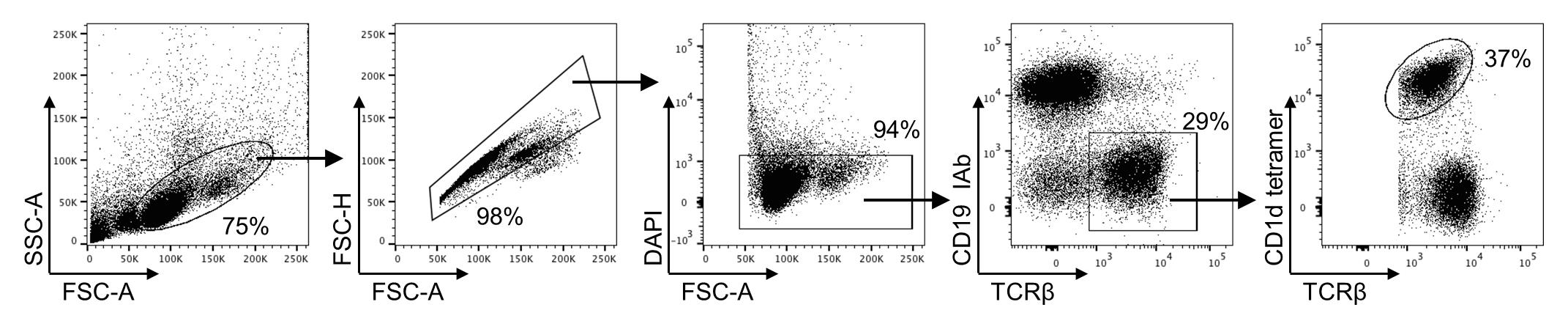
Figure 3. Gating strategy for analysis of invariant natural killer T (iNKT) cells purification steps. Representative dot plots of gating strategy for analysis of ex vivo splenic iNKT cells used to evaluate each purification step.
iNKT cell activation and transduction
Activate purified iNKT cells with mouse T activator anti-CD3/CD28 magnetic beads in 1:1 ratio.
Centrifuge the iNKT cell positive fraction at 450× g for 5 min at 4 °C.
Meanwhile, transfer the appropriate volume of mouse T activator anti-CD3/CD28 magnetics beads to a 15 mL tube, add an equal volume of PBS, vortex for 5 s, place the tube on the magnet (DynaMagTM-15 Magnet) for 1 min, and discard the supernatant.
Remove the tube from the magnet and resuspend the washed magnetic beads in the proper volume of complete RPMI (see Recipe 6) to have 5 × 105 iNKT cells in 1 mL. Use this suspension to resuspend the iNKT cell pellet.
Plate 1 mL of the cell suspension (5 × 105 iNKT cells) + anti-CD3/CD28 magnetic beads in a 48-well plate with 20 U/mL human recombinant (hr)IL-2 and incubate at 37 °C.
Transduce iNKT cells after 48 h in culture.
Note: All handling of retroviral production and transduction is carried out in at least class II biosafety cabinets.
Centrifuge the 48-well plate at 450× g for 7 min at RT.
Carefully remove the cell media by pipetting, starting from the top of the well to avoid disturbing the cells.
Add 200 μL of complete RPMI, plus the appropriate volume of concentrated RV (Section A), and 8 μg/mL of polybrene to the wells.
Note: Always leave one well for the non-transduced (NT) control by adding complete RPMI and 8 μg/mL polybrene only.
Centrifuge the plate at 800× g for 2 h at 30 °C.
Add 800 μL of prewarmed media to each well with original culture conditions [complete RPMI and 20 U/mL of (hr)IL-2] and incubate at 37 °C.
Analyze iNKT cell transduction five days post-infection as described below (Section E).
iNKT cell expansion
Split the cells 1:2 when they reach 80%–90% confluency (usually starting from day 3–4 after activation)
Maintain the cell in complete RPMI and 20 U/mL of (hr)IL-2 until day 5 in culture.
After five days in culture, add 10 ng/mL of mouse IL-7.
In these culture conditions [complete RPMI, 20 U/mL of (hr)IL-2, 10 ng/mL of mouse IL-7], iNKT cells can be expanded for up to 15 days, when in vivo or in vitro assays can be performed.
Note: For iNKT adoptive cell transfer, beads have to be removed from the cell culture by loading the cell suspension on the magnet (incubate for 3 min). Cells are washed three times with PBS before injection.
Analysis of iNKT cell transduction
At five days post-infection, iNKT cell transduction can be assessed by flow cytometry.
Note: Work under a class II biosafety cabinet, since at this time point some RV particles can still be present in cell supernatant.
Collect 200 μL of cell suspension and transfer to a U-bottom 96-well plate for staining.
Centrifuge at 450× g for 5 min at RT.
Remove the supernatant by pipetting (change the tips and bleach them).
Add 50 μL of PBS + 0.5 μL of Fc blocker per sample and incubate for 15 min at RT.
Centrifuge at 450× g for 5 min at 4 °C.
Remove the supernatant, add 50 μL of PBS + 0.1 μL of Live/Dead Fixable Violet per sample, and incubate for 20 min at RT.
Centrifuge at 450× g for 5 min at 4 °C.
Remove the supernatant and stain the cells with CD1d-tetramer-PE, according to the antibody titration, in 50 μL of PBS-FBS per sample. Mix well and incubate for 30 min in the dark on ice.
Centrifuge at 450× g for 5 min at 4 °C.
Meanwhile, prepare the flow cytometry mix for surface staining:
100 μL of PBS-FBS.
1 μL of TCRβ-APC and 1 μL of Vβ5.2-PE/Cy7 (for OT-I TCR staining) per sample.
Note: If the RV contains a GFP reporter, avoid the use of FITC-conjugated antibodies for the cell staining.
Remove the supernatant and add 100 μL of flow cytometry mix. Mix well and incubate for 15 min in the dark on ice.
Centrifuge at 450× g for 5 min at 4 °C.
Remove the supernatant and add 200 μL of 2% (w/v) PFA in PBS. Incubate for 10 min in the dark on ice.
Centrifuge at 600× g for 7 min at 4 °C.
Remove the supernatant and wash twice with PBS: add 200 μL of PBS, centrifuge at 450× g for 5 min at 4 °C, and repeat.
Resuspend the cells in 200 μL of PBS and transfer to flow cytometry tubes for FACS analysis.
Use gating strategy in Figure 4 to identify transduced iNKT cells.
iNKT cell transduction can also be assessed later on over cell culture. Starting from day 7 after infection, fixation with PFA is no longer required.
Data analysis
Use gating strategy lymphocytes/single cell/viable cells/ CD19- IAb- TCRβ+/TCRβ+ CD1d tetramer+ for analysis of ex vivo splenic iNKT cells (Figure 3).
Use gating strategy lymphocytes/single cell/viable cells for analysis of transduced iNKT cells (Figure 4). Here, iNKT cells were transduced to express the OT-I TCR, and transduction efficiency can be evaluated by the expression of the Vβ5.2, which is the specific Vβ chain of the OT-I TCR. The co-expression of the endogenous invariant TCR can be evaluated by staining iNKT cells also with the CD1-tetramer. In case the RV also encodes a GFP reporter, the transduction efficiency can simply be evaluated by the GFP expression.
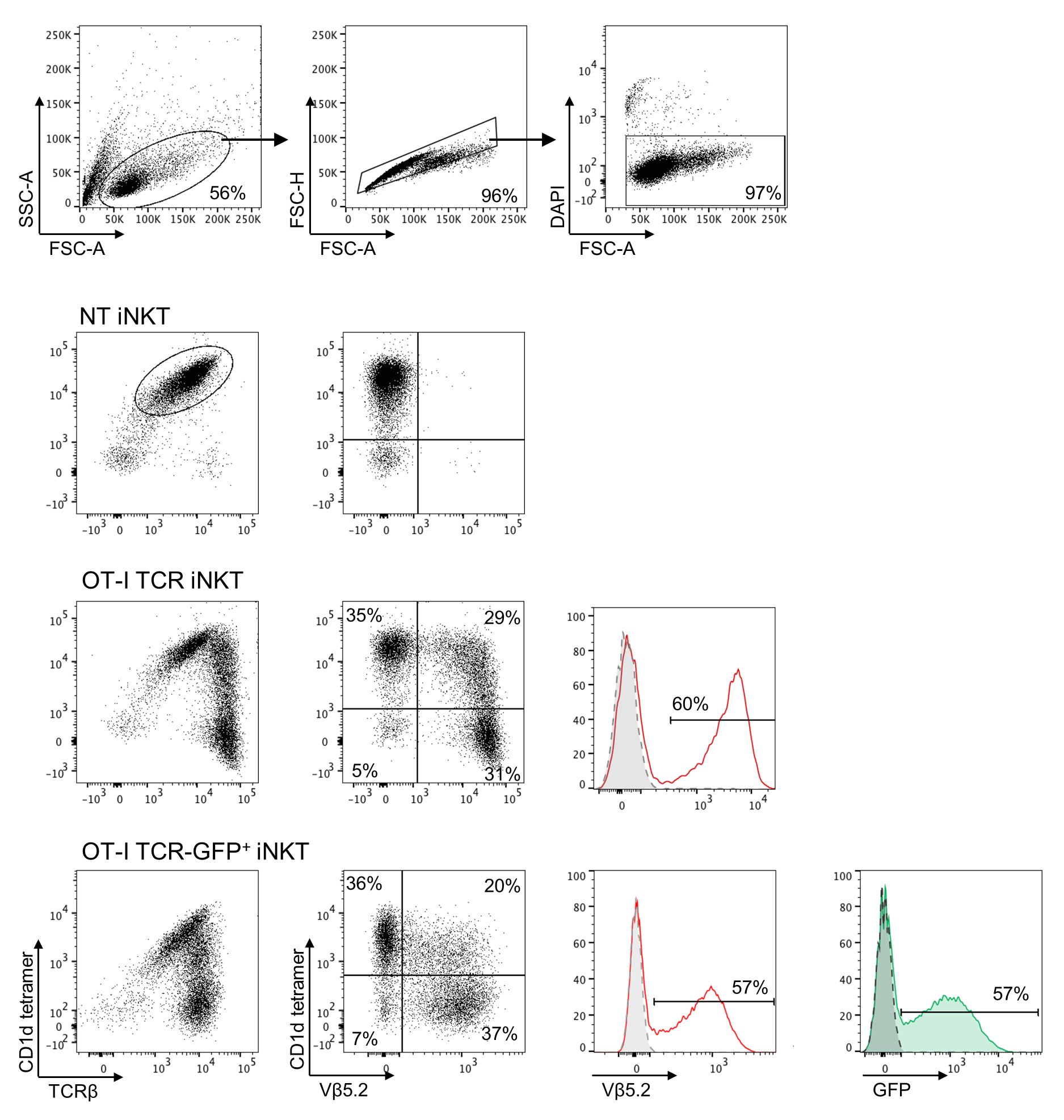
Figure 4. Gating strategy for analysis of transduced invariant natural killer T (iNKT) cells. Representative dot plots of gating strategy for analysis of control non-transduced (NT), OT-I TCR, and OT-I TCR GFP-transduced iNKT cells. The expression of the transgene is assessed on viable cells by staining with anti-Vβ5.2 monoclonal antibody (mAb). Dotted grey lines in the histograms represent the NT cells. When the RV also expresses a GFP reporter (bottom line), iNKT cells transduction can also be evaluated by the GFP expression.
Notes
iVα14 tg mice are commercially available; however, their availability could be a limitation for the reproducibility of the current protocol. In absence of these mice, we envisage the possibility of using a large number of C57BL/6 mice, but the protocol needs to be set up accordingly due to the paucity of iNKT cells in C57BL/6 mice [iNKT cell frequencies in C57BL/6 and iVα14 tg are shown in Supplementary Figure 1 of Delfanti et al. (2022)].
For unequivocal detection of iNKT cells in the present protocol, we used mouse PBS-57-CD1d-tetramers provided by NIH. PBS-57 is an analog of the prototypical antigen recognized by iNKT cells αGal-Cer, with improved solubility (Liu et al., 2006). The NIH Tetramer Facility provides PBS-57 ligand complexed to CD1d tetramers. However, we envisage the possibility to adjust the protocol for the use of commercially available CD1d dimers/tetramers/dextramers that can be loaded with lipid antigens as αGal-Cer.
For optimal iNKT cell transduction, it is recommended to determine the RV titer. To do so, actively proliferating mouse hybridoma cells (i.e., 58α-β- cells) can be transduced as described in Section C steps 5–6, with serial dilution of the RV (from 1:50 up to 1:100,000). Five days after transduction, the transgene expression can be assessed by flow cytometry (fix cells with 2% PFA), and RV titer (expressed as transducing units/milliliter, TU/mL) is calculated according to the formula [N × (F/100) × D]/0.2, where N is the number of the transduced cells (usually 2.5 × 105), F is the percentage of cells expressing the transgene, and D is the fold dilution of virus used in transduction. The RV titer allows the calculation of the multiplicity of infection (MOI) with the following formula: MOI = V(mL) × TU/mL/N.
Transduction efficacy can vary depending on the construct used. In the case of the TCR transfer (Delfanti et al., 2022), we obtained a good (60%) transduction using MOI 4 of our RV. However, there are two possible strategies to increase iNKT cell transduction efficacy: by increasing the MOI employed, or by transducing iNKT cells 48 h after activation and repeating the transduction also 72 h after the activation.
Fresh, non concentrated RV supernatants can also be employed for iNKT cell transduction. In this case, the maximum volume of RV that can be added to the 48-well plate is 1.5 mL. We recommend always adding 8 μg/mL of polybrene and performing spin inoculation as described above.
Recipes
IMDM + 10% Hyclone
IMDM with L-Glutamine
10% Hyclone
1× Penicillin/streptomycin solution
TE 0.1-H2O
Dilute TE buffer 1:10 in dH2O to obtain TE 0.1; then, mix two volumes of TE 0.1 and one volume of dH2O.
2× HEPES buffered saline (HBS)
281 mM NaCl
100 mM HEPES
1.5 mM Na2HPO4
Dissolve in dH2O, pH 7.14, and 0.22 μm filter.
Store at -20 °C and avoid re-freezing.
ACK (ammonium-chloride-potassium) lysing buffer
0.15 M NH4Cl
10 mM KHCO3
0.1 mM N2EDTA
Dissolve in dH2O, pH 7.2–7.4, and 0.22 μm filter.
Store at 4 °C. ACK is also commercially available.
MACS separation buffer
0.5% BSA
2 mM EDTA stock solution
In PBS, pH 7.2, 0.22 μm filtered
Store at 4 °C. MACS buffer is also commercially available.
Complete RPMI
RPMI 1640-GlutaMAX
10% heat-inactivated FBS
1× Penicillin/streptomycin solution
1× non-essential amino acids
1 mM sodium pyruvate
50 μM 2-mercaptoethanol
Acknowledgments
This work was funded by Associazione Italiana Ricerca sul Cancro (AIRC) project grant IG2017-ID.20081 (to G.C.), AIRC “under-5-per-Mille” 2019-ID.22737 (to P.D.), Italian Healthy Ministry project on CAR T RCR-2019-23669115 (to P.D.), Worldwide Cancer Research project grant 19-0133 (to G.C.). G.D. was supported by FIRC-AIRC Fellowship number 2019-22604 and by the “young researchers mobility programme” fellowship by Associazione Giovanna Tosi per la lotta contro i tumori. The authors acknowledge the NIH tetramer facility for providing the mouse CD1d tetramers. The protocol presented here was adapted from previously published works (Delfanti et al., 2021 and 2022).
Competing interests
The authors declare no conflict of interest.
Ethical considerations
All procedures were reviewed and approved by the San Raffaele Scientific Institute Institutional Animal Care and Use Committee (678 and 1067) and by the Italian Ministry of Health (Rome, Italy) and were conducted in compliance with national laws and policies.
References
- Bendelac, A., Savage, P. B. and Teyton, L. (2007). The biology of NKT cells. Annu Rev Immunol 25: 297-336.
- Chiba, A., Cohen, N., Brigl, M., Brennan, P. J., Besra, G. S. and Brenner, M. B. (2009). Rapid and reliable generation of invariant natural killer T-cell lines in vitro. Immunology 128(3): 324-333.
- Cortesi, F., Delfanti, G., Grilli, A., Calcinotto, A., Gorini, F., Pucci, F., Lucianò, R., Grioni, M., Recchia, A., Benigni, F., et al. (2018). Bimodal CD40/Fas-Dependent Crosstalk between iNKT Cells and Tumor-Associated Macrophages Impairs Prostate Cancer Progression. Cell Rep 22(11): 3006-3020.
- Delfanti, G., Cortesi, F., Perini, A., Antonini, G., Azzimonti, L., de Lalla, C., Garavaglia, C., Squadrito, M. L., Fedeli, M., Consonni, M., et al. (2022). TCR-engineered iNKT cells induce robust antitumor response by dual targeting cancer and suppressive myeloid cells. Sci Immunol 7(74): eabn6563.
- Delfanti, G., Perini, A., Zappa, E. and Fedeli, M. (2021). Purification and Expansion of Mouse Invariant Natural Killer T Cells for in vitro and in vivo Studies. J Vis Exp (168). doi: 10.3791/62214.
- De Santo, C., Arscott, R., Booth, S., Karydis, I., Jones, M., Asher, R., Salio, M., Middleton, M. and Cerundolo, V. (2010). Invariant NKT cells modulate the suppressive activity of IL-10-secreting neutrophils differentiated with serum amyloid A. Nat Immunol 11(11): 1039-1046.
- De Santo, C., Salio, M., Masri, S. H., Lee, L. Y., Dong, T., Speak, A. O., Porubsky, S., Booth, S., Veerapen, N., Besra, G. S., et al. (2008). Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest 118(12): 4036-4048.
- Dowling, P., Gargan, S., Zweyer, M., Henry, M., Meleady, P., Swandulla, D. and Ohlendieck, K. (2020). Protocol for the Bottom-Up Proteomic Analysis of Mouse Spleen. STAR Protoc 1(3): 100196.
- Godfrey, D. I., Le Nours, J., Andrews, D. M., Uldrich, A. P. and Rossjohn, J. (2018). Unconventional T Cell Targets for Cancer Immunotherapy. Immunity 48(3): 453-473.
- Gorini, F., Azzimonti, L., Delfanti, G., Scarfo, L., Scielzo, C., Bertilaccio, M. T., Ranghetti, P., Gulino, A., Doglioni, C., Di Napoli, A., et al. (2017). Invariant NKT cells contribute to chronic lymphocytic leukemia surveillance and prognosis. Blood 129(26): 3440-3451.
- Griewank, K., Borowski, C., Rietdijk, S., Wang, N., Julien, A., Wei, D. G., Mamchak, A. A., Terhorst, C. and Bendelac, A. (2007). Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27(5): 751-762.
- Heczey, A., Courtney, A. N., Montalbano, A., Robinson, S., Liu, K., Li, M., Ghatwai, N., Dakhova, O., Liu, B. and Raveh-Sadka, T., et al. (2020). Anti-GD2 CAR-NKT cells in patients with relapsed or refractory neuroblastoma: an interim analysis. Nat Med 26(11): 1686–1690.
- Janakiram, N. B., Mohammed, A., Bryant, T., Ritchie, R., Stratton, N., Jackson, L., Lightfoot, S., Benbrook, D. M., Asch, A. S., Lang, M. L., et al. (2017) Loss of natural killer T cells promotes pancreatic cancer in LSL-KrasG12D/+ mice. Immunology 152(1): 36-51.
- Landoni, E., Smith, C. C., Fucá, G., Chen, Y., Sun, C., Vincent, B. G., Metelitsa, L. S., Dotti, G. and Savoldo, B. (2020). A High-Avidity T-cell Receptor Redirects Natural Killer T-cell Specificity and Outcompetes the Endogenous Invariant T-cell Receptor. Cancer Immunol Res 8(1): 57–69.
- Liu, D., Song, L., Wei, J., Courtney, A. N., Gao, X., Marinova, E., Guo, L., Heczey, A., Asgharzadeh, S., Kim, E., et al. (2012). IL-15 protects NKT cells from inhibition by tumor-associated macrophages and enhances antimetastatic activity. J Clin Invest 122(6): 2221-2233.
- Liu, Y., Goff, R. D., Zhou, D., Mattner, J., Sullivan, B. A., Khurana, A., Cantu, C., 3rd, Ravkov, E. V., Ibegbu, C. C., Altman, J. D., et al. (2006). A modified alpha-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods 312(1-2): 34-39.
- Rotolo, A., Caputo, V. S., Holubova, M., Baxan, N., Dubois, O., Chaudhry, M. S., Xiao, X., Goudevenou, K., Pitcher, D. S., Petevi, K., et al. (2018). Enhanced Anti-lymphoma Activity of CAR19-iNKT Cells Underpinned by Dual CD19 and CD1d Targeting. Cancer Cell 34(4): 596-610.e11.
- Song, L., Asgharzadeh, S., Salo, J., Engell, K., Wu, H. W., Sposto, R., Ara, T., Silverman, A. M., DeClerck, Y. A., Seeger, R. C., et al. (2009). Valpha24-invariant NKT cells mediate antitumor activity via killing of tumor-associated macrophages. J Clin Invest 119(6): 1524-1536.
- Wolf, B. J., Choi, J. E. and Exley, M. A. (2018). Novel Approaches to Exploiting Invariant NKT Cells in Cancer Immunotherapy. Front Immunol 9: 384.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Delfanti, G., Dellabona, P. and Casorati, G. (2023). Primary Mouse Invariant Natural Killer T (iNKT) Cell Purification and Transduction. Bio-protocol 13(13): e4707. DOI: 10.21769/BioProtoc.4707.
- Delfanti, G., Cortesi, F., Perini, A., Antonini, G., Azzimonti, L., de Lalla, C., Garavaglia, C., Squadrito, M. L., Fedeli, M., Consonni, M., et al. (2022). TCR-engineered iNKT cells induce robust antitumor response by dual targeting cancer and suppressive myeloid cells. Sci Immunol 7(74): eabn6563.
Category
Immunology > Immune cell isolation > Leukocyte
Cell Biology > Cell isolation and culture > Cell isolation > Dynabead
Cell Biology > Cell engineering
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link