- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Autolysin Production from Chlamydomonas reinhardtii
Published: Vol 13, Iss 13, Jul 5, 2023 DOI: 10.21769/BioProtoc.4705 Views: 1682
Reviewed by: Ansul LokdarshiSamed DelicNoelia Foresi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2852 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1966 Views
Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1756 Views
Abstract
Chlamydomonas reinhardtii is a model organism for various processes, from photosynthesis to cilia biogenesis, and a great chassis to learn more about biofuel production. This is due to the width of molecular tools available, which have recently expanded with the development of a modular cloning system but, most importantly, with CRISPR/Cas9 editing now being possible. This technique has proven to be more efficient in the absence of a cell wall by using specific mutants or by digesting Chlamydomonas cell wall using the mating-specific metalloprotease autolysin (also called gametolysin). Multiple protocols have been used and shared for autolysin production from Chlamydomonas cells; however, they provide very inconsistent results, which hinders the capacity to routinely perform CRISPR mutagenesis. Here, we propose a simple protocol for autolysin production requiring transfer of cells from plates into a dense liquid suspension, gametogenesis by overnight incubation before mixing of gametes, and enzyme harvesting after 2 h. This protocol has shown to be highly efficient for autolysin production regardless of precise control over cell density at any step. Requiring a minimal amount of labor, it will provide a simple, ready-to-go approach to produce an enzyme critical for the generation of targeted mutants.
Graphical overview
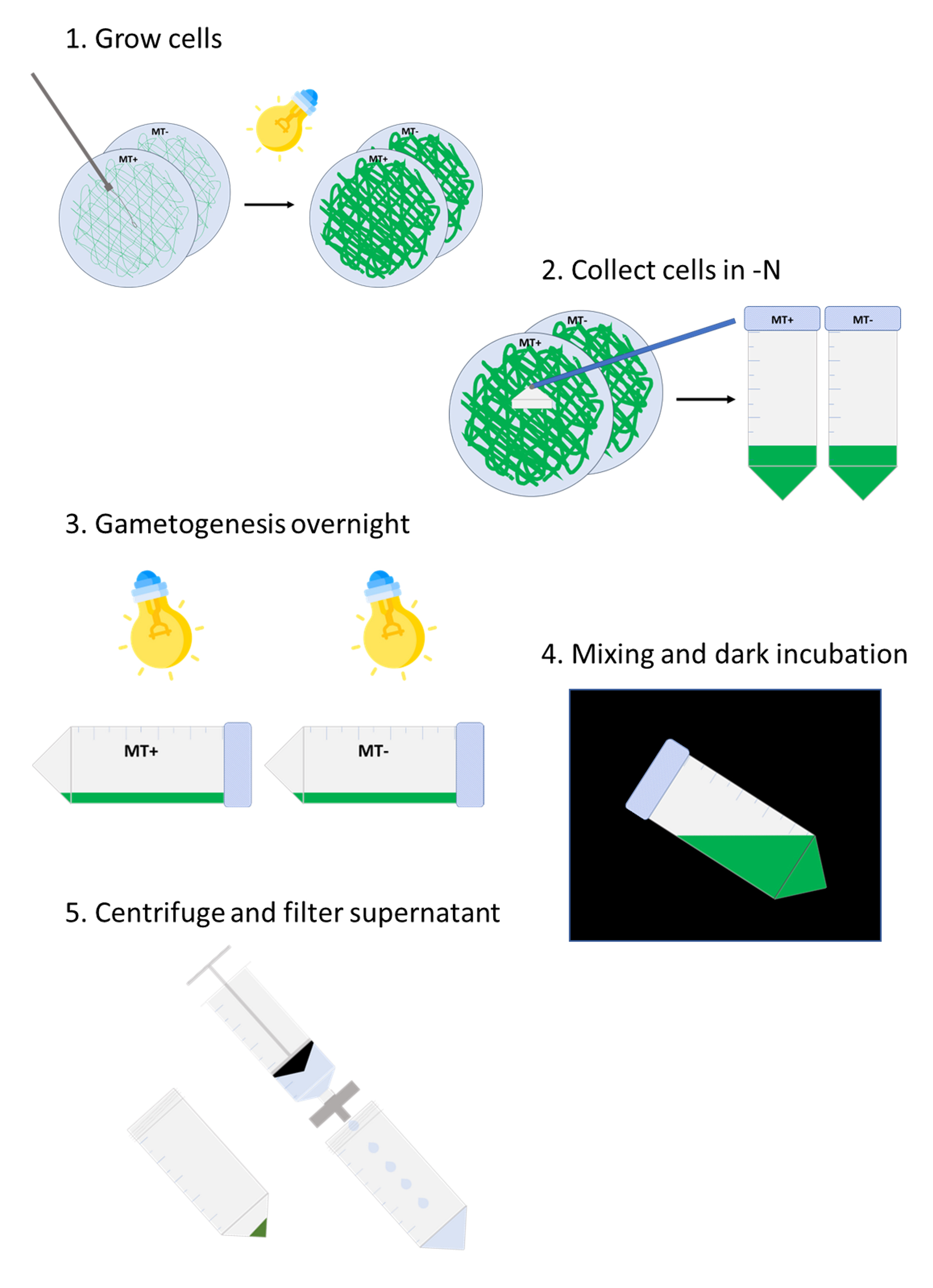
Workflow for autolysin production from Chlamydomonas reinhardtii
Background
Chlamydomonas’ genetic transformation has been possible for random insertional mutagenesis for over three decades (Kindle, 1990). In the last six years, methods have been developed for the precise editing of Chlamydomonas genome using the CRISPR technology by transfection of the ribonucleoprotein complex (RNP) directly through electroporation (Shin et al., 2016). Results have made clear that the cell wall is a strong barrier to entry of the RNP, as editing efficiency is usually much higher in cell wall–less strains or after digestion of the cell wall. Cell wall degradation can be performed using a commercially available enzyme called subtilisin/alcalase, but it has been shown to be only half as efficient as autolysin (Hwang et al., 2018), an enzyme produced by Chlamydomonas during mating in order to digest the cell wall and allow cell fusion of the two partners (Matsuda and Kubo, 2004). Autolysin can be extracted from a mixture of mating cells by centrifugation, preserved by freezing at -80 °C, and used later on any given walled strain to prepare for transformation. Several protocols are available in the literature as well as on the Chlamydomonas Resource Center website, but their efficiency has proven inconsistent in our hands. Here, we present a very simple protocol that requires minimal monitoring of cell growth and density, with overall > 70% efficiency. This will allow researchers interested in using the CRISPR technology to produce good quality autolysin and obtain mutated strains in their gene of interest, regardless of their favorite wildtype strain.
Materials and reagents
90 mm Petri dishes
Inoculating loop
Sterile 50 mL centrifuge tubes
Cell scraper
Pipette tips
1.7 mL microcentrifuge tubes
50 mL conical tubes
50 mL Luer-lock syringe
0.45 μm PES filter (VWR, catalog number: 76479-020)
Permanent marker pen
Microscope slides with eight wells (75 mm × 25 mm) (MP Biomedicals, catalog number: 096040805)
Plastic spectrophotometer cuvette (VWR, catalog number: 97000-586)
1 L filtration unit (VWR, catalog number: 10040-440)
Mating type (MT)+ wildtype strain CC-125 (available from the Chlamydomonas Resource Center, www.chlamy.org)
MT- wildtype strain CC-124 (available from the Chlamydomonas Resource Center, www.chlamy.org)
Note: Other pairs of MT+/MT- strains can be used; CC-620/621 are usually recommended due to their high mating efficiency.
Cell wall–less wildtype strain CC-503 (cw92, available from the Chlamydomonas Resource Center, www.chlamy.org)
Ammonium chloride (NH4Cl) (Sigma, catalog number: A9434)
Sodium chloride (NaCl) (Sigma, catalog number: 746398)
Calcium chloride dihydrate (CaCl2·2H2O) (Sigma, catalog number: C3881)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma, catalog number: 63140)
Tris base (J.T. Baker, catalog number: 4099-07)
Potassium phosphate monobasic (KH2PO4) (J.T. Baker, catalog number: 3246-07)
Potassium phosphate dibasic (K2HPO4) (J.T. Baker, catalog number: 3252-07)
Ethylenediaminetetraacetic acid disodium salt dihydrate (Na2EDTA·2H2O) (Sigma, catalog number: E5134)
Iron (II) sulfate heptahydrate (FeSO4·7H2O) (Sigma, catalog number: F-8048)
Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sigma, catalog number: Z0251)
Boric acid (H3BO3) (Sigma, catalog number: B6768)
Manganese (II) chloride tetrahydrate (MnCl2·4H2O) (Sigma, catalog number: 221279)
Copper (II) chloride dihydrate (CuCl2·2H2O) (Sigma, catalog number: 467847)
Sodium molybdate dihydrate (Na2MoO4·2H2O) (J.T. Baker, catalog number: 3764)
Cobalt (II) chloride hexahydrate (CoCl2·6H2O) (Sigma, catalog number: C2644)
Glacial acetic acid (Sigma, catalog number: 320099)
Triton X-100 (Sigma, catalog number: T-9284)
Granulated agar (Fisher Scientific, catalog number: BP9744-5)
0.1% Triton X-100 (see Recipes)
Sucrose (Sigma, catalog number: S0389)
Stock solutions
Salts +N solution (20×) (see Recipes)
Salts -N solution (20×) (see Recipes)
2 M Tris Base (100×)
Phosphate buffer (200×) (see Recipes)
Hutner’s trace elements (200×) (see Recipes, also available from the Chlamydomonas Resource Center, www.chlamy.org)
TAP (Tris-Acetate-Phosphate) liquid medium (see Recipes)
TAP 1.5% agar medium (see Recipes)
TAP -N liquid medium (see Recipes)
TAP sucrose 40 mM (see Recipes)
Equipment
Pipettes
Centrifuge and microcentrifuge
Light microscope
Laminar flow hood
Glass beaker
Spectrophotometer
Vacuum pump
Procedure
Cell growth
Spread MT+ (CC-125) and MT- (CC-124) cells on TAP 1.5% agar medium using an inoculating loop (approximately two loop-full of cells spread over the whole surface).
Incubate for 7–10 days in bright light (lid facing down, 22 °C, ~100 μmoles m-2·s-1) (Figure 1).
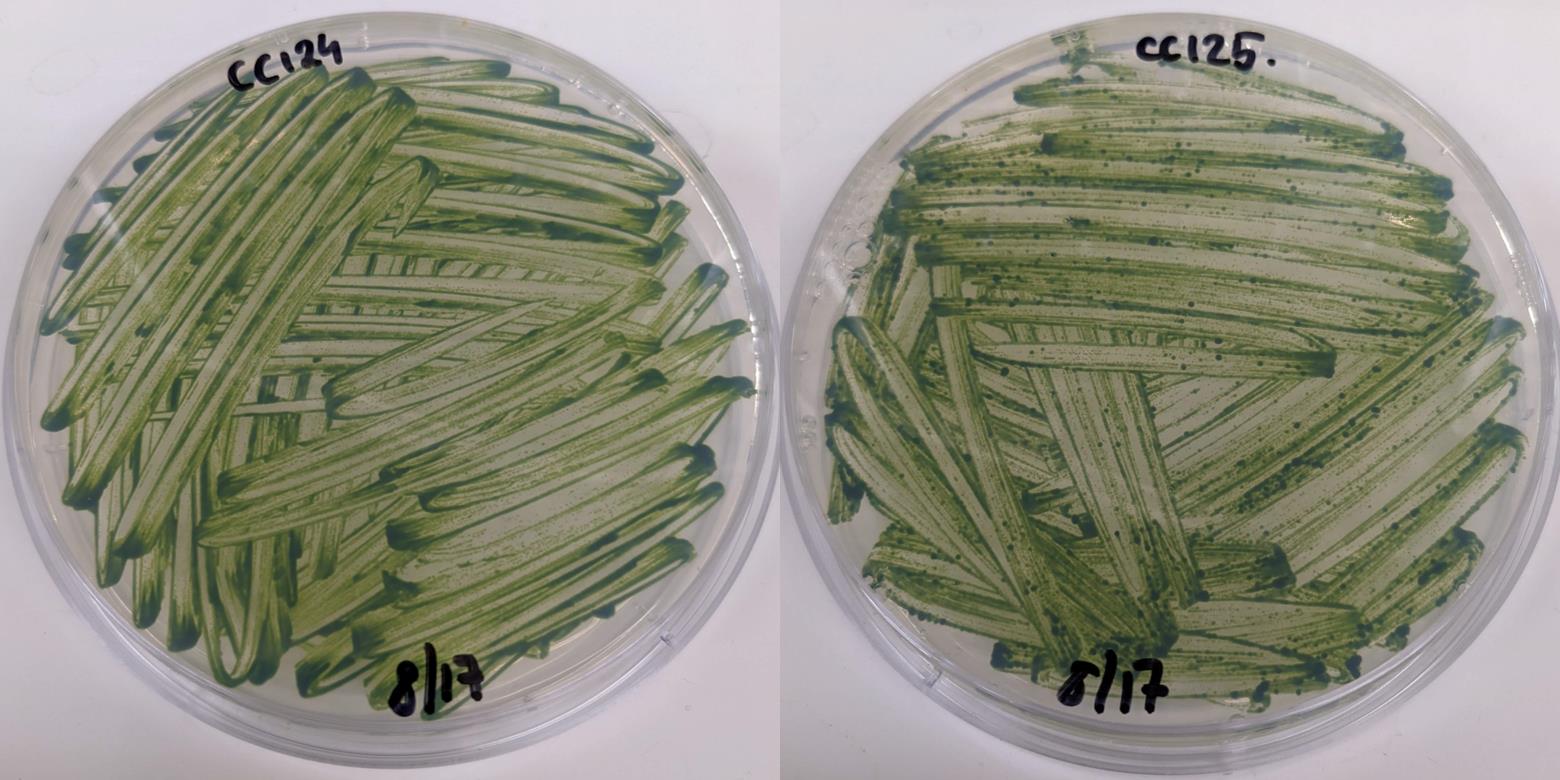
Figure 1. Plates of Chlamydomonas CC-124 (MT-) and CC-125 (MT+) cells grown on TAP agar plate at harvesting time. Two loop-full of cells were spread over the plate and grown under bright light (100 μmoles m-2·s-1) for seven days.
Gametogenesis
Start this step in the late afternoon (5:00–6:00 PM).
For each plate, add 10 mL of TAP -N liquid medium to a sterile 50 mL centrifuge tube (use one tube per plate).
Using a cell scraper, transfer the cells from the agar to the liquid.
Use a pipette to resuspend cell clumps as much as possible (Figure 2).

Figure 2. Cell suspension after transfer from TAP agar plates to TAP -N liquid medium. Cells were scraped from plate and resuspended in liquid medium (here, MT- CC-124 on the left and MT+ CC-125 on the right). Pipetting is used to break cell clumps as much as possible.Leave horizontally in bright light (~100 μmoles m-2·s-1) overnight without shaking.
Mating and autolysin production
In the morning, transfer 10 μL of cells to a microscope slide and check for motility.
Note: Over 50% of cells should be moving actively.
Mix the MT+ and MT- strains in the 50 mL centrifuge tubes and incubate for 2 h in the dark without shaking.
Centrifuge at 3,000× g for 5 min.
Transfer supernatant to a clean glass beaker and discard the tube containing the pellet.
Filter the supernatant into a sterile 50 mL centrifuge tube using a 50 mL syringe and a 0.45 μm PES filter.
Set aside 2 mL of filtered supernatant for Section D and place the rest to freeze immediately in a -80 °C freezer.
Testing autolysin preparation efficiency
Four days prior to autolysin production, inoculate CC-125 (cell walled) and CC-503 (cell wall–less) strains in 5 mL of TAP liquid medium in a 50 mL conical tube.
Note: Any cell wall–less strain (cw15 or cw92 mutant) can be used as a control for lysis. After detergent treatment (step D10), it should show a white pellet and a green supernatant. CC-5325/CC-4533, which is annotated cw15, synthesizes a cell wall and should not be used for that purpose (Zhang et al., 2022).
Harvest the 5 mL of late log CC-125 and CC-503 cells and adjust each OD750nm to 0.3 with fresh TAP -N medium.
To evaluate lysis efficiency by chlorophyll absorbance, transfer 1 mL to a cuvette and measure OD662nm of the initial cell suspensions (OD1, zero with TAP -N medium).
Centrifuge 4 × 1 mL of CC-125 and 2 × 1 mL of CC-503 at 1,500× g for 5 min in 1.7 mL centrifuge tubes.
Discard the supernatants.
Resuspend the two pellets of CC-503 in 1 mL each of TAP -N (positive control), two pellets of CC-125 in 1 mL each of TAP -N liquid medium (negative control), and the two other CC-125 pellets in 1 mL each of the autolysin preparation supernatant set aside in step C6. We now have six tubes for three conditions: cell wall–less CC-503, cell-walled CC-125 in TAP -N, and cell-walled CC-125 treated with the prepared autolysin.
Incubate horizontally for 45 min in bright light (~100 μmoles m-2·s-1) without shaking.
Centrifuge all the tubes at 1,500× g for 5 min.
Discard the supernatants.
For each condition, resuspend tube 1 in 1 mL of water and tube 2 in 1 mL of 0.1% Triton X-100.
Vortex for 2 min.
Centrifuge at 1,500× g for 5 min.
Autolysin efficiency can be quickly assessed from the color of the pellets and supernatants (Figure 3). After Triton X-100 treatment, the supernatant in the CC-125 cells treated with efficient autolysin is green, and we can proceed with the quantification of lysis efficiency. If it is still translucent, the autolysin preparation failed, and all tubes should be discarded.
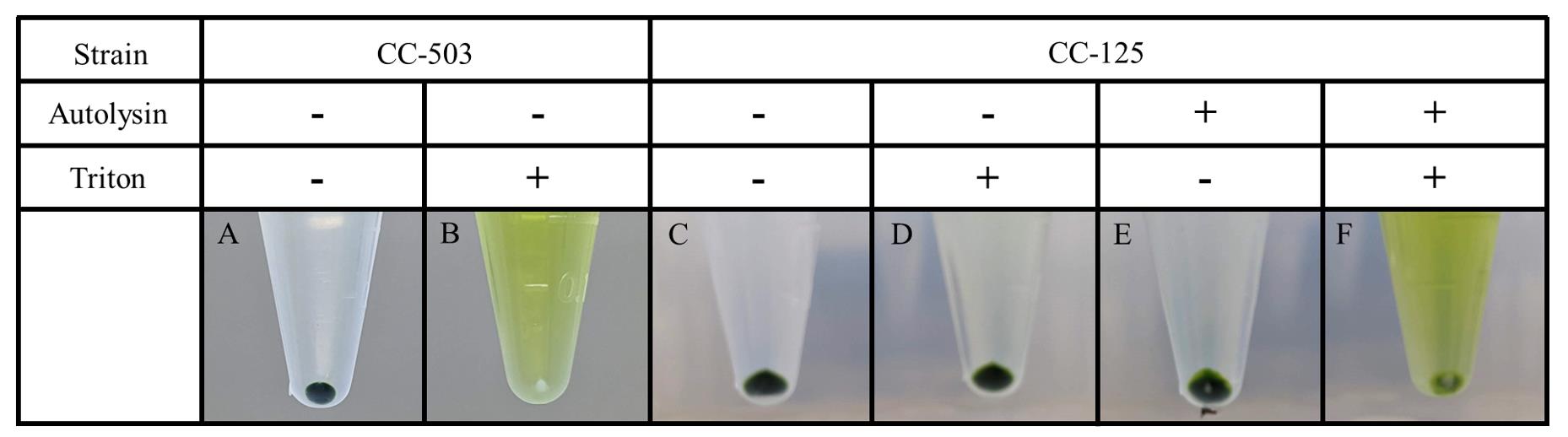
Figure 3. Expected results from autolysin and detergent treatment. Cells incubated with the prepared autolysin or in control conditions were treated with the detergent Triton X-100 to induce cell lysis. Cell wall–less cells (CC-503) display a green supernatant showing cell lysis and release of chlorophyll, while the pellet becomes white (B). Cell-walled cells (CC-125) incubated in control conditions are resistant to the detergent and the cell pellet stays green (D). Cells treated with good autolysin undergo at least partial lysis and display a green supernatant (F).Transfer the supernatant to a cuvette and measure OD662nm (OD2, zero with 0.1% Triton X-100).
Calculate gross efficiency by the ratio between the OD662nm after (OD2) and before (OD1) detergent treatment:
Efficiency (%) = OD2662nm/OD1662nm
Using the autolysin preparation
Thaw the frozen autolysin in a beaker filled with water at room temperature for ~1 h.
Note: We recommend using the freshest preparation possible. Avoid using autolysin stored for more than two months, as it will significantly lose activity over time.
Centrifuge cells to transform at 1,500× g for 5 min.
Discard the supernatant.
Resuspend the pellet in autolysin solution at ~5 × 106 cells/mL.
Incubate in bright light (~100 μmoles m-2·s-1) without shaking for 45 min.
Centrifuge at 1,500× g for 5 min.
Discard the supernatant.
Resuspend the pellet in 25 mL of TAP sucrose 40 mM.
Centrifuge at 1,500× g for 5 min.
Discard the supernatant.
Proceed with the transformation protocol.
Data analysis
All samples not treated with Triton X-100 are expected to show a clear supernatant and a dark green pellet (Figure 3A, 3C, and 3E). After Triton X-100 treatment, the cell wall–less strain CC-503 will show a green supernatant and a white pellet (Figure 3B). On the contrary, the CC-125 control cells that were incubated in TAP -N display a clear supernatant and a dark green pellet like the untreated cells (Figure 3D). This is because the control cells are still protected by their cell wall, while the CC-503 strain is highly susceptible to lysis due to the absence of the cell wall and releases its chlorophyll content in the supernatant after detergent treatment. If the preparation works, the CC-125 cells treated with autolysin and Triton X-100 will show a similar result to the cell wall–less strain. Depending on the quality of the autolysin preparation, part of the pellet can still be green; in this case, serial treatment of cells to be transformed can be necessary. If no white pellet is observed, discard the frozen autolysin tube.
Note: Sometimes, the pellet from autolysin-treated cells will appear partly white even without detergent treatment because vortexing is harsh enough to lyse cell wall–less cells.
To ensure the best quality of autolysin, cell lysis efficiency can also be quantified by measuring chlorophyll release in the supernatant. The best result will be reached when the ratio of OD662nm after lysis over OD662nm before lysis is the closest to the value obtained with the cell wall–less strain. In Table 1, we provide an example of efficiency of chlorophyll release obtained from cell wall–less cells (CC-503) and from untreated cell-walled cells (CC-125), showing the difference observed between lysed and intact cells.
Table 1. Quantification of chlorophyll release by detergent treatment. Example of quantification of chlorophyll release in the supernatant from cell wall–less (CC-503) and cell-walled (CC-125) cells treated with Triton X-100.
OD662nm of the | OD662nm of the | Efficiency (OD1/OD2, %) | Average efficiency (%) | |
| CC-503 | 0.27 | 0.183 | 67.7 | 67.7 |
| 0.261 | 0.187 | 71.6 | ||
| 0.276 | 0.176 | 63.8 | ||
| CC-125 control | 0.21 | 0.033 | 15.7 | 15.9 |
| 0.208 | 0.032 | 15.4 | ||
| 0.212 | 0.035 | 16.5 |
Validation of protocol
Autolysin prepared using this protocol has been used to generate targeted mutants by CRISPR/Cas9 in the TPT2 and TPT3 genes of Chlamydomonas reinhardtii (Huang et al., 2023). This protocol was also used for preparation of CC-124 cells prior to insertional mutagenesis and phenotypic screen for starch catabolism mutants (Tunçay et al., 2013).
Recipes
0.1% Triton X-100 (for 100 mL)
100 μL of Triton X-100
Complete with distilled water
Store at room temperature
Stocks solutions
Salts +N solution (for 1 L, 20×)
150 mM NH4Cl
16.6 mM MgSO4·7H2O
6.8 mM CaCl2·2H2O
Complete with distilled water
Store at room temperature
Salts -N solution (for 1 L, 20×)
150 mM NaCl
16.6 mM MgSO4·7H2O
6.8 mM CaCl2·2H2O
Complete with distilled water
Store at room temperature
2 M Tris base (for 1 L, 100×)
2 M Tris base
Complete with distilled water
Store at room temperature
Phosphate buffer (for 1 L, 200×)
687 mM KH2PO4
362 mM K2HPO4
Complete with distilled water
Store at room temperature
Hutner’s trace elements (for 1 L, 200×)
800 mL of distilled water
27 mM Na2EDTA·2H2O
Adjust pH to 6.5 with NaOH
3.6 mM FeSO4·7H2O
15 mM ZnSO4·7H2O
37 mM H3BO3
5.2 mM MnCl2·4H2O
1.3 mM CuCl2·2H2O
1.1 mM Na2MoO4·2H2O
1.3 mM CoCl2·6H2O
Complete with distilled water
Autoclave for 30 min at 120 °C
The solution’s color is light green after mixing all the powders. Let the solution oxidize for a week in the dark. It will turn orange, brown, and then purple
Store at room temperature in obscurity
TAP medium (for 1 L)
800 mL of distilled water
50 mL of +N Salt solution
10 mL of tris base
5 mL of phosphate buffer
5 mL of Hutner’s trace elements
1 mL of glacial acetic acid
Check pH range 7–7.2
Complete to 1 L with distilled water
Autoclave for 30 min at 120 °C
TAP 1.5% agar medium (for 1 L)
800 mL of distilled water
50 mL of salt +N solution
10 mL of Tris Base
5 mL of phosphate buffer
5 mL of Hutner’s trace elements
1 mL of glacial acetic acid
Check pH range 7–7.2
Complete to 1 L with distilled water
15 g of agar
Autoclave for 30 min at 120 °C
TAP -N medium (for 1 L)
800 mL of distilled water
50 mL of salt -N solution
10 mL of tris base
5 mL of phosphate buffer
5 mL of Hutner’s trace elements
1 mL of glacial acetic acid
Check pH range 7–7.2
Complete with distilled water
Autoclave for 30 min at 120 °C
TAP sucrose (for 1 L)
40 mM sucrose
Complete with TAP liquid medium
Filter sterilize
Acknowledgments
I am thankful to Neda Fakhimi, Sai Madireddi, and Carolyne Stoffel for feedback on this protocol. I acknowledge David Dauvillée from whom I adapted this protocol. This work was supported by the Carnegie Institution for Science, Department of Plant Biology.
Competing interests
I declare no competing interests.
References
- Huang, W., Krishnan, A., Plett, A., Meagher, M., Linka, N., Wang, Y., Ren, B., Findinier, J., Redekop, P., Fakhimi, N., et al. (2023). Chlamydomonas mutants lacking chloroplast TRIOSE PHOSPHATE TRANSPORTER3 are metabolically compromised and light-sensitive. Plant Cell. doi: 10.1093/plcell/koad095.
- Hwang, H. J., Kim, Y. T., Kang, N. S. and Han, J. W. (2018). A Simple Method for Removal of the Chlamydomonas reinhardtii Cell Wall Using a Commercially Available Subtilisin (Alcalase). J Mol Microbiol Biotechnol 28(4): 169-178.
- Kindle, K. L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 87(3): 1228-1232.
- Matsuda, Y. and Kubo, T. (2004). Gametolysin. In: Handbook of Proteolytic Enzymes (2nd Edition). Academic Press, 592-595.
- Shin, S. E., Lim, J. M., Koh, H. G., Kim, E. K., Kang, N. K., Jeon, S., Kwon, S., Shin, W. S., Lee, B., Hwangbo, K., et al. (2016). CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci Rep 6: 27810.
- Tunçay, H., Findinier, J., Duchêne, T., Cogez, V., Cousin, C., Peltier, G., Ball, S. G. and Dauvillée, D. (2013). A forward genetic approach in Chlamydomonas reinhardtii as a strategy for exploring starch catabolism. PLoS One 8(9): e74763.
- Zhang, N., Pazouki, L., Nguyen, H., Jacobshagen, S., Bigge, B. M., Xia, M., Mattoon, E. M., Klebanovych, A., Sorkin, M., Nusinow, D. A., et al. (2022). Comparative Phenotyping of Two Commonly Used Chlamydomonas reinhardtii Background Strains: CC-1690 (21gr) and CC-5325 (The CLiP Mutant Library Background). Plants 11(5): 585.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Findinier, J. (2023). Autolysin Production from Chlamydomonas reinhardtii. Bio-protocol 13(13): e4705. DOI: 10.21769/BioProtoc.4705.
Category
Plant Science > Plant molecular biology > DNA > DNA modification
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








