- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A New Methodology for the Quantification of Neutrophil Extracellular Traps in Patient Plasma
Published: Vol 13, Iss 12, Jun 20, 2023 DOI: 10.21769/BioProtoc.4701 Views: 3025
Reviewed by: Alessandro DidonnaKomuraiah MyakalaMarco Di Gioia

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
In vitro Demonstration and Quantification of Neutrophil Extracellular Trap Formation
Dongsheng Jiang [...] Karin Scharffetter-Kochanek
Jul 5, 2017 17299 Views
Measuring Myeloperoxidase Activity as a Marker of Inflammation in Gut Tissue Samples of Mice and Rat
Nikita Hanning [...] Benedicte Y. De Winter
Jul 5, 2023 2615 Views
Utilizing EdU to Track Leukocyte Recruitment to the Brain
Zoie K. Lipfert [...] David P. Sullivan
Dec 5, 2025 1362 Views
Abstract
Neutrophil extracellular traps (NETs) are web-like structures made up of decondensed chromatin fibers along with neutrophil granular proteins that are extruded by neutrophils after activation or in response to foreign microorganisms. NETs have been associated with autoimmune and inflammatory diseases such as systemic lupus erythematosus (SLE), rheumatoid arthritis, coronavirus disease 2019 (COVID-19), and others. There are reliable methods available to quantitate NETs from neutrophils, but their accurate quantification in patient plasma or serum remains a challenge. We developed a highly sensitive ELISA to detect NETs in serum/plasma and designed a novel smear immunofluorescence assay to detect NETs in as little as 1 μL of serum/plasma. We further validated our technology on plasma samples from SLE patients and healthy donors that carry interferon regulatory factor 5 genetic risk. The multiplex ELISA combines the use of three antibodies against myeloperoxidase (MPO), citrullinated histone H3 (CitH3), and DNA to detect the NET complexes with higher specificities. The immunofluorescence smear assay can visually detect intact structures of NETs in 1 μL of serum/plasma and provide similar results that correlate with findings from the multiplex ELISA. Furthermore, the smear assay is a relatively simple, inexpensive, and quantifiable method of NET detection for small volumes.
Graphical overview
Background
Neutrophils are the most abundant leukocyte in the human blood and are the first cells to respond to an external stimulus such as a pathogen and/or inflammatory trigger (Kaplan and Radic, 2012; Matta et al., 2022). They migrate to the site of infection where they protect the host by phagocytosing, killing, and digesting pathogens (Kaplan and Radic, 2012; Matta et al., 2022). Another powerful tool neutrophils use to defend the body is the release of neutrophil extracellular traps (NETs). NETs can trap microorganisms and kill them by releasing granular antimicrobial peptides and proteins such as neutrophil elastase (NE), myeloperoxidase (MPO), and cathepsin G(3); however, when this process gets dysregulated or NET clearance is diminished, it can lead to tissue damage, autoimmunity, and other inflammatory conditions (Lande et al., 2011; Kaplan and Radic, 2012; Corsiero et al., 2016; Bruschi et al., 2020; Matta et al., 2022).
Formation of NETs involves the citrullination of histones, chromatin decondensation, and disintegration of the nuclear membrane followed by the release of nuclear content along with proteins such as MPO and NE (Kaplan and Radic, 2012; Matta et al., 2022). Traditionally, NETs are detected by using either anti-MPO or anti-citrullinated histone H3 (CitH3), or neutrophil elastase as coating antibody separately followed by anti-DNA detecting antibody (Zuo et al., 2020 and 2021). We have developed and validated a more sensitive ELISA, where we use a cocktail of anti-MPO and anti-CitH3 antibodies as coating antibodies, followed by anti-DNA as the detecting antibody. We further optimized the best blocking and substrate combination suited for increased sensitivity for ELISA. To the best of our knowledge, we are the first one to develop the smear assay to visualize NETs in small amounts of patient plasma. This method requires minimal time, reagents, specialized equipment, and/or cost. These techniques can be applied to detection of NETs in serum/plasma of patients with autoimmune diseases and other inflammatory conditions where NETs have been shown to play a crucial role. But apart from that, we propose that the application of these techniques can be extended to other bodily fluids as well, where accumulation of NETs has been shown to play a role in disease pathogenesis. For example, NETs can be quantified in the synovial fluid of joints to study rheumatoid arthritis (Corsiero et al., 2016) or excretion of NETs in urine in certain inflammatory conditions such as urinary tract infections (Yu et al., 2019). Since the smear assay requires only 1 μL of sample, this assay can be applied to samples that can be obtained only in smaller quantities; for example, NETs have shown to be involved in many ocular diseases such as glaucoma, age-related macular degeneration, dry eye, and diabetic retinopathy for which only a very small volume of ocular fluid can be obtained (Martínez-Alberquilla et al., 2022). Another example is bronchiolar lavage, where NETs have been shown to be elevated in patients with pneumonia-related acute respiratory distress syndrome (Bendib et al., 2019), primary graft dysfunction after lung transplantation (Sayah et al., 2015), and COVID-19 (Yaqinuddin et al., 2020). Moreover, these techniques can also be applied to pediatric autoimmune and inflammatory diseases such as pediatric lupus, where there are limitations on sample volume (Garcia-Romo et al., 2011).
Materials and reagents
Poly-L-Lysine adhesive microscope slides (Newcomer Supply, catalog number: 5010)
ImmulonTM MicrotiterTM 96-well plates (Thermo Scientific, catalog number: 3855)
Mouse monoclonal antibody [2C7] to myeloperoxidase (MPO) (Abcam, catalog number: 25989)
Rabbit polyclonal antibody to histone H3 (anti-CitH3 citrulline R2 + R8 + R17) (Abcam, catalog number: 5103)
Sytox Green nucleic acid stain (Invitrogen, catalog number: S7020)
Cell Death Detection ELISA PLUS Anti-DNA POD kit (Roche, catalog number: 11774425001)
Goat anti-mouse Alexa Fluor 488 (2 mg/mL) (Invitrogen, catalog number: A11029)
Goat anti-rabbit Alexa Fluor 594 (2 mg/mL) (Molecular Probes, catalog number: A11012)
DAPI (4’,6-Diamidino-2-Phenylindole, dihydrochloride) (Sigma-Aldrich, catalog number: D9542-1MG)
TMB (3,3’,5,5’-tetramethylbenzidine) enhanced one component HRP membrane substrate (Sigma-Aldrich, catalog number: T9455)
2N sulfuric acid stop solution (Reagents, catalog number: CS106300-500A)
Normal rat serum control (Fisher Scientific, catalog number: 10-710-C)
Tween 20 (Thermo Fisher, catalog number: J20605.AP)
Triton X-100 (Thermo Fisher, catalog number: HFH10)
Bovine serum albumin (BSA) (Fisher catalog number: BP9703-100)
Formaldehyde (Sigma, catalog number: 252549)
Na2CO3 (Sigma, catalog number: 497-19-8)
NaHCO3 (Sigma, catalog number: 144-55-8)
VectaMount AQ mounting medium (Vector Labs, catalog number: H-5501)
Dulbecco’s Phosphate Buffer saline (PBS without calcium and magnesium chloride (Sigma, catalog number: D8537-500ML)
Wash buffer (see Recipes)
Blocking buffer (see Recipes)
Dilution buffer (see Recipes)
Coating buffer (see Recipes)
Smear dilution buffer (see Recipes)
Smear blocking buffer (see Recipes)
Fixing buffer (see Recipes)
Equipment
Pipettes, multi-channel pipettes for washings and tips
Synergy Neo2 Multi-Mode microplate reader (BioTek, model: BTNEO2)
EVOS M7000 imaging system (Invitrogen, catalog number: AMF7000)
ZEISS Confocal LSM 880 Airyscan
NanoDrop 2000 (Thermo Fisher Scientific, catalog number: ND-2000)
Software
ImageJ—Java-based program (V1.8) (https://imagej.nih.gov/ij/download.html)
GraphPad Prism 8 (https://www.graphpad.com/scientific-software/prism/)
MyAssays online program (https://www.myassays.com/download-and-install-myassays-desktop.html)
Procedure
NETs ELISA
Coat 96-well ultra-high-binding plates with 50 μL of anti-MPO and anti-CitH3 unlabeled primary antibodies diluted to a concentration of 5 μg/mL each in coating buffer. Cover the plate with the plate sealer. Leave plate overnight at 4 °C.
The next morning, wash wells three times with 200 μL of PBS at room temperature.
Block plate with 200 μL of blocking buffer for 2 h at room temperature.
Wash the plate five times with 200 μL of PBS.
Add 50 μL of undiluted plasma sample to each coated well and incubate at room temperature for 2 h. Set up wells for blanks where no plasma samples are added.
Carefully wash the plate five times with 200 μL of wash buffer; do not invert plate during washes.
Add 50 μL of anti-DNA-POD antibody (from Cell Death Detection ELISA PLUS Anti-DNA POD kit) prepared 1:100 in dilution buffer to each well and incubate at room temperature for 2 h for the detection of NETs.
Wash the plate five times with 200 μL of wash buffer.
Add 100 μL of TMB (allow to come to room temperature before using) to each well and incubate plate in the dark for 3 min.
Add 100 μL of 2 N sulfuric acid stop solution to each well and measure absorbance using a Synergy Neo2 Multi-Mode microplate reader, or any ELISA microplate reader set to a wavelength of 450 nm.
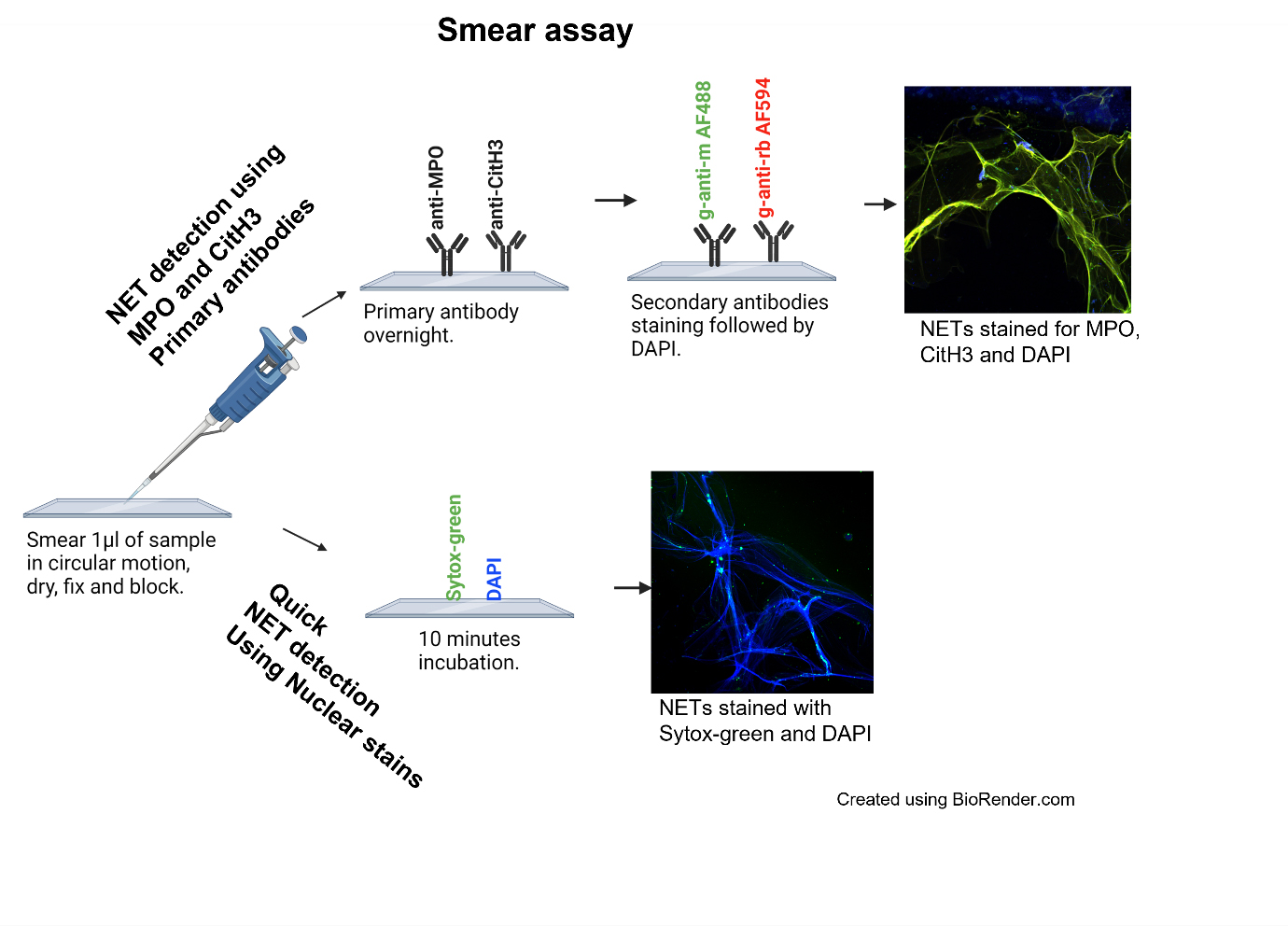
Smear assay
Draw a circle using a histology liquid repellent pen on a poly-L-Lysine glass slide (2–3 circles per slide).
Drop 1 μL of plasma sample to each circle and smear in a circular motion until the volume is equally distributed. Let plasma completely dry.
Fix samples with 100 μL of smear fixing buffer (4% formaldehyde) for 10 min.
Carefully tilt the slide to remove the liquid to be collected in the formaldehyde waste container, dab the sides of the slides with a paper towel, and carefully wash each circle three times with 100 μL of PBS.
Block the slides with 100 μL of smear blocking buffer (5% BSA/PBS) for 1 h at room temperature.
Add 50–100 μL of anti-MPO and anti-CitH3 primary antibodies diluted to smear dilution buffer (1 μg/mL in 0.3% Triton X-100 + 0.1% BSA solution) to each circle and leave overnight in a moist slide chamber.
The following morning, wash slides three times with PBS and add 50–100 μL of goat anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 594 secondary antibodies diluted 1:500 in dilution buffer to each circle and incubate in the dark for 1 h.
Wash three times with 200 μL of PBS and stain slides with 5 mg/mL of DAPI diluted 1:200 in PBS for 15 min in the dark.
In another set of slides for quick nonspecific visualization, after fixing and blocking, NETs can be stained directly with nuclear dyes Sytox Green diluted 1:200 and 5 mg/mL of DAPI diluted 1:200 in PBS for 15 min in the dark.
Wash an additional three times with 200 μL of wash buffer after staining. Add a drop of VectaMount AQ mounting medium and secure a glass coverslip onto the slide making sure that there are no bubbles.
Allow the slides to dry before imaging on EVOS M7000 imaging system and/or ZEISS Confocal LSM 880.
Stain secondary antibody–only controls the similar way, except that no primary antibodies are added.
Data analysis
For ELISA data analysis, a standard curve is prepared using serial dilutions of three pooled phorbol 12-myristate 13-acetate (PMA)-stimulated healthy donor plasma samples. DNA is quantified using the NanoDrop 2000. NETs in serially diluted PMA-induced plasma samples were quantified using CitH3 and MPO + CitH3 ELISA methodology. Four-parameter logistic standard curves were created for each antibody coating, and data were extrapolated using MyAssays online software. A detailed protocol can be found in the published manuscript (Matta et al., 2022) in methodology under section “Preparing in-house standard for the quantification of NET remnants by ELISA.”
For quantification of NETs in smear assay we used ImageJ Java-based program (V1.8) to convert the images to grayscale 8-bit images, whose intensities were then quantified and graphed. A detailed protocol can be found in the published manuscript (Matta et al., 2022) in methodology under section “NET Quantification using ImageJ.”
All statistical analyses should be performed and graphed using GraphPad Prism 8. A two-tailed parametric t-test should be used for comparisons between samples with normal distribution. P < 0.05 should be considered statistically significant. A two-tailed correlation analysis should be performed using Pearson correlation coefficients assuming Gaussian distribution.
Notes
There is a disparity between ELISA and smear assay when using fresh vs. frozen plasma samples. ELISA can detect NETs in circulation from samples stored at -80 °C for long periods of time (> 6 months), as it can detect both intact and fragmented NETs. But as the smear assay is visualizing intact NETs, we found that NET structures are generally maintained in plasma samples up to six months when stored at -80 °C. Beyond six months, we could detect fragmented NETs with smear assay that compromised their quantification. According to our preliminary studies, we suggest adding EDTA/EGTA immediately after plasma isolation to prolong the stability of intact NET structures for quantification. Although we used the anti-DNA-POD antibody from Cell Death Detection ELISA PLUS Anti-DNA POD kit for the detection of NETs in ELISA, we believe that any other anti-DNA-peroxidase-conjugated antibody can be used for detection.
Recipes
Wash buffer
1% BSA + 0.05% Tween 20 in PBS
Blocking buffer
5% BSA + 5% normal rat serum in PBS
Dilution buffer
5% BSA + 0.05% Tween 20 in PBS
Coating buffer
15 mM Na2CO3, 35 mM NaHCO3, pH 9.6
To 50 mL of deionized water, add 80 mg of Na2CO3 and 150 mg of NaHCO3
Smear dilution buffer
0.3% Triton X-100 + 0.1% BSA in PBS
Smear blocking buffer
5% BSA in PBS
Fixing buffer
4% formaldehyde in PBS
Acknowledgments
This work was supported by grants from the National Institutes of Health NIAMS 1 R01 AR 076242-03, Department of Defense CDMRP LRP W81XWH-18-1-0674, and Lupus Research Alliance to BJB.
We would also like to thank members of “The NETwork to Target Neutrophils in COVID-19” for inspiration to develop new methodologies to detect NETosis.
This protocol was derived from the original work of Matta et al. (2022).
Graphical overview made with the help of https://biorender.com/.
Competing interests
The authors declare no conflicts of interest.
Ethics
The studies involving human participants were reviewed and approved by The Feinstein Institutes for Medical Research IRB. The patients/participants provided their written informed consent to participate in this study.
References
- Bendib, I., de Chaisemartin, L., Granger, V., Schlemmer, F., Maitre, B., Hue, S., Surenaud, M., Beldi-Ferchiou, A., Carteaux, G., Razazi, K., et al. (2019). Neutrophil Extracellular Traps Are Elevated in Patients with Pneumonia-related Acute Respiratory Distress Syndrome. Anesthesiology 130(4): 581-591.
- Bruschi, M., Bonanni, A., Petretto, A., Vaglio, A., Pratesi, F., Santucci, L., Migliorini, P., Bertelli, R., Galetti, M., Belletti, S., et al. (2020). Neutrophil Extracellular Traps Profiles in Patients with Incident Systemic Lupus Erythematosus and Lupus Nephritis. J Rheumatol 47(3): 377-386.
- Corsiero, E., Pratesi, F., Prediletto, E., Bombardieri, M., and Migliorini, P. (2016). NETosis as source of autoantigens in rheumatoid arthritis. Front immunol 7: 485.
- Garcia-Romo, G. S., Caielli, S., Vega, B., Connolly, J., Allantaz, F., Xu, Z., Punaro, M., Baisch, J., Guiducci, C., Coffman, R. L., et al. (2011). Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med 3(73): 73ra20.
- Kaplan, M. J. and Radic, M. (2012). Neutrophil extracellular traps: double-edged swords of innate immunity. J Immunol 189(6): 2689-2695.
- Lande, R., Ganguly, D., Facchinetti, V., Frasca, L., Conrad, C., Gregorio, J., Meller, S., Chamilos, G., Sebasigari, R., Riccieri, V., et al. (2011). Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med 3(73): 73ra19.
- Martínez-Alberquilla, I., Gasull, X., Pérez-Luna, P., Seco-Mera, R., Ruiz-Alcocer, J. and Crooke, A. (2022). Neutrophils and neutrophil extracellular trap components: Emerging biomarkers and therapeutic targets for age-related eye diseases. Ageing Res Rev 74: 101553.
- Matta, B., Battaglia, J., and Barnes, B. J. (2022). Detection of neutrophil extracellular traps in patient plasma: Method development and validation in systemic lupus erythematosus and healthy donors that carry IRF5 genetic risk. Front immunol 13:951254.
- Sayah, D. M., Mallavia, B., Liu, F., Ortiz-Munoz, G., Caudrillier, A., DerHovanessian, A., Ross, D. J., Lynch, J. P., 3rd, Saggar, R., Ardehali, A., et al. (2015). Neutrophil extracellular traps are pathogenic in primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med 191(4): 455-463.
- Yaqinuddin, A., Kvietys, P. and Kashir, J. (2020). COVID-19: Role of neutrophil extracellular traps in acute lung injury. Respir Investig 58(5): 419-420.
- Yu, Y., Kwon, K. and Pieper, R. (2019). Detection of Neutrophil Extracellular Traps in Urine. Methods Mol Biol 2021: 241-257.
- Zuo, Y., Yalavarthi, S., Shi, H., Gockman, K., Zuo, M., Madison, J. A., Blair, C., Weber, A., Barnes, B. J., Egeblad, M., et al. (2020). Neutrophil extracellular traps in COVID-19. JCI Insight 5(11): e138999.
- Zuo, Y., Zuo, M., Yalavarthi, S., Gockman, K., Madison, J. A., Shi, H., Woodard, W., Lezak, S. P., Lugogo, N. L., Knight, J. S., et al. (2021). Neutrophil extracellular traps and thrombosis in COVID-19. J Thromb Thrombolysis 51(2): 446-453.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Matta, B., Battaglia, J. and Barnes, B. J. (2023). A New Methodology for the Quantification of Neutrophil Extracellular Traps in Patient Plasma. Bio-protocol 13(12): e4701. DOI: 10.21769/BioProtoc.4701.
Category
Immunology > Immune cell function > Neutrophil
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









