- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Selection of Molecules with Immunological Potential from Excretory and Secretory Products from the Nematode Haemonchus placei by Cell Proliferation and Gene Expression Assays
Published: Vol 13, Iss 12, Jun 20, 2023 DOI: 10.21769/BioProtoc.4702 Views: 1272
Reviewed by: Jorge Francisco GonzálezAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Primary Mouse Invariant Natural Killer T (iNKT) Cell Purification and Transduction
Gloria Delfanti [...] Giulia Casorati
Jul 5, 2023 2114 Views
Detection of Cytoplasmic and Nuclear Circular RNA via RT-qPCR
Ke-En Tan [...] Yat-Yuen Lim
Sep 5, 2023 3283 Views
Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
Ruben Warkentin and Anna Marie Pyle
Oct 20, 2025 1317 Views
Abstract
The nematode Haemonchus placei is a pathogenic parasite, the most seriously affecting ruminant’s health and being responsible for enormous economic losses all over the world. The present protocol describes different in vitro techniques to select potential candidate antigens with immune-protective activity from excretory and secretory products (ESP) from H. placei transitory infective larvae (xL3). ESP from xL3 were obtained from the in vitro infective larvae (L3) maintained in Hank’s medium at 37 °C with 5% CO2 for 48 h. Then, the presence of ESP proteins was confirmed by SDS-PAGE to be used in an in vitro proliferation assay with bovine peripheral blood mononuclear cells (PBMCs). The ESP were exposed to the PBMCs during two different periods (24 and 48 h). Genes associated with immune response against the nematode were analyzed using relative gene expression and bioinformatic tools. These are simple, economic, and helpful tools to identify potential immune-protective molecules under in vitro conditions for confirming the efficacy of future in vivo assays.
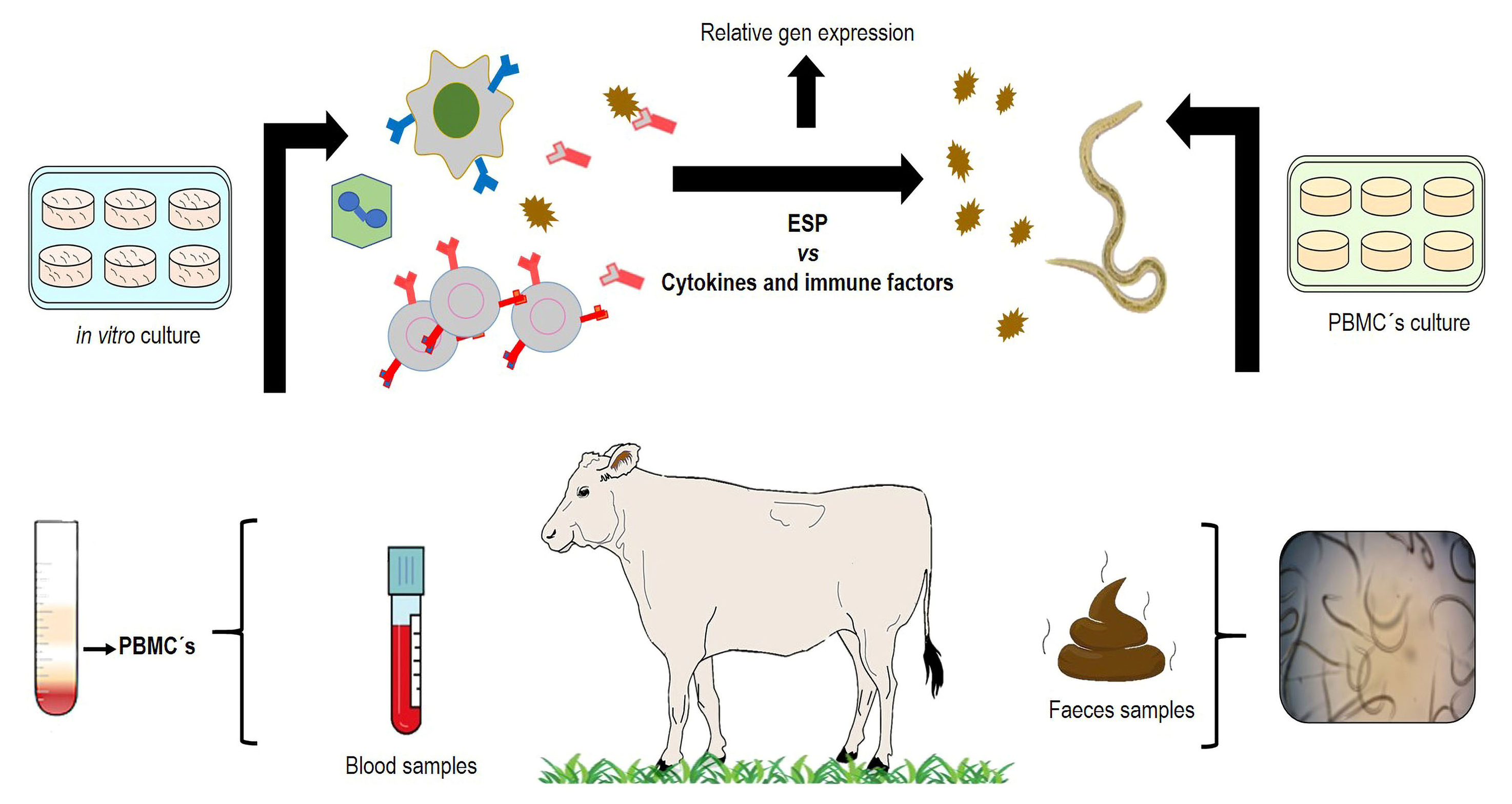
Graphical overview
Background
Domestic ruminants are severely affected by the complex of gastrointestinal nematodes (GINs). Some parasites belonging to this group (i.e., Ostertagia and Haemonchus) are particularly highly pathogenic due to their blood-feeding habits. The genus Haemonchus spp. is the most pathogenic parasitic nematode, particularly in tropical regions where its prevalence reaches > 80% (Liébano-Hernández et al., 2011). Traditionally, the control of GINs is based on the use of anthelmintic drugs; however, anthelmintic resistance has dramatically increased during the last decades, risking animal health and production. In order to reduce the use of chemical anthelmintic drugs, different strategies of control and prevention have been explored. One of them is assessing the use of specific proteins as potential immunizing agents, mainly those that are involved in the host–parasite interaction. The genus Haemonchus has been used as a biological model for understanding the invasive process to the host tissues. During invasion of the gastric mucosa tissues, different excretory and secretory products (ESP), mainly proteins, are produced; some of them could have important implications as potential immune protective antigens. The in vitro development from Haemonchus spp. infective larvae (L3) to the transitory infective larvae (xL3) is an easy and economic technique that allows the release of potential antigenic molecules with important immunoprotective value. The invasion mechanism of larvae to the host tissue and their further development require the activity of enzymes to use nutrients and thus achieve the adult stage for surviving in the abomasum (stomach) of ruminants. Recently, numerous studies have intensified the identification of ESP to induce immune protection, thus hampering the larvae’s invasive process. In addition, the importance of ESP during tissue invasion by parasitic nematodes is associated to inflammatory and regulatory immune mechanisms such as cytokines (e.g., IL4, IL5, IL6, IL8, IL10), immunoglobulins (IgA, IgG, and IgE), and other important immune cells like eosinophils and neutrophils. The study of antigens and immune cells requires the use of different experimental in vitro and in vivo processes, to improve the identification of ESP with possible biological functions. The present methodology describes reproducible experimental procedures for obtaining and studying the nematode’s ESP and the host’s immune response. The following protocol describes methods related to parasitology, immunology, and molecular procedures to obtain ESP from xL3of the nematode speciesHaemonchus placeiand to evaluate them in vitro activity.
Materials and reagents
T-75 cell culture flask (Thermo Fisher, catalog number: 156800)
Gauze (any brand can be used)
McMaster egg counting chamber (JorVetTM, catalog number: J0335M)
Plastic container, 18 cm × 38 cm (any brand can be used)
Foam rubber (any brand can be used)
Aluminum foil (any brand can be used)
Distilled water (any brand can be used)
Polypropylene funnel 150 mm diameter (Corning®, catalog number: 6120P-150)
10 mL culture tube (Pyrex®, catalog number: 99445-13)
Plastic hose assembly (Fisher Scientific, catalog number: 02-594-1F)
Pasteur plastic pipette, 1 mL (MediLab, catalog number: 121227/01)
Optical lens wipes (Carolina Biol. Supply Co., catalog number: 634000)
15 mL plastic centrifuge tube (Corning®, catalog number: 430052)
50 mL plastic centrifuge tube (Corning®, catalog number: 430828)
6-well plate, flat-bottom, clear and sterile, TC-treated (Corning®, catalog number: 3506)
Petri dishes 100 mm × 15 mm, sterile (Falcon®, catalog number: 351029)
Diameter syringe filters 0.2 μm pore (Corning®, Catalog number: 431219)
Vacutainer tubes with EDTA (Becton Dickinson, catalog number: 367863)
Hypodermic green needles, 21 G × 32 mm (Nipro®, catalog number: A21)
Pasteur pipettes (Sigma, catalog number: S5893)
0.2 mL PCR Tubes with flat cap (Axygen®, catalog number: PCR-02-C)
96 well-microplates, flat-bottom, clear and sterile, TC-treated (Corning®, catalog number: 3628)
24-well plate, flat-bottom, clear and sterile, TC-treated (Corning®, catalog number: 3527)
1.5 mL microcentrifuge plastic tuber (Axygen®, catalog number: MCT-150-A)
Plastic pestle (Bel-ArtTM, catalog number: F19923-0001)
Recently collected fecal samples from infected cattle withH. placei(300–500 g)
Sodium chloride (NaCl) of commercial food grade (any brand can be used)
Sucrose (Sigma-Aldrich, catalog number: MFCD00006626)
Sodium hypochlorite (NaClO) (Cloralex-Grupo AlEn-México)
Antibiotic-antimycotic (100×) (Thermo Fisher Scientific, GibcoTM, catalog number: 15240062)
Hanks’ Balanced Salts without sodium bicarbonate (Sigma, catalog number: H6136)
Ethanol molecular grade (Hycel, catalog number: 1822-500)
Peripheral blood sample from three non-infected cattle (36 mL)
LymphoprepTM (Alere Technologies, Axis-Shield, catalog number: 1114547)
RPMI 1640 medium with L-glutamine and HEPES (Gibco, catalog number: 23400062)
Fetal bovine serum (FBS) (By Products, catalog number: 90020)
Trypan blue stain (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061)
Neubauer Improved bright line (MARIENFELD, catalog number: 0640011)
CellTiter 96® Aqueous One Solution Cell Proliferation assay (Promega, catalog number: G3582)
QIAzol lysis reagent (Qiagen®, catalog number: 79306)
Chloroform (J.T. Baker, catalog number: 9180)
Isopropyl alcohol (Sigma-Aldrich, catalog number: W292907)
Nuclease-free water (Promega, catalog number: P1193)
Agarose (Bio-Rad, catalog number: 1613100)
Ethidium bromide 10 mg/mL (Bio-Rad, catalog number: 1610433)
RQ1 RNase-Free DNase kit (Promega, catalog number: M6101)
ImProm-II Reverse Transcription System kit (Promega, catalog number: A3800)
GoTaq® qPCR Master Mix 2× (Promega, catalog number: A6001)
Custom RT2 Profiler PCR array (Qiagen, catalog number: CAPB13410R)
Trizma base (Sigma, catalog number: 93362)
Glacial acetic acid (Meyer, catalog number: 0040)
EDTA 0.5 M pH 8.0 (Invitrogen, catalog number: AM9260G)
Potassium chloride (KCl) (Sigma, catalog number: P3911)
Sodium phosphate dibasic (Na2HPO4) (Sigma, catalog number: RDD038)
Potassium phosphate monobasic (KH2PO4) (Sigma, catalog number: 221309)
1× sterile phosphate buffered saline (PBS), pH 7.4 (see Recipes)
Buffer 50× TAE (see Recipes)
3% agarose gel (see Recipes)
0.187% NaClO solution (see Recipes)
Hank’s balanced salts (medium) (see Recipes)
40% sucrose solution (see Recipes)
Saturated sodium chloride solution (see Recipes)
Equipment
Centrifuge (Thermo Fisher Scientific, Sorvall ST 8R, catalog number: 75007204)
Microscopy (Zeizz, Primo Star Hal/Led Full-Köhler ERc5s, catalog number: 415500-0057-000)
0.1–10 μL, 2–20 μL, 20–200 μL, and 100–1,000 μL micropipettes (any brand can be used)
Shaker (Labnet, OrbitTM 1000, catalog number: S2030-1000-B)
Incubator (ECOSHEL, CI-80, CO2incubator)
Mixer (Benchmark Scientific, VortexTM, catalog number: BV101-P)
Microplate absorbance reader (Bio-Rad, iMarkTM, catalog number: 1681135)
Nanophotometer (Implen, NP80)
PowerPAc chamber (Bio-Rad, 300, catalog number: 165-5050)
Thermal cycler 6000 (Qiagen, Corbett Rotor-Gene 6000)
Transilluminator Imagine System (UVP, EC3, catalog number: 81-020901)
Nucleic acid electrophoresis chamber (any brand can be used)
Granatary scale 610 × 0.1 g (any brand can be used)
Freezer of -80 °C (any brand can be used)
pH meter (any brand can be used)
Software
Rotor-Gene Q—Pure detection Software (version 1.7)
GeneGlobe Data Analysis Center of Qiagen® (https://geneglobe.qiagen.com/analyze/)
Procedure
Obtaining H. placei L3 antigens
Recovery of H. placei’s L3 stages using parasitological techniques:
Note: Basic knowledge of copro-parasitological techniques and at least one isolated H. placei strain is required to perform this methodology. Any further information can be consulted in Thienpont et al. (2003).
McMaster technique: Perform the parasitological diagnosis of infected bovine with H. placei by McMaster technique to estimate the number of eggs per gram (EPG) from fecal samples (Liébano-Hernández et al., 2011; Cedillo-Borda et al., 2020).
i. Collect 10 g of fecal samples from infected bovine by rectal route using a clean plastic bag. Using nitrile gloves, introduce the index finger into the anal sphincter of the bovine, performing circular movements in a clockwise direction. The rectal stimulus will allow the transit of the feces from the rectum, until they cross through the anal sphincter. Collect the expelled feces with a plastic bag.
ii. Weigh 2 g of feces on a granatary scale and place them in a 50 mL plastic tube.
iii. Add 28 mL of saturated sodium chloride solution at 1:24 density (see Recipes) and homogenize the feces with a plastic paddle. Put a piece of gauze (5 cm × 5 cm) on the surface of the homogenized feces.
iv. Take out 5 mL of the homogenized sample through the folds formed in the introduced gauze with a Pasteur pipette. Then, fill both compartments of the McMaster chamber.
v. Perform egg count through the McMaster chamber. Consider the number of EPG into the McMaster chamber lines from top to bottom and from left to right between chambers.
vi. Quantify the total EPG using the formula shown in Figure 1.
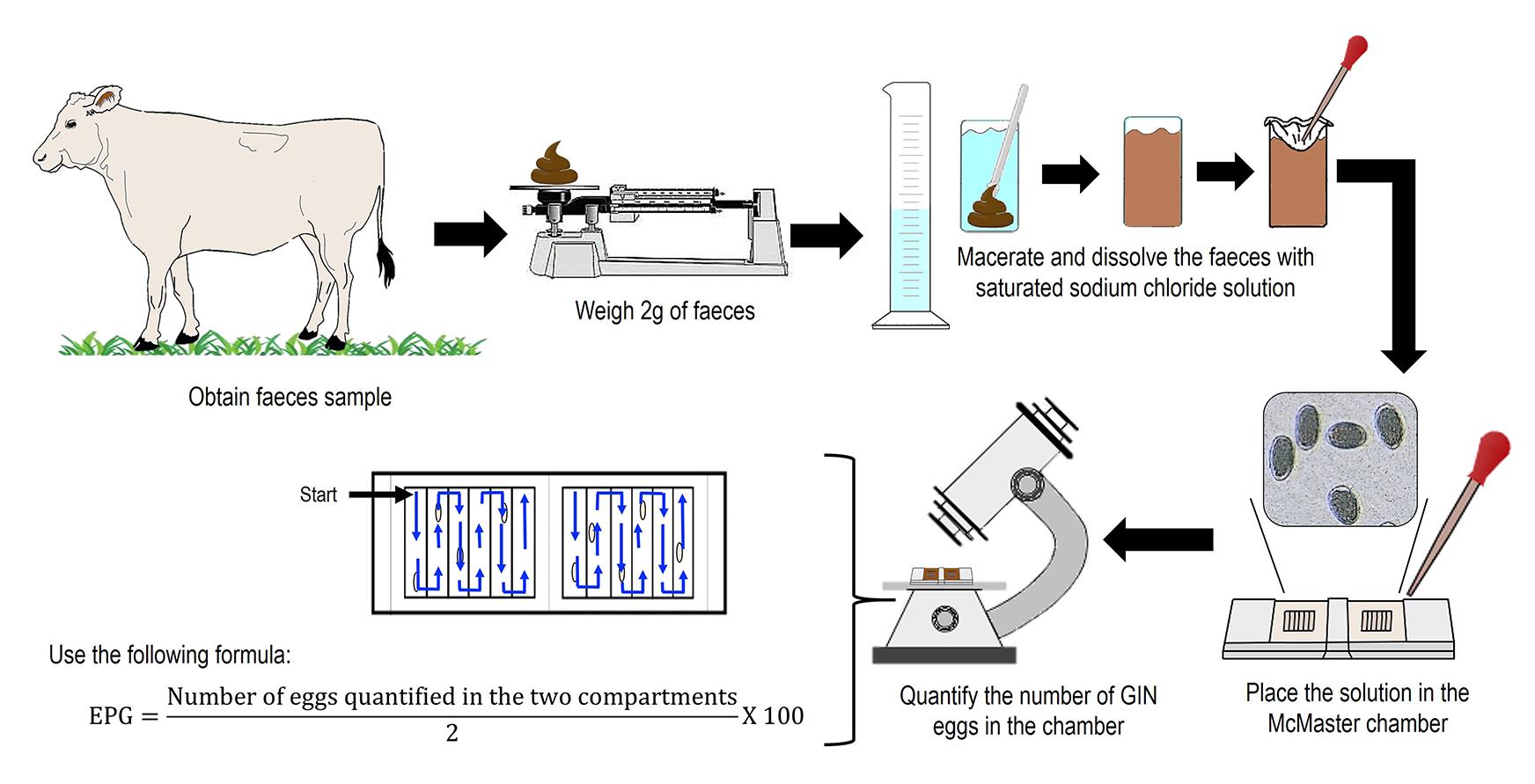
Figure 1. Representative figure of McMaster techniqueCoproculture and Baermann technique (Liébano-Hernández et al., 2011; Cedillo-Borda et al., 2020):
i. Collect bovine fecal samples directly from the rectum (from 300 to 500 g) using nitrile gloves as indicated in step A.1.a.i.
ii. Place the fecal sample in a plastic container (38 cm diameter by 18 cm depth), add distilled water (dH2O) at 25 °C to cover the sample (~350 mL), and macerate the feces with a pistil (20 cm long × 5 cm wide) until obtaining a paste.
iii. Add foam rubber and homogenize with the fecal sample. Cover with aluminum foil and incubate at room temperature (25 °C) for seven days. Mix the feces culture every 48 h to ease the development from egg to L3.
iv. After seven days, place 100 g of fecal/foam tuber mixture in a piece of non-sterile gauze (15 cm × 15 cm) and wrap in a ball.
v. Place each gauze ball using the hands into a Baermann funnel (plastic funnel of 14 cm diameter connected with PVC hose to a 10 mL tube) and add dH2O (at 25 °C) to cover the gauze ball.
vi. After 12 h, remove all tubes with the larvae on the bottom and store them at 4 °C for 2 h to precipitate the L3.
vii. Remove the supernatant using plastic Pasteur pipettes and recover the L3 pellets.
viii. Mix all the L3in new plastic tubes. Taking advantage from the hydrotropism and phototropism of the L3, filter the L3using the optical lens paper over the Baermann funnel for their precipitation (Thienpont et al., 2003).
ix. After 24 h at room temperature, remove the tubes from the Baermann funnels, recover L3 at the bottom of the tube using a Pasteur pipette (3 mL), and place the pellet in a T-75 cell culture flask with 15 mL of dH2O. Store in a refrigerator at 4 °C (Figure 2).
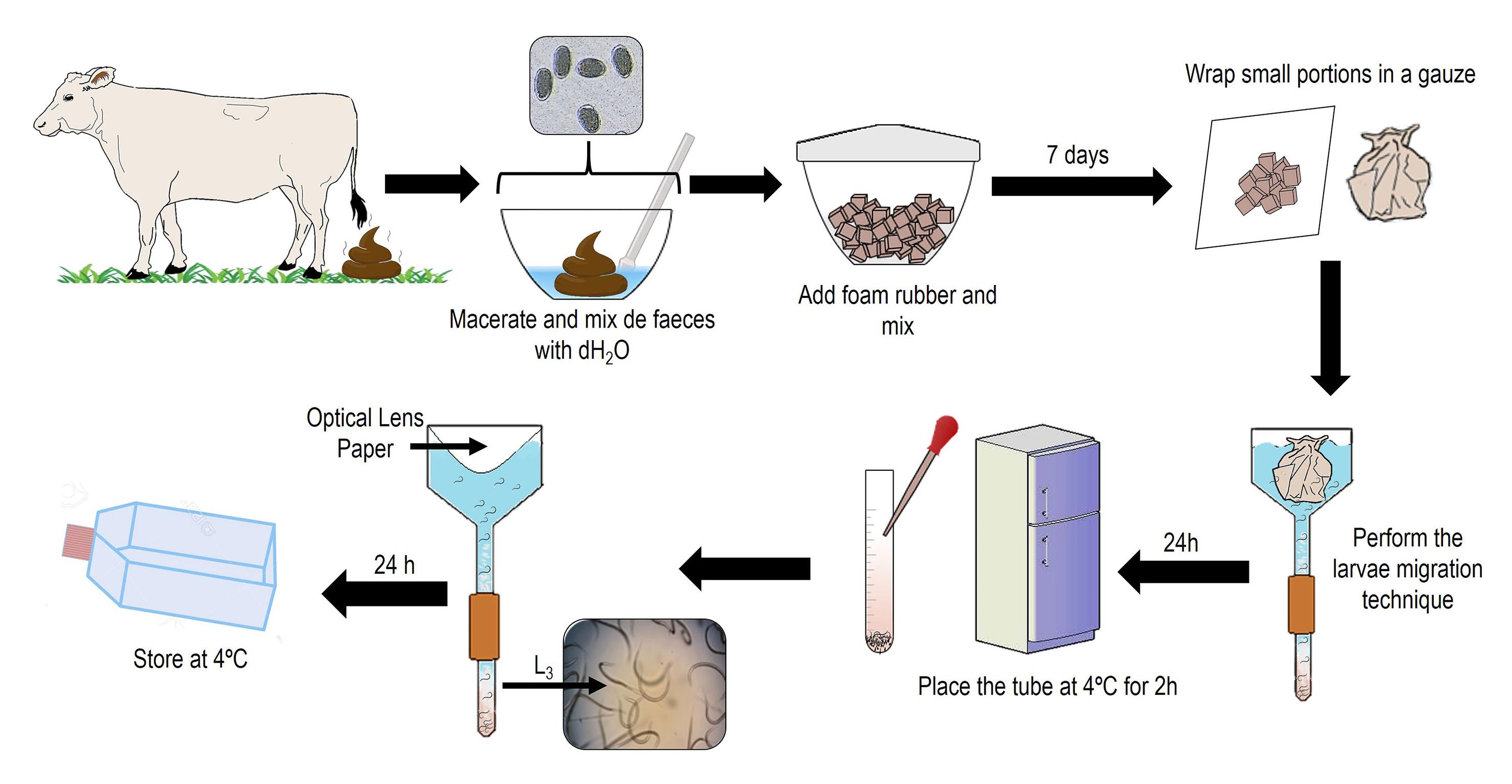
Figure 2. Representative figure of the fecal culture and Baermann techniqueCleaning of H. placei L3 by density gradient and centrifugation (Liébano-Hernández et al., 2011; Cedillo-Borda et al., 2020):
i. Centrifuge the L3 at 1,000× g for 3 min at room temperature. Discard the supernatant and add 2 mL of dH2O.
ii. Add 6 mL of 40% sucrose solution (see Recipes) in a 15 mL plastic tube.
iii. Place carefully the L3 pellet in the tube containing the sucrose solution. Centrifuge the plastic tube at 1,000× g for 5 min.
iv. A larval ring will be formed in the plastic tube. Carefully transfer the larval ring (maximum 2 mL) to a new plastic tube using a glass Pasteur pipette.
v. Add 10 mL of dH2O and centrifuge at 1,000× g for 3 min at room temperature. Discard the supernatant and repeat this step twice.
vi. Suspend the larval pellet in dH2O. Store the L3 at room temperature or remove the second molt of L3 (Figure 3).
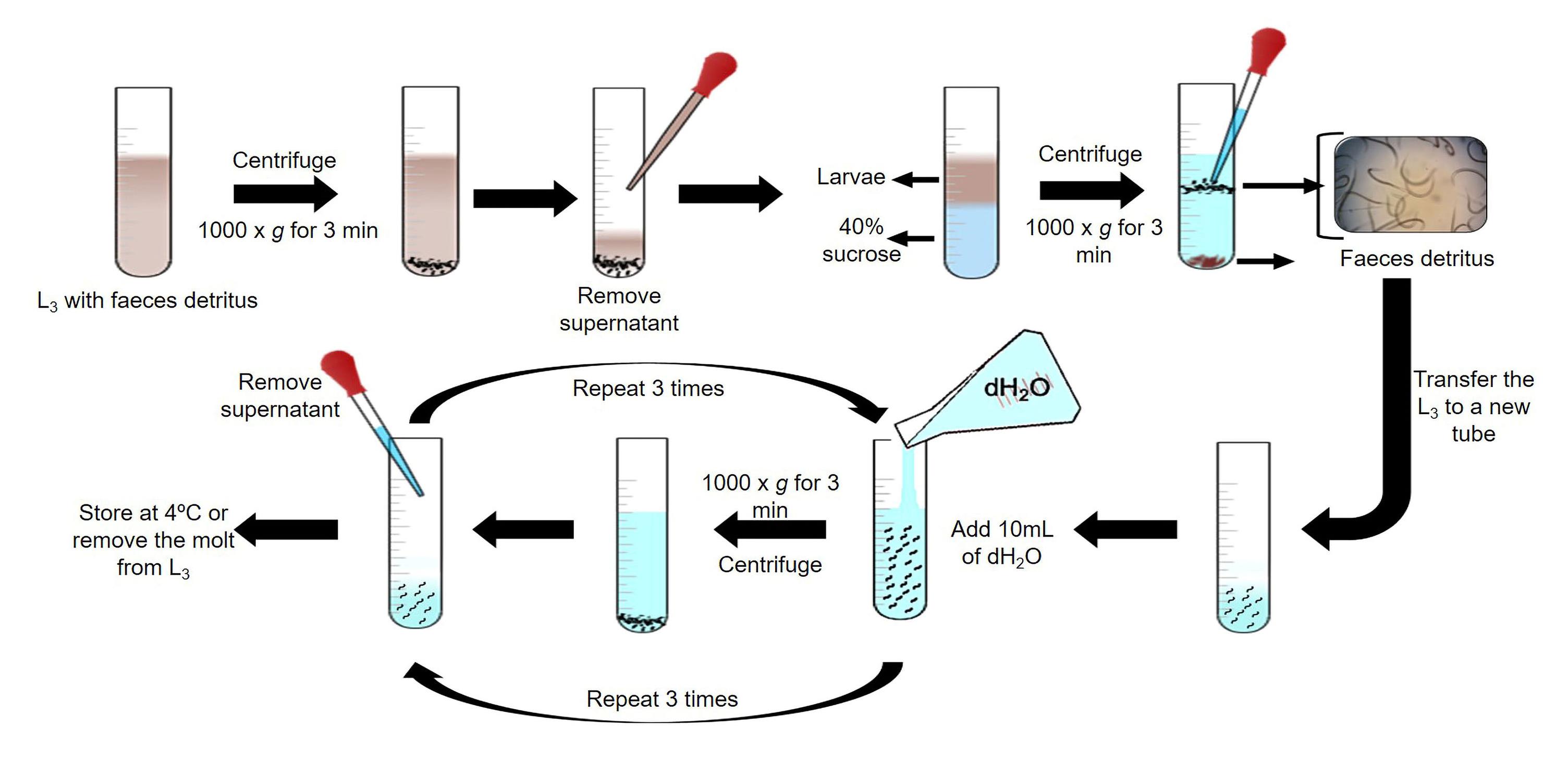
Figure 3. Schematic representation of L3 cleaning by density gradientRemoval of H. placei L3 sheath (Liébano-Hernández et al., 2011; Cedillo-Borda et al., 2020):
i. Centrifuge the L3 pellet at 1,000× g for 3 min. Discard the supernatant.
ii. Prepare 6 mL of 0.187% NaClO solution (see Recipes) in a 15 mL plastic tube. Homogenize L3 from 5 to 10 min.
iii. Confirm the elimination of H. placei L3 sheath by observing a 10 μL aliquot under an optical microscope (10×).
iv. Add 7 mL of dH2O and centrifuge at 1,000× g for 3 min. Discard the supernatant and repeat this step twice or more.
v. Suspend the unsheathed larval pellet in dH2O (Figure 4).
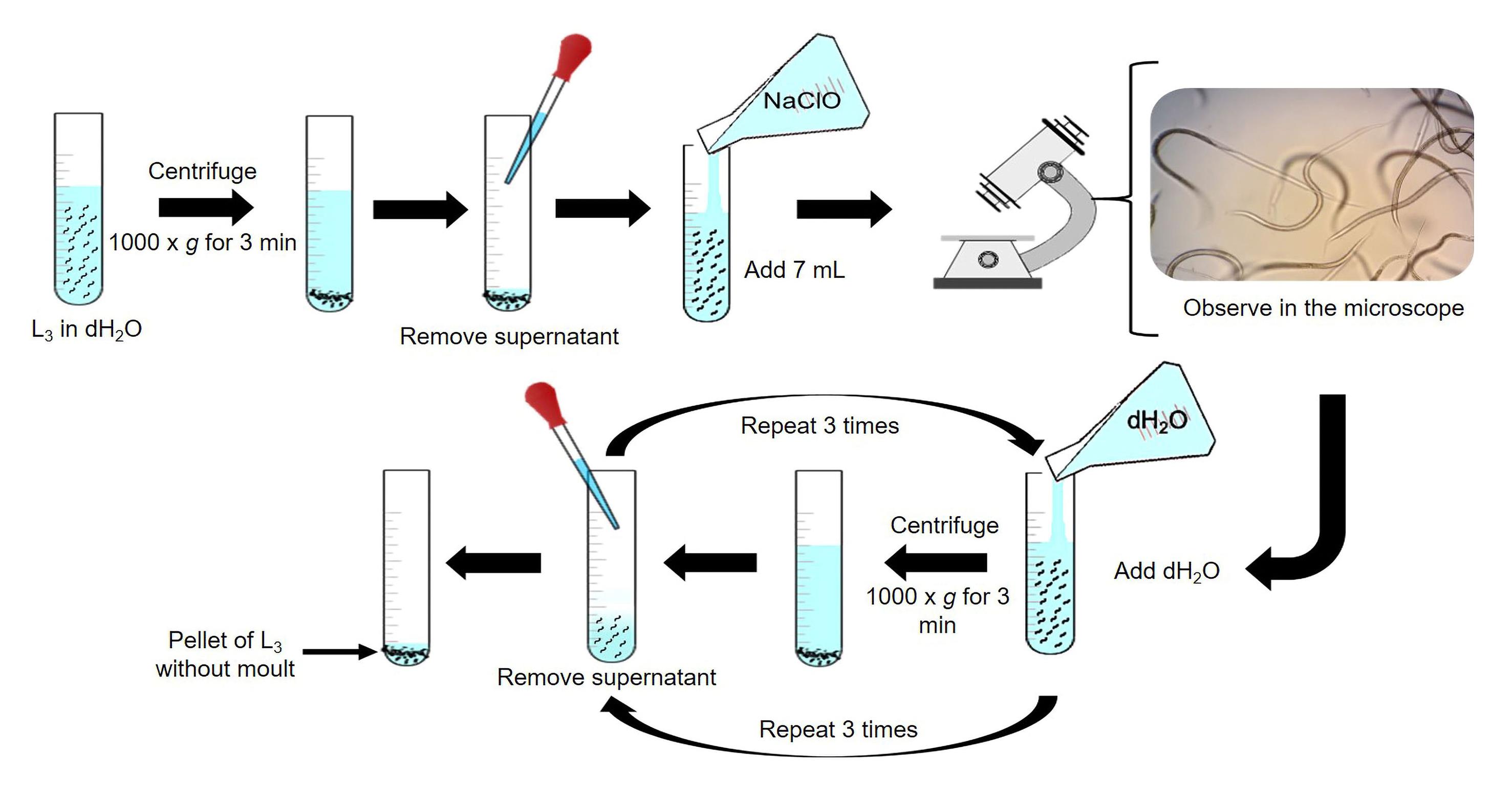
Figure 4. Representation of the technique for removing H. placei L3 sheathObtaining excreted-secreted products (ESP) from H. placei:
Note: Count the third-infective larvae considering 10 aliquots of 5 μL. Then, make a conversion taking the total volume from the aliquots and the final volume of the larvae stock.
Example:
50 μL (total aliquots volume) = 300 L3 (counted larvae)
25 mL (final volume) = X (Total larvae stock) = 150,000 L3
Obtaining ESP from in vitro culture (Figure 5)
i. Add 18,000 unsheathed L3 to a 50 mL tube with 20 mL of 1× PBS (see Recipes) supplemented with 1% antibiotic-antimycotic 100×.
ii. Then, place the 50 mL tube with L3 vertically and incubate for 30 min at room temperature.
iii. Centrifuge the L3 at 1,000× g for 5 min. Remove the supernatant and recover the L3 at the bottom of the tube.
iv. Place 3,000 L3 in a 6-well plate with 3 mL of Hank’s balanced salt medium (see Recipes) per well.
v. Incubate the 6-well plate at 37 °C with 5% CO2 for five days, in order to stimulate and collect the ESP into the culture medium.
vi. Collect 1 mL per well of the supernatant from the plate; the first collection of ESP should be at 16 h and then every 24 h. Replace the volume removed with new Hank’s medium.
ESP concentration and confirmation
i. Centrifuge the ESP from H. placei xL3 at 12,000× g for 40 min at 4 °C to recover the supernatant containing the ESP; then, filter the ESP using 0.2 μm syringe filters and store at -80 °C until use.
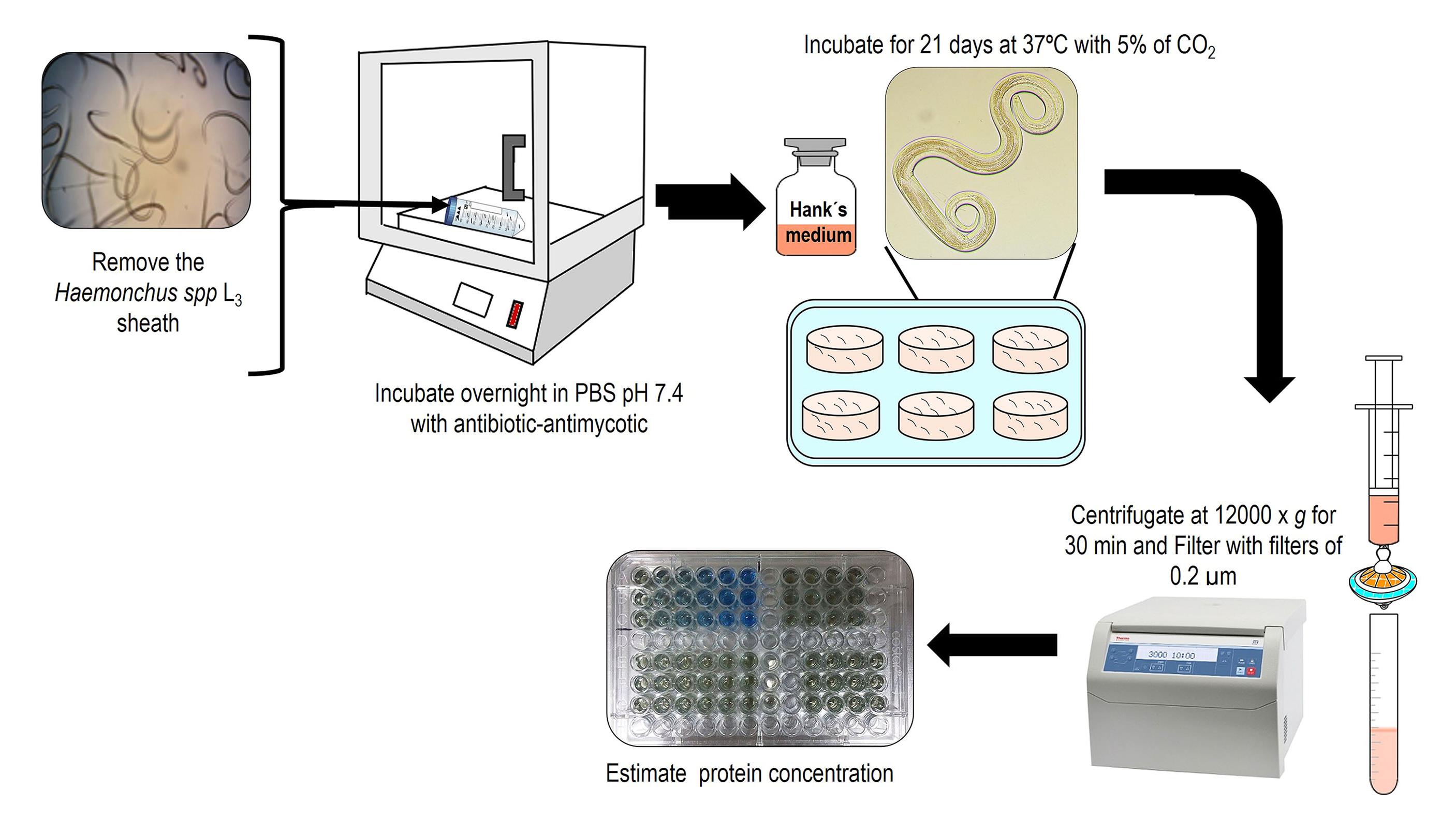
Figure 5. Schematic representation of obtaining excreted and secreted products from H. placeiii. Confirm ESP through SDS-PAGE (Sambrook and Russell, 2001) at 5%–12% and protein estimation (Bradford, 1976) (Figure 6).
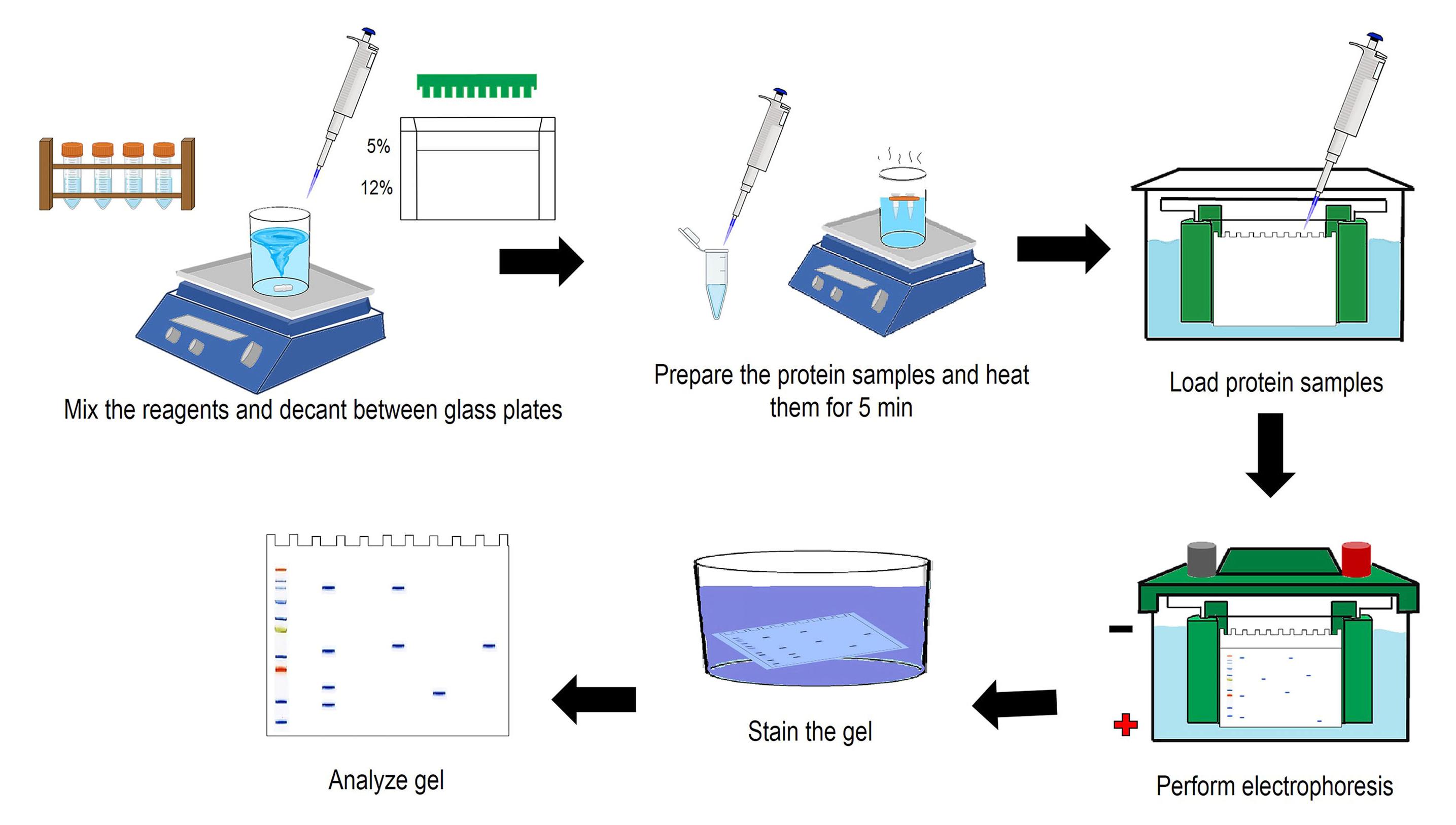
Figure 6. Summary of SDS-PAGE assays
Peripheral blood mononuclear cells (PBMCs) proliferation and relative gene expression assays
Obtaining PBMCs:
Blood samples
i. Select three young cattle (male or female) of similar age (between six to eight months) and housed under worm-free conditions.
ii. Identify the jugular vein and clean the puncture site with dH2O and 70% alcohol.
iii. Collect ~12 mL of blood from each bovine in EDTA tubes and mix by inverting the tubes repeatedly (Figure 7).
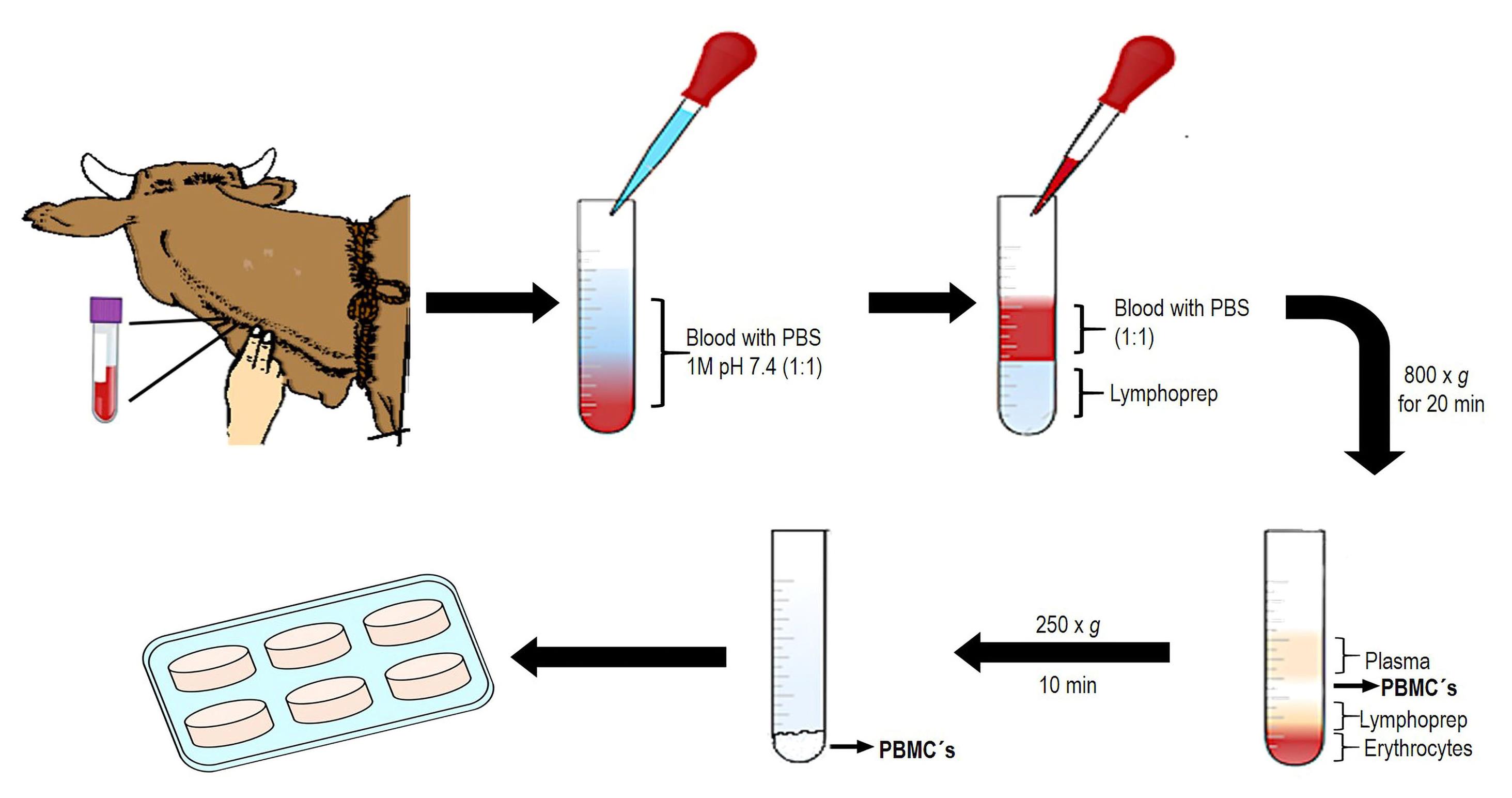
Figure 7. Representative figure of PBMCs isolationPBMCs isolation (protocol modified from human mononuclear cells)
i. Dilute each blood sample in PBS 1× pH 7.4 in a 1:1 proportion (3 mL of blood sample to 3 mL of PBS) and mix slowly by inversion to avoid the lysis of PBMCs.
ii. Place 3 mL of Lymphoprep reagent in a sterile 15 mL plastic tube and carefully add 6 mL of mixed blood/PBS. Avoid mixing the Lymphoprep reagent with the mixed blood/PBS by placing the tip of the micropipette on the side of the tube close to the Lymphoprep reagent and carefully adding the mix. Two phases should be seen.
iii. Centrifuge at 800× g for 20 min at room temperature in a Sorvall ST 8R centrifuge with oscillating rotor and slow deceleration. A white PBMCs ring will be formed in the plasma/Lymphoprep reagent interface.
iv. Remove the white PBMCs ring using a 3 mL glass Pasteur pipette and transfer all cells in 15 mL tubes with PBS. Immediately dilute the PBMCs with PBS 1× pH 7.4 in a 1:2 proportion and homogenize the mix slowly.
PBMCs quantification
i. Determine the volume of PBMCs suspension in RPMI 1640+HEPES supplemented media, according to the total number of flat-bottom plates per trial, including replicates of each treatment. Mix the PBMCs suspension carefully by inversion. Consider that each well should contain 50 μL as final volume if using 96-well plates.
ii. Dilute 45 μL of 0.4% trypan blue and 5 μL of PBMCs (1:10 proportion) in a 0.2 mL tube and mix slowly using a glass Pasteur pipette.
iii. Place 20 μL of PBMCs/trypan blue mix into a Neubauer chamber and quantify under an optical microscope (40×).
iv. Count the cells using four quadrants (Figure 8). Cells localized on the outer margins of each corner should not be included. Quantify dead (blue) and living cells (white) separately in each quadrant.
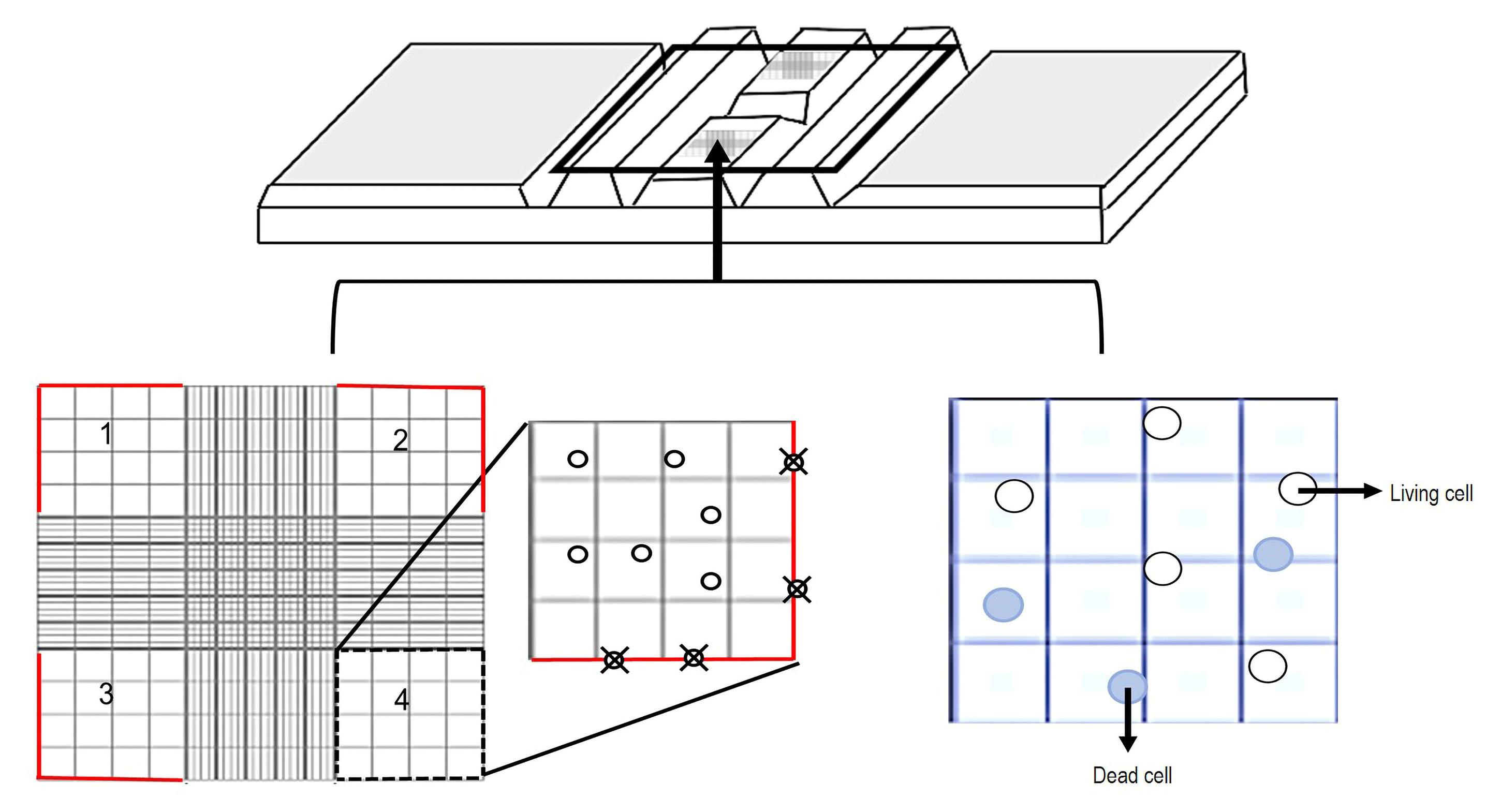
Figure 8. Schematic figure of the PBMCs quantificationv. Obtain the PBMCs concentration per milliliter using the following formula:
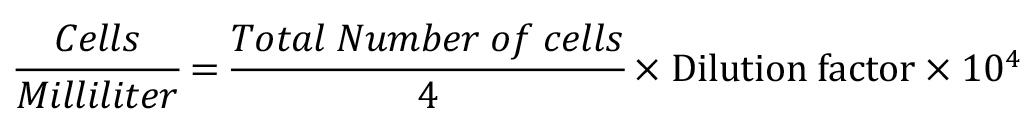
Proliferation assays and H. placei ESP from larval stages:
Proliferation assays
Notes:
1) Use a concentration from 2.5 to 5 μg/mL of positive controls (i.e., phytohemagglutinin, lipopolysaccharide, or Concanavalin A) and from 0.02 to 2 μg/mL for ESP suspended with culture medium. Untreated cells as negative controls are required, and a final volume of 100 μL should be adjusted per treatment.
2) An evaluation with different periods of incubation post-treatment (i.e., 6, 12, 24, 48, and 72 h) is recommended.
3) Perform at least three repetitions and replicates from each donor animal.
i. Place 300,000–500,000 PBMCs containing 50 μL of medium per well in flat-bottom 96-well plates. Add 50 μL of serial dilutions (previously prepared) of each treatment (e.g., controls and ESP). Incubate the 96-well plates at 37 °C with 5% CO2 (Figure 9).
ii. To count the PBMCs proliferation, add 20 μL of CellTiter 96® Aqueous One Solution to each well. Incubate at 37 °C with 5% CO2 for 2 h.
iii. Cell quantification is carried out at 490 and 690 nm using a microplate reader for data analysis.
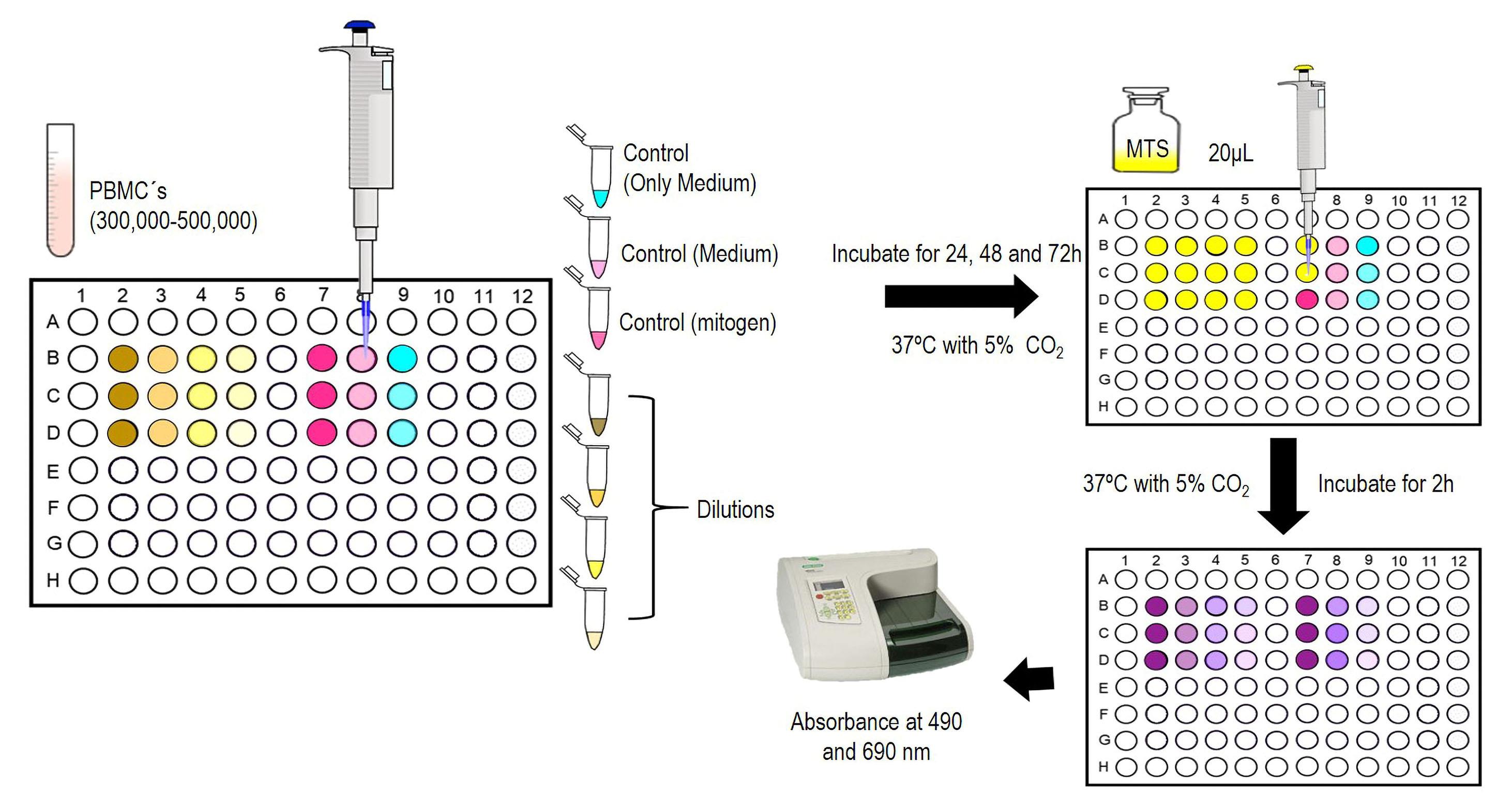
Figure 9. Proliferation assays representationData analysis of proliferation assays
i. Subtract the absorbance of 490 nm from the 690 nm value. This data will allow the elimination of background from the excess of cell debris, fingerprints, and other non-specific absorbance.
ii. Obtain the proliferation values through the normalization of untreated cells, expressed as percentage, as follows:
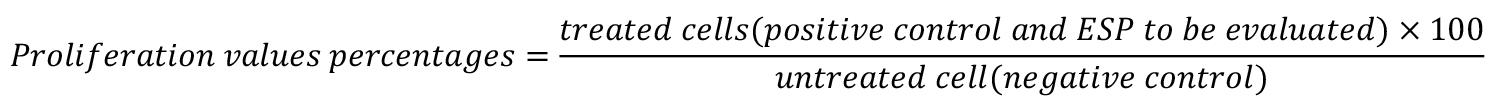
Relative gene expression method and analysis of immune genes:
Notes:
1) Perform the study with at least three repetitions per replicate of each donor animal.
2) Determine ESP concentrations according to the proliferation assays.
3) Evaluate different periods of incubation post-treatments (i.e., 6, 12, 24, 48, and 72 h).
4) The evaluation of immune genes activated by ESP using relative gene expression requires untreated cells as negative controls.
RNA purification (protocol modified from TRizolTM):
i. Place from 3 × 106 to 5 × 106 PBMCs in a 24-well plate containing 1.5 mL of RPMI 1640 + HEPES culture medium per well. Add each treatment—ESP or control groups (negative and positive)—using a micropipette. Incubate at 37 °C with 5% CO2.
ii. With a micropipette, collect the stimulated PBMCs from each well treated after incubation. Place the cells in 1.5 mL plastic tubes and centrifuge at 250× g for 5 min at room temperature. Remove the supernatant.
iii. Add 1 mL of TRizol reagent to 1.5 mL tubes with PBMCs and homogenize slowly at 4 °C with plastic pestle.
iv. Mix the samples using a vortex and incubate for 5 min at room temperature.
v. Add 0.2 mL of cold chloroform per 1 mL of TRizol reagent and mix by inverting the tubes repeatedly for 15 s. Incubate for 5 min at room temperature.
vi. Centrifuge samples at 12,000× g for 20 min at 4 °C. Transfer the aqueous phase corresponding to the RNA to a new tube. This phase will have a transparent appearance.
vii. Add 0.5 mL of isopropyl alcohol to the aqueous phase and mix slowly. Incubate for 15 min at room temperature.
viii. Centrifuge at 12,000× g for 15 min at 4 °C. Total RNA will remain as a pellet at the bottom of the tube.
ix. Carefully discard the supernatant with a micropipette or by decantation.
x. Suspend the RNA with 75% of cold ethanol.
xi. Homogenize the samples using a vortex and centrifuge at 7,500× g for 5 min at 4 °C. Discard the supernatant with a micropipette.
xii. Leave the RNA to air dry for 20 min inside the fume hood.
xiii. Add 30 μL of RNase-free water by carefully pipetting.
xiv. Estimate the RNA purity and concentration using a Nanophotometer at a ratio A260/A280 value. Set 1–2 μL of nuclease-free water as blank and then a similar quantity of RNA. Use a 0.5–10 μL micropipette. The result corresponds to the RNA sample purity between 1.8–2.0 values, where lower ratios indicate contamination with proteins.
xv. Confirm the integrity of RNA by setting a volume of 8 μL on electrophoresis with a 3% agarose gel (see Recipes) and carry out at 60 V for 30 min in buffer 1× TAE (see Recipes). Then, visualize the RNA integrity (28S and 18S rRNA) using a transilluminator Imagine System (Sambrook and Russell, 2001; Cedillo-Borda et al., 2020).
RNA decontamination:
i. Perform RNA decontamination in 0.2 mL plastic tubes using the reaction DNase as follows (Table 1):
Table 1. Components of the RNA decontamination reaction
Component Volume (μL) RNA in water (300 ng is recommended) 3 RQ1 RNase-Free DNase 10× Reaction Buffer 1 RQ1 RNase-Free DNase 1 Nuclease-free water 5 ii. Incubate at 37 °C for 30 min.
iii. Add 1 μL of RQ1 DNase Stop Solution and incubate at 65 °C for 10 min to stop the reaction.
Reverse Transcription reaction (protocol modified from ImProm-II Reverse Transcription System Kit®):
i. Add 1 μL of random primers and incubate at 70 °C for 10 min. Immediately place the tube with the sample on ice.
ii. Working on ice, add the following components (Table 2) to the same PCR tube.
Table 2. Components for the reverse transcription reaction
Component Volume (μL) imProm-IITM 5× Reaction Buffer 4 MgCl2(final concentration 1.5–8.0 mM) 2 dNTP mix (final concentration 0.5 mM each dNTP) 1 Recombinant RNasin® Ribonuclease Inhibitor 0.3 ImProm-IITM Reverse Transcriptase 1 Nuclease-free water (to 15 μL) 6.7 iii. Incubate at 25 °C for 5 min, at 42 °C for 60 min, and then at 70 °C for 15 min.
qPCR assay:
i. Perform qPCR assays using the commercial custom RT2 Profiler PCR and NCBI primer BLAST. The genes are selected from the National Centre of Biotechnology (https://www.ncbi.nlm.nih.gov/) show in Table 3.
Table 3. Gene information used in the PCR design
Custom PCR array design (CAPB13410R) Genes GenBank Access IL2 NM_180997 IL4 NM_173921 IL5 NM_173922 IL6 NM_173923 IL8 NM_173925 IL10 NM_174088.1 IL13 NM_174089.1 IFNγ NM_174086.1 FCεR1A NM_001100310.1 TGFβ1 NM_001166068.1 Housekeeping design using the NCBI Primer BLAST Genes GenBank Access β-2 microglobulin
Fw
Rv
XM_002691119.4
CCATCCAGCGTCCTCCAAAGATTC
CTGCTCCGATTTAATCTTCTCCCCA
β-actin
Fw
Rv
XM_027528015.1
CATCGCGGACAGGATGCAGAAA
CCTGCTTGCTGATCCACATCTGCT
gapdh
Fw
Rv
NM_001190390.1
TTGTCTCCTGCGACTTCAACAGCG
CACCACCCTGTTGCTGTAGCCAAAT
ii. Perform the PCR assay using the reagents indicated in Table 4 (Estrada-Reyes et al., 2017).
Table 4. Components of qPCR assays
Component Volume (μL) cDNA (from 300 ng of RNA) 2 GoTaq® qPCR Master Mix 10.3 Nuclease-free water 12.68 iii. Place the commercial custom RT2 Profiler PCR in the Rotor-Gene 6000 and perform the PCR with the following conditions (Table 5):
Table 5. Conditions for real-time PCR
Cycles Stages Temperature (°C) Time 1 Initial denaturation 95 10 min 40 Denaturation 95 15 s Annealing-elongation 60 45 s 1 Dissociation temperature 65–95 Rising ramp Data analysis:
i. Obtain the threshold cycle (CT) from each gene using a threshold value of 0.05.
ii. Record the CTvalue in an Excel spreadsheet and analyze on the Qiagen® Gene Globe Data Analysis Center web platform, to normalize through the ΔΔCT method (Double delta CT).
iii. Enter https://geneglobe.qiagen.com/mx/analyze/ and select the PCR analysis tool option. A registered user account is required to access the web platform (Figure 10).
iv. Select and upload the data recorded in a datasheet with the CT values.
v. Select the control group, groups under study, and housekeeping genes. Select at least two housekeeping genes to the expression analysis.
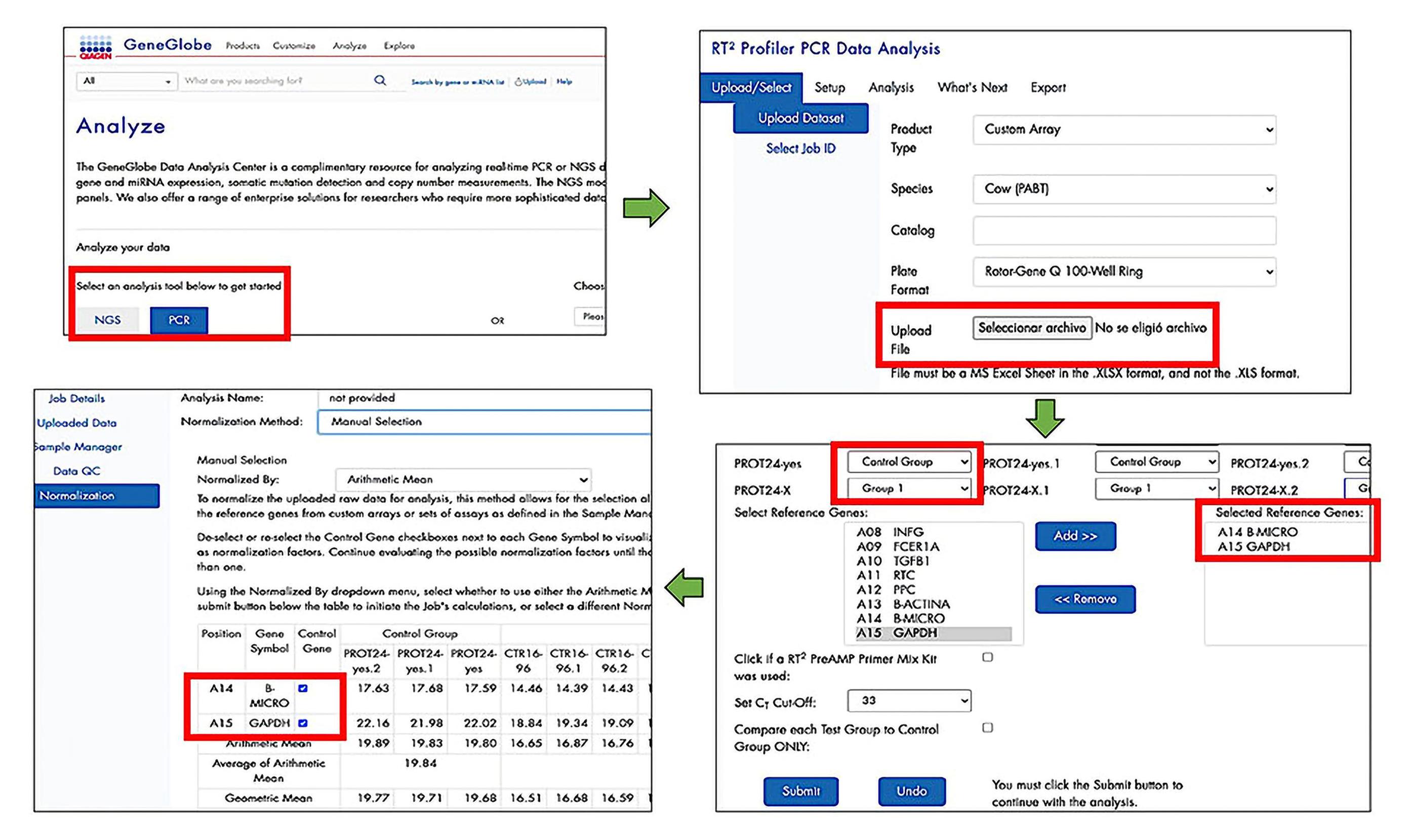
Figure 10. Process to use the Qiagen® GeneGlobe Data Analysis Center web platformvi. The Qiagen® Gene Globe Data Analysis Center platform shows the fold change values. Optionally, download graphs to represent the fold change values. Figure 11 shows an example of the relative expression interpretation.
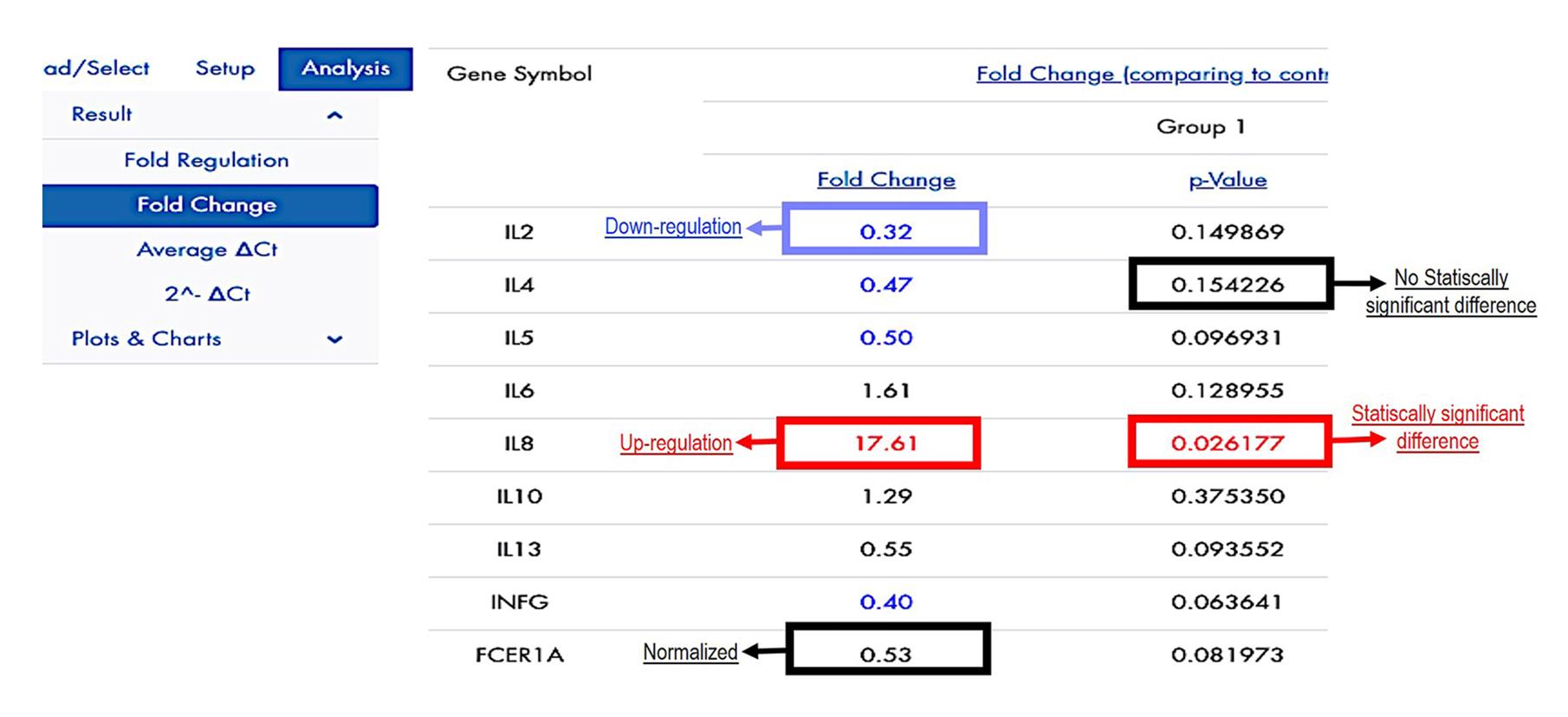
Figure 11. Fold change values on the Qiagen® GeneGlobe Data Analysis Center web platform
Recipes
Note: All the solutions should be adjusted with HCl and NaOH 1 N.
Buffer 50× TAE
242 g of Trizma base (FW = 121.14)
57.1 mL of glacial acetic acid
100 mL of 0.5 M EDTA (pH 8.0)
Adjust the final volume to 1 L with deionized water.
3% agarose gel (size 10 cm × 7 cm)
1.06 g of agarose
45 mL of buffer 1× TAE
1.5 μL of ethidium bromide
0.187% NaClO solution
0.183 mL of NaClO
5.813 of distilled water
Hank’s balanced salts (medium)
49.2 mL (9.8 g/L) of Hanks’ medium
246 mL of antibiotic-antimycotic
8 μL of bovine erythrocyte (previously treated with VyM) (Rojas Martinez et al., 2016)
40% sucrose solution
20 g of sucrose
50 mL of distilled water
Phosphate buffered saline, pH 7.4
8 g of NaCl 137 mM
0.2 g of KCl 2.7 mM
1.44 g of Na2HPO4 10 mM
0.24 g of KH2PO4 2 mM
1,000 mL of distilled water
Saturated sodium chloride solution
400 g of NaCl
Adjust the final volume to 1 L with distilled water.
Acknowledgments
This protocol was derived from an original research paper (Maza-Lopez et al., 2021, https://doi.org/10.1016/j.vetpar.2021.109512), which received financial support from Consejo Nacional de Ciencia y Tecnología - Secretaría de Educación Pública (CONACYT-SEP, Grant number 287598).
Competing interests
The authors declare no conflict of interest.
Ethics
The criteria for care and handling of experimental animals were set forth in the Official Mexican Standard NOM-033-Z00-1995, NOM-051-ZOO-1995, and NOM-062-ZOO-1999.
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Cedillo-Borda, M., López-Arellano, M. E. and Reyes-Guerrero, D. E. (2020). In vitro assessment of ivermectin resistance and gene expression profiles of P-glycoprotein genes from Haemonchus contortus (L3). Bio-101: e3851.
- Estrada-Reyes, Z., López-Arellano, M. E., Torres-Acosta, F., López-Reyes, A., Lagunas-Martínez, A., Mendoza-de-Gives, P., González-Garduño, R., Olazarán-Jenkins, S., Reyes-Guerrero, D. and Ramírez-Vargas, G. (2017). Cytokine and antioxidant gene profiles from peripheral blood mononuclear cells of Pelibuey lambs after Haemonchus contortus infection. Parasite Immunol 39(6). doi: 10.1111/pim.12427.
- Liébano-Hernández, E., López-Arellano, M. E., Mendoza-De-Gives G. P. and y Aguilar-Marcelino, L. (2011). Manual de diagnóstico para la identificación de larvas de nematodos gastrointestinales de rumiantes. Publicación Especial Vol. 2. México: Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP): 1-44.
- Maza-López, J., Contreras-Ochoa, C. O., Reyes-Guerrero, D. E., Encarnación-Guevara, S., Hernández-Ortíz, M., Olmedo-Juárez, A. and López-Arellano, M. E. (2021). Analysis of the immunomodulatory activity of excreted and secreted products from Haemonchus placei transition infective larvae (xL3). Vet Parasitol 298: 109512.
- Rojas Martínez, C., Rodríguez-Vivas, R. I., Figueroa Millán, J. V., Acosta Viana, K. Y., Gutiérrez Ruiz, E. J. And Álvarez Martínez, J. A. (2016). In vitro culture of Babesia bovis in a bovine serum-free culture medium supplemented with insulin, transferrin, and selenite. Exp Parasitol 170: 214-219.
- Sambrook, J. and Russel, D. W. (2001) Molecular cloning, a laboratory manual. 3rd edition. Cold Spring Harbor Laboratory Press. Vol. 1. P. pp. 5.4-5.13; Vol. 3. Appendix A8.40-A.849.
- Thienpont, D., Rochette, F. and Vanparijs, O.F.J. (2003) Diagnosing helminthiasis by coprological examination. 3rd edition. Janssen Animal Health. P. 110. pp. 17-43.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Maza-Lopez, J., Camas-Pereyra, R., López-Arellano, M. E. and Contreras-Ochoa, C. O. (2023). Selection of Molecules with Immunological Potential from Excretory and Secretory Products from the Nematode Haemonchus placei by Cell Proliferation and Gene Expression Assays. Bio-protocol 13(12): e4702. DOI: 10.21769/BioProtoc.4702.
Category
Immunology > Immune cell isolation > Leukocyte
Medicine
Molecular Biology > RNA > qRT-PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










