- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Recombinant Human Amphiphysin 1 and its N-BAR Domain
(*contributed equally to this work) Published: Vol 13, Iss 12, Jun 20, 2023 DOI: 10.21769/BioProtoc.4699 Views: 1983
Reviewed by: David PaulAnindita SarkarAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Expression and Purification of the Human Voltage-Gated Proton Channel (hHv1)
Emerson M. Carmona [...] Luis G. Cuello
Jun 20, 2025 1984 Views
Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2366 Views
Isolation of Antigen-Specific Nanobodies From Synthetic Libraries Using a Protein Selection Strategy That Combines MACS-Based Screening of YSD and FLI-TRAP
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
Jan 20, 2026 430 Views
Abstract
Bin/Amphiphysin/Rvs (BAR) proteins are known as classical membrane curvature generators during endocytosis. Amphiphysin, a member of the N-BAR sub-family of proteins that contain a characteristic amphipathic sequence at the N-terminus of the BAR domain, is involved in clathrin-mediated endocytosis. Full-length amphiphysin contains a ~ 400 amino acid long disordered linker connecting the N-BAR domain and a C-terminal Src homology 3 (SH3) domain. We express and purify recombinant amphiphysin and its N-BAR domain along with an N-terminal glutathione-S-transferase (GST) tag. The GST tag allows extraction of the protein of interest using affinity chromatography and is removed in the subsequent protease treatment and ion-exchange chromatography steps. In the case of the N-BAR domain, cleavage of the GST tag was found to cause precipitation. This issue can be minimized by adding glycerol to the protein purification buffers. In the final step, size exclusion chromatography removes any potential oligomeric species. This protocol has also been successfully used to purify other N-BAR proteins, such as endophilin, Bin1, and their corresponding BAR domains.
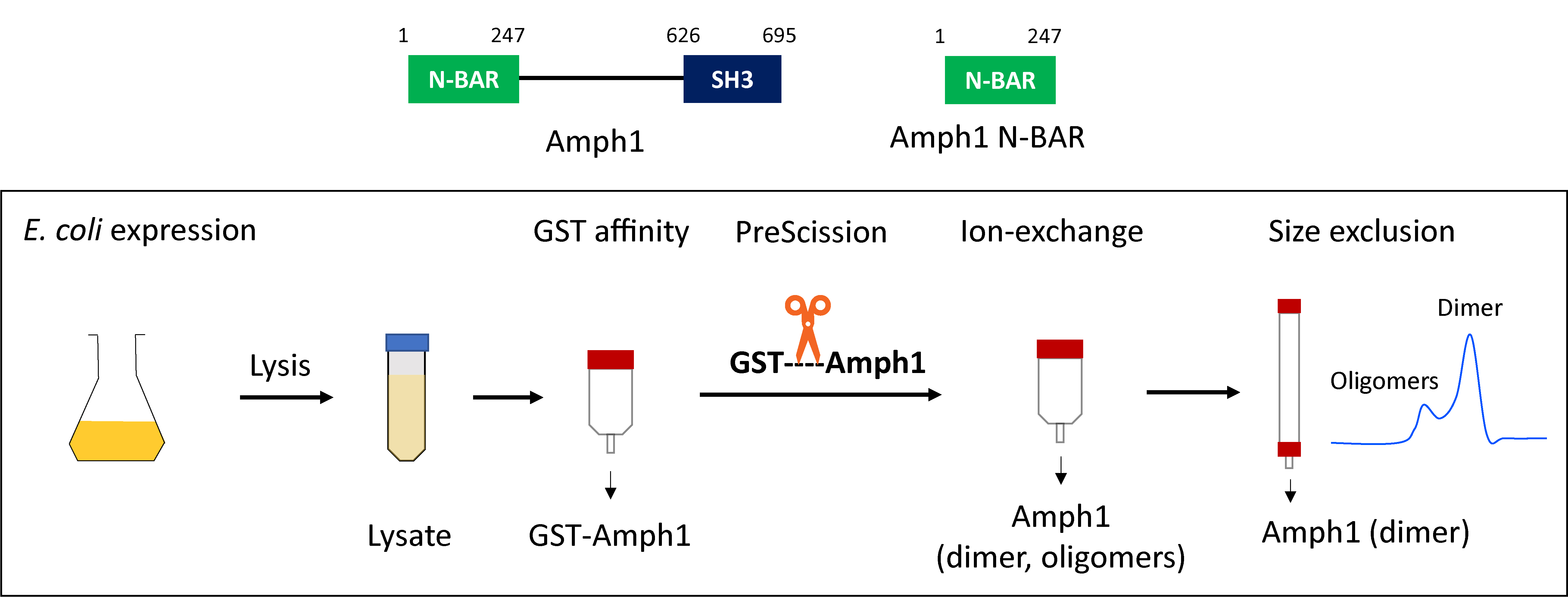
Graphical overview
Background
Bin/Amphiphysin/Rvs (BAR) family proteins have drawn wide interest due to their roles in several membrane trafficking pathways (Frost et al., 2009). During clathrin-mediated endocytosis, the N-BAR proteins endophilin and amphiphysin are recruited to the neck of endocytic buds and facilitate membrane scission (Takei et al., 1999; Milosevic et al., 2011). The N-BAR domain forms a homodimer in solution and adopts a crescent-like shape known for generating, sensing, and stabilizing highly curved endocytic membrane nanostructures (Gallop et al., 2006). Our group's recent in vitro reconstitution study has shown that endophilin can undergo phase separation via self-association and, alternatively, via multivalent interactions with its binding partners (Mondal et al., 2022). Unlike endophilin, amphiphysin (full length protein) did not show phase separation behavior, although its N-BAR domain did phase separate. These recent observations suggest that spatiotemporal organization on the membrane and curvature generation properties of BAR proteins could be regulated by a phase separation–dependent mechanism.
Purification of recombinant amphiphysin and endophilin as GST fusion proteins was first reported by DeCamilli and coworkers (David et al., 1996; Ringstad et al., 1997). The GST tag facilitates the purification of the protein of interest from E. coli lysate by affinity chromatography. Moreover, it is recognized for its ability to improve protein solubility and enhance expression efficiency (Smith and Johnson, 1988). Additionally, Gallop et al. (2005) demonstrated that using a GST tag instead of a polyhistidine tag resulted in more efficient purification of the endophilin N-BAR domain. A general strategy to purify the GST-tagged BAR proteins involves extraction of the fusion protein from bacterial cell lysate by GST affinity chromatography, removal of the GST tag by PreScission protease cleavage followed by ion exchange chromatography, and finally, size exclusion chromatography. Later on, several other groups adopted this basic purification strategy to purify various N-BAR proteins (Gallop et al., 2006; Capraro et al., 2013; Ambroso et al., 2014). Our present protocol allows the purification of various full-length N-BAR proteins, their N-BAR domains, and their mutants, with minor or no further optimization. We found that additional optimization is needed when purifying the amphiphysin N-BAR domain, since the protein tends to precipitate upon removal of the GST tag and for that reason the N-BAR domain loses its membrane curvature generation activity. The protein stability in aqueous solutions can be enhanced by the addition of cosolvents such as glycerol. Vagenende et al. (2009) reported that glycerol prevents protein aggregation by inhibiting protein unfolding and by interaction with hydrophobic surface regions of the protein to form amphiphilic interfaces. The addition of glycerol to buffers for all the chromatography steps was found to minimize precipitation, and the protein retained its curvature generation abilities. Here, we provide a step-by-step guide to purifying full-length amphiphysin and the N-BAR domain. Steps that are common and different for full-length proteins and the N-BAR domain are indicated.
Materials and reagents
Pipette tips 10 μL (Thermo Scientific, catalog number: 2139)
Pipette tips 200 μL (Fisher Scientific, catalog number: 02-707-500)
Pipette tips 1 mL (Fisher Scientific, catalog number: 02707508)
Disposable cuvettes (Fisher Scientific, catalog number: 14955127)
Polypropylene round-bottom tube (Corning, catalog number: 352059)
Petri dishes (Corning, catalog number: 353003)
0.22 μm filter (Merck Millipore, catalog number: SLGVR33RS)
10 mL syringes (Becton, Dickinson and Company, catalog number: 301029)
Collection tubes (Fisher Scientific, catalog number: 14-961-26)
Syringe (Henke Sass Wolf, catalog number: 4850001000)
Gel-loading tips (Fisher Scientific, catalog number: 05-408-150)
E. coli BL21 DE3 codon plus (Agilent Technologies, catalog number: 230280)
Plasmids: full length human amphiphysin 1 (1-695) and its N-BAR domain (1-247) sequences contained in pGEX-6P vectors were generously provided by Pietro DeCamilli’s lab. Plasmids are available from the Baumgart lab upon request.
PreScission protease (Genscript, catalog number: Z02799)
SOC medium (Corning, catalog number: 46-003-CR)
LB agar, Miller (Fisher BioReagents, catalog number: BP1425-500)
Ampicillin (Fisher BioReagents, catalog number: BP1760-25)
Chloramphenicol (Acros Organics, catalog number: 227920250)
200 proof ethanol (Decon Laboratories, catalog number: 2716)
Parafilm (Bemis, catalog number: PM-999)
LB broth, Miller (Sigma-Aldrich, catalog number: L3152)
Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Lab Scientific, catalog number: I-555)
Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: 11359061001)
Glycerol (Sigma-Aldrich, catalog number: G7893)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-3)
Tris(hydroxymethyl)aminomethane (Tris) (Fisher BioReagents, catalog number: BP152-1)
Ethylenediaminetetraacetic acid (EDTA) (Fisher Scientific, catalog number: BP120-500)
Dithiothreitol (DTT) (Fisher BioReagents, catalog number: BP172-5)
Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A144-500)
Glutathione (GSH) (Acros Organics, catalog number: 120001000)
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Fisher Scientific, catalog number: BP310-500)
Tris(2-carboxyethyl)phosphine (TCEP) (Thermo Scientific, catalog number: PG82089)
Sodium dihydrogen phosphate (Fisher Scientific, catalog number: BP329-500)
Disodium hydrogen phosphate (Fisher Scientific, catalog number: S374-500)
Sodium hydroxide (Fisher Scientific, catalog number: S318-500)
Acrylamide, 30% (Bio-Rad Laboratories, catalog number: 1610158)
Sodium dodecyl sulfate (SDS) (Acros Organics, catalog number: 327311000)
NovexTM sharp prestained protein ladder (Invitrogen, catalog number: 57318)
Loading buffer for SDS-PAGE (0.3 M Tris, 60% v/v glycerol, 12% w/v SDS, 0.3 M DTT, 0.6% bromophenol blue)
GelCodeTM Blue safe protein stain (ThermoFisher Scientific, catalog number: 24596)
Amicon® ultra centrifugal filters (Millipore Sigma, catalog numbers: UFC901024, and UFC903024)
LB agar plates (see Recipes)
Luria Bertani (LB) media (see Recipes)
PMSF stock solution (see Recipes)
Buffers (see Recipes)
GST lysis buffer
GST wash buffer
GST elution buffer
Cation exchange buffer A
Cation exchange buffer B
Size exclusion buffer
SDS-PAGE running buffer
Equipment
Incubator and shaker (Eppendorf, catalog number: M1335-0004)
UV Vis spectrophotometer (Agilent Technologies, catalog number: G1103A)
Sonicator (QSonica, catalog number: Q700)
Centrifuge (Sorvall, catalog number: SO-RC5B)
PTI F10S-6 × 500 rotor (Piramoon Technologies, catalog number: I590496)
SS34 8 × 50 rotor (Sorvall, catalog number: SO-34)
Centrifuge bottles, 500 mL (Nalgene, Thermo Scientific, catalog number: 3120-9500)
High-speed centrifuge tubes, 50 mL (Nalgene, Thermo Scientific, catalog number: 3114-0050)
Tabletop centrifuge (Sorvall, catalog number: SO-RT7)
Swing buckets (Sorvall, catalog number: 00436)
pH sensor (Fisher Scientific, catalog number: 13-620-631)
Akta Purifier (GE Healthcare) or Akta Pure (Cytiva)
GST column (Cytiva, catalog number: 17528202)
Cation exchange column (Cytiva, catalog number: 17115201)
Superdex 200 column (Cytiva, catalog number: 28989335)
Hot water bath (Precision Scientific, catalog number: 66643)
Autoclave (Steris, model: Amsco Lab 250)
Mini gel tank (Life Technologies, catalog number: A25977)
Electrophoresis power supply (Fisher Scientific, catalog number: 55860-4)
NanoDrop (Thermo Scientific, catalog number: ND-ONE-W)
Gel imagers: ChemiDocTM MP Imaging System (Bio-Rad, catalog number: 12003154) and Typhoon FLA 7000 (GE Healthcare)
Software
Unicorn: version 5.20 for Akta purifier (GE Healthcare) and version 7.6 for Akta Pure (Cytiva)
Procedure
Transformation of BL21 codon plus (DE3) RIPL cells
Thaw chemically competent BL21 codon plus (DE3) RIPL cells and the plasmid (pGEX-6P-1) on ice for 10 min.
Add 50 μL of the bacterial suspension into a polypropylene round-bottom sterile tube. Further, add 1 μL (~100 ng) of the desired plasmid [either full-length amphiphysin 1 (Amph1) or the N-BAR domain encoded into a pGEX-6P-1 vector] directly into the bacterial suspension. Incubate the mixture on ice for 20 min. Do not vortex.
Incubate the bacteria at 42 °C in a water bath for precisely 45 s and put the tube directly on ice for 20 min.
Add 1 mL of SOC medium and incubate at 37 °C for 1 h in a shaker at 250 rpm.
Spread 100 μL of the transformation reaction mixture onto a prewarmed LB agar plate containing ampicillin (0.1 mg/mL; see Recipes).
Incubate the plate at 37 °C for 16 h.
After transformation, the agar plates can be stored for one week at 4 °C.
Expression of full-length Amph1/Amph1 N-BAR in E. coli
Prepare the starter culture by inoculating a single colony of the transformed bacteria into 100 mL of autoclaved LB media supplemented with ampicillin (0.1 mg/mL) and chloramphenicol (0.035 mg/mL) (see Recipes). Incubate the culture at 37 °C in a shaker at 250 rpm for 12–16 h.
Note: We prepare stock solutions of ampicillin (25 mg/mL) in Milli Q water and chloramphenicol (35/mL) in absolute ethanol and store them in a -20 °C freezer. Required volume of the stock solution to the culture media can be added to achieve final working concentration of ampicillin (0.1 mg/mL) and chloramphenicol (0.035 mg/mL).
Inoculate 4 × 1 L of autoclaved LB broth containing ampicillin (0.1 mg/mL) and chloramphenicol (0.035 mg/mL) with 25 mL of starter culture. Grow the secondary culture at 37 °C in a shaker at 225 rpm.
Using a UV Vis spectrophotometer and disposable cuvettes, monitor the optical density at the wavelength of 600 nm every 1 h. Once the OD600 reaches a value of ~0.5 (approximately 3 h), reduce the incubator temperature to 18 °C and wait (another 20 min) for the OD600 to reach approximately 0.8.
Make an aqueous stock solution of IPTG by dissolving 300 mg of IPTG in 10 mL of autoclaved water. Add 2.5 mL of the stock solution to each 1 L of culture to obtain 0.3 mM final IPTG concentration. Also, add 1 mL from the 25 mg/mL stock solution of ampicillin and 0.25 mL from the 35 mg/mL stock solution of chloramphenicol to each 1 L culture. Incubate the flasks at 18 °C with shaking (225 rpm) for 12–16 h.
Divide the culture between multiple centrifuge bottles. Harvest the cells by centrifugation using a Sorvall SO-RC5B centrifuge equipped with a PTI F10S rotor, operated at 6,000× g for 20 min at 4 °C. Discard the supernatant and proceed with the next step. The pellet can be stored at -80 °C after transferring into 50 mL conical tubes and flash freezing with liquid nitrogen. Thaw the frozen bacterial cell pellet obtained from the post-induction of secondary culture on ice for 2–3 h.
Purification of full-length Amph1/Amph1 N-BAR
Cell Lysis: Resuspend the pellet in 60–80 mL of GST lysis buffer [see Recipes; use buffer without glycerol for full-length Amph1 and buffer containing 10% glycerol (v/v) for Amph1 N-BAR] supplemented with 1 mM of PMSF (see Recipes). Break any clumps by pipetting up and down. Transfer the suspension into a metallic beaker.
Sonicate the suspension using a tip sonicator at an amplitude value set between 30 and 50 for 5 min with a 1 s pulse on and 4 s pulse off cycle (25 min total sonication time). The maximum power output of the sonicator we use is 700 watts and, for a 1 s pulse at amplitude of 50, approximately 3.5 J is delivered to the probe. The sonication should be performed on ice to prevent heating of the suspension. Ensure the sonicator tip remains well immersed in the solution throughout the entire sonication process to prevent frothing. At the end of the sonication cycle, the lysed suspension should turn from light yellow to orangish and less viscous than the starting mixture. Continue the sonication for an additional 1–2 min if mixture still looks yellowish or seems to be similarly viscous as before.
Transfer the suspension into high-speed centrifuge tubes and centrifuge at 30,000× g for 1 h at 4 °C, using an SS34 8 × 50 rotor to pellet cell debris.
After centrifugation, filter the supernatant (hereafter cell lysate) through the 0.22 μm syringe filter to remove the particulate matter.
Note: Collect the cell pellet and cell lysate (~20 μL) samples for SDS-PAGE analysis.
GST affinity chromatography: Perform GST affinity chromatography using a fast protein liquid chromatographic (FPLC) system (ÄKTA purifier) with 2 × 5 mL GSTrapTM FF columns connected in a series. Pre-equilibrate the GSTrapTM FF column with five column volumes (CV) of GST lysis buffer [use buffer without glycerol for full-length Amph1 and buffer containing 10% glycerol (v/v) for Amph1 N-BAR9] prior to sample application.
Apply the filtered cell lysate from Step C4 onto the column using an injection loop at a flow rate of 1 mL/min and monitor the absorbance at 280 nm (Figure 1A and Figure 2A). Collect the flowthrough 1 (FT1) and inject it onto the column, and again save flowthrough 2 (FT2).
Wash the column with lysis buffer in order to remove the non-specifically bound protein. Washing should be performed until the UV absorption at 280 nm (UV_280) reaches a flat baseline (typically requires the passing of 100–150 mL of lysis buffer).
Wash the column with GST wash buffer (30–50 mL; see Recipes) in order to bring the salt concentration down to 150 mM (can be monitored from the conductance of the mobile phase; a rapid reduction in the conductance indicates buffer switching).
Elute the protein by passing elution buffer [see Recipes; use buffer without glycerol for full-length Amph1 and buffer containing 10% glycerol (v/v) for Amph1 N-BAR]. Start collecting elution as soon as the UV_280 signal starts increasing and collect until the signal reaches a flat baseline (~10–15 mL typically).
Note: Typically, we maintain a flow rate of 1 mL/min for loading the cell lysate, 2 mL/min for loading FT1 and for the washing steps, and 1 mL/min for the elution step. One can also maintain 1 mL/min flow rate throughout all steps.
Cleavage of GST tag by protease treatment: Thaw an aliquot (125 μg) of PreScission protease from -80 °C storage and add the protease to the eluted protein solution. Keep the mixture for overnight shaking at 4 °C in order to cleave the GST affinity tag.
Notes:
Keep a small (~20 μL) aliquot of the GST elution for running SDS-PAGE.
Typically, addition of 125 μg of PreScission protease has shown complete cleavage of GST-tag when we purified proteins at a scale of 4 × 1 L expressions. In case there is a band of protein with uncleaved GST-tag still present after overnight protease treatment, one more aliquot of protease can be added.
Collect samples (~20 μL) of FT2 and pre- and post-cleavage protein samples for SDS-PAGE analysis.
Running SDS-PAGE analysis: Mix the samples with loading buffer in a 1:5 ratio. Further, heat all the samples on a heating block set at approximately 95 °C for 5 min. Then, using gel-loading tips, load the samples into a 12% acrylamide gel along with pre-stained protein ladder. Run the gel at 200 V for 1 h using a running buffer (see Recipes). Stain the gel with GelCodeTM Blue safe protein stain and then destain with water to visualize the bands (Figure 1B and Figure 2D).
Note: Molecular weights of full-length Amph1 and its N-BAR domain are 76 kDa and 29 kDa, respectively. GST has a molecular weight of 26 kDa. The full-length protein appeared slightly closer to the 110 kDa band and the N-BAR appeared closer to the 30 kDa band of the protein marker.
Cation exchange chromatography: Equilibrate the cation exchange column (5 mL) with cation exchange buffer A [see Recipes; without glycerol for full-length Amph1 and with 10% glycerol (v/v) for the N-BAR] and cation exchange buffer B [see Recipes; without glycerol for full-length Amph1 and with 10% glycerol (v/v) for the N-BAR]. A typical column equilibration program starts with 100 % buffer A and 0% buffer B (five column volumes); then, switches to 100% buffer B and 0% buffer A (five column volumes); and finally switches back to 100% buffer A and 0% buffer B (five column volumes). Also, equilibrate the injection loop with two loop volumes of buffer A.
Inject the cleaved protein sample onto the column using an injection loop at a flow rate of 1 mL/min and collect the FT1. Repeat the same and save FT2.
Wash the column with buffer A until the absorbance at 280 nm reaches baseline.
Elute the protein with a linear gradient from 0% buffer B, 100% buffer A to 45% buffer B, and 55% buffer A over the course of a 100 mL elution volume and collect the fractions (Figure 1C and Figure 2B).
Run the FT2 and fractions on SDS-PAGE (Figure 1E and Figure 2D).
Pool the fractions containing the protein and concentrate using Amicon® ultra centrifugal filters (use molecular weight cutoff of 10 kDa for N-BAR and 30 kDa for full-length Amph1) up to a final volume of 10 mL.
Size exclusion chromatography: Equilibrate the size exclusion column and the injection loop with size exclusion buffer (see Recipes; without glycerol for full-length Amph1 and with 10% glycerol for the N-BAR).
Inject the pooled protein fractions post-cation exchange chromatography and after concentrations into the size exclusion column (Superdex 200) at a flow rate of 0.8 mL/min.
Elute the proteins and collect the fractions (Figure 1D and Figure 2C). Analyze the fractions using SDS-PAGE (Figure 1F and Figure 2E).
Note: The BAR domain proteins often elute as two separate bands in size exclusion (Figure 2C). Samples from both these size exclusion bands usually appear as the same molecular weight species in SDS-PAGE, indicating that these bands are actually different oligomeric species. The dimer usually elutes at the end due to its smaller size than the larger oligomers. We recommend that the fractions under the different elution bands should be collected and concentrated separately to avoid mixing between various oligomers.
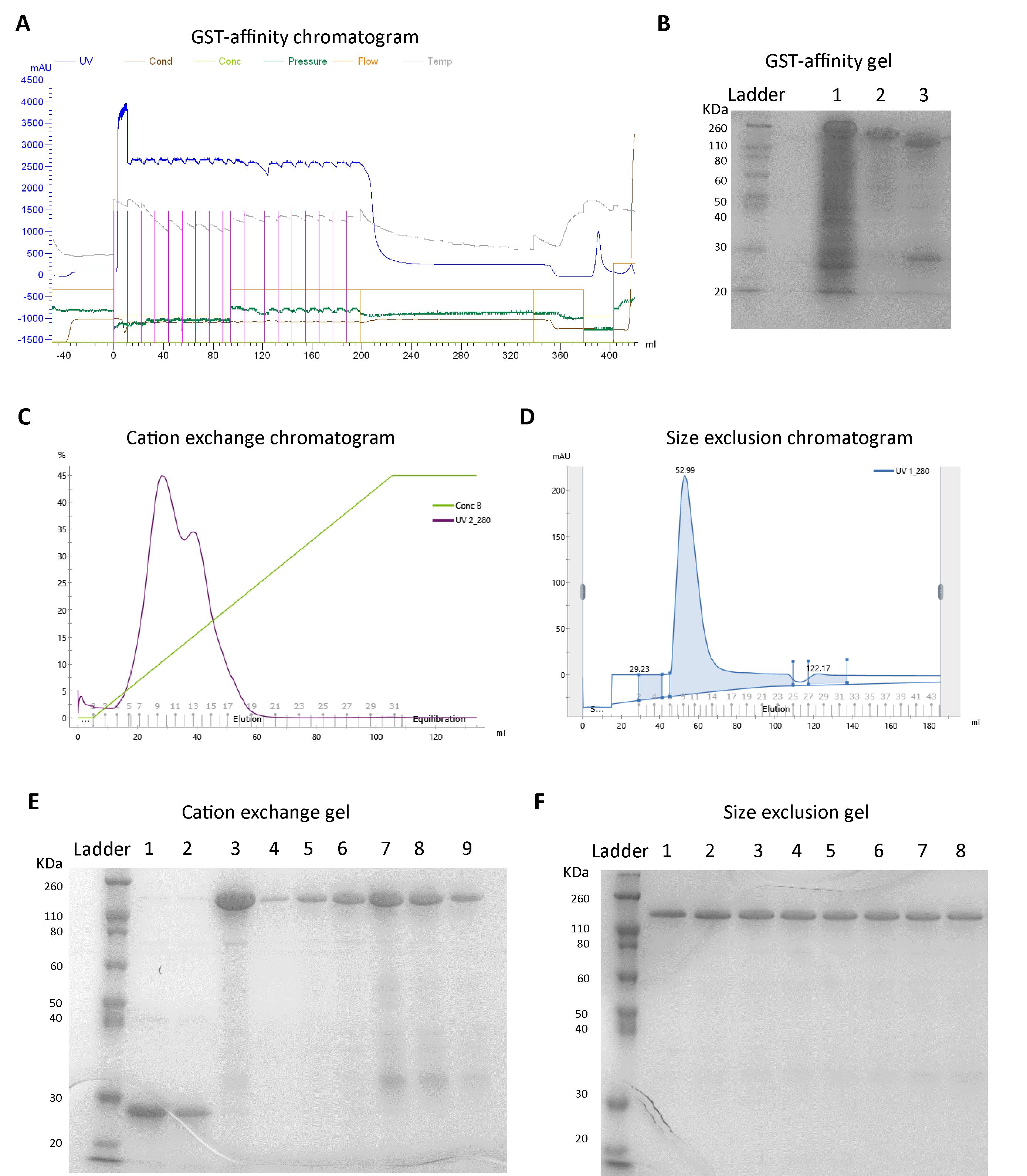
Figure 1. Representative FPLC chromatograms and SDS-PAGE images from full-length Amph1 purification. A. Chromatogram showing loading of cell lysate (supernatant) onto GST affinity column and elution. Line colors represent the chromatograms corresponding to the parameters mentioned on top. The chromatogram between 0 and 200 mL represents the sample loading step, 200–340 mL indicates washing with GST lysis buffer, 340–380 mL indicates washing with GST wash buffer, and 380–400 mL indicates the elution step. B. SDS-PAGE after GST affinity purification. Lane 1: flowthrough 2 (FT2); lane 2: elution before protease cleavage; lane 3: elution after PreScission treatment. C. Cation exchange elution chromatogram (purple line represents UV_280 profile and green line represents the concentration of buffer B indicating the protein of interest elutes around 50 mL). D. UV_280 profile from the size exclusion chromatography step showing the elution band between 45 and 70 mL. E. SDS-PAGE after cation exchange chromatography. Lanes 1 and 2: flowthrough from cation exchange column; lanes 3–9: fractions collected under the elution peak shown in Figure 1C. F. SDS-PAGE after size exclusion chromatography. Lanes 1–8: fractions collected under the peak shown in Figure 1D.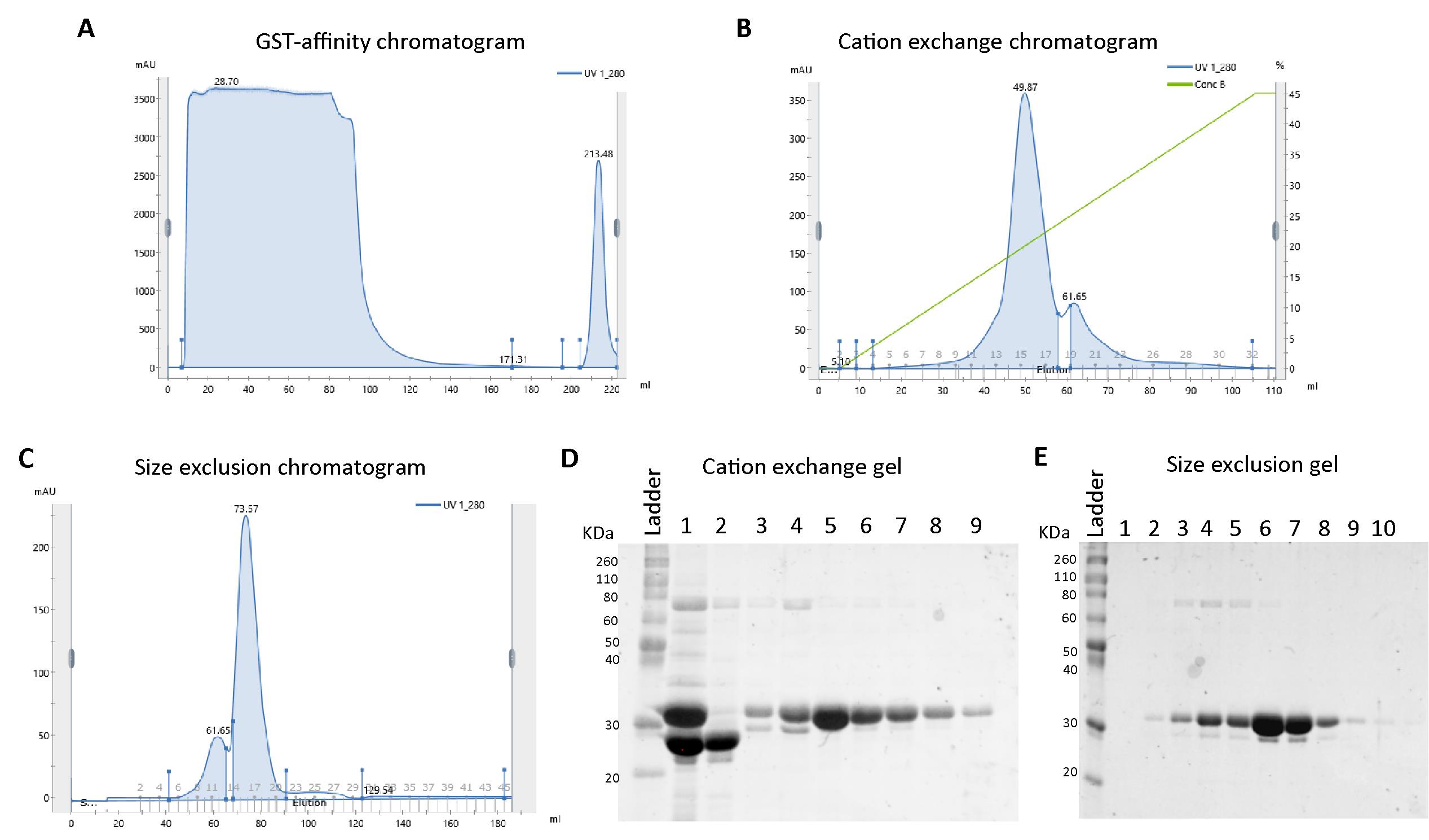
Figure 2. Representative FPLC chromatograms and SDS-PAGE image from Amph1 N-BAR purification. A. Chromatogram from the GST affinity purification showing the UV_280 profile during loading (0–100 mL), column wash with lysis buffer (90–170 mL), column wash with GST wash buffer (170–200 mL), and elution (200–220 mL) steps. B. Cation exchange elution chromatogram (blue line represents UV_280 profile and green line represents the concentration of buffer B) indicating the protein of interest elutes around 50 mL. C. Size exclusion chromatogram showing a peak around 60 mL corresponding to the oligomeric species, and a peak around 75 mL corresponding to the homodimeric species. D. Representative SDS-PAGE image after cation exchange chromatography of Amph1 N-BAR. Lane 1: PreScission protease-treated sample before loading to cation exchange column showing the bands for cleaved GST tag (between 20 and 30 kDa markers) and N-BAR domain (~30 kDa); lane 2: flowthrough 2 (FT2) from the cation exchange column; lanes 3–9: fractions collected under the elution peaks shown in Figure 2B. E. SDS-PAGE for Amph1 N-BAR after size exclusion chromatography. Lanes 1–10: fractions collected under the elution peaks (elution volume 50–90 mL) shown in Figure 2C.Pool and concentrate the fractions containing protein using Amicon ultra centrifugal filters (use molecular weight cutoff of 10 kDa for N-BAR and 30 kDa for full-length Amph1).
Concentration determination and storage: Determine the protein concentration using NanoDrop by measuring the absorbance at 280 nm. The extinction coefficients of full-length Amph1 and N-BAR are 55,000 M-1 cm-1 and 28,500 M-1 cm-1, respectively.
Transfer the proteins into small aliquots, flash freeze into liquid nitrogen, and store at -80 °C.
Notes
Always handle live bacterial cultures under a flame or in a biosafety cabinet, in order to maintain aseptic conditions.
Dispose of E. coli cultures as recommended by biosafety protocols.
The flash-frozen cell pellets can be stored at -80 °C for up to six months.
Recipes
LB agar plates
Add 3.7 g of LB agar in 100 mL of Milli Q water and sterilize by autoclaving. Allow the medium to reach a lukewarm temperature and add the required antibiotic ampicillin (0.1 mg/mL). Further, pour approximately 25 mL into sterile Petri dishes. Allow to solidify and then seal with parafilm. The plates can be stored at 4 °C for one month.
Luria Bertani (LB) media
Prepare 4 L of media by dissolving LB in Milli Q water (25 g/L) and autoclave to sterilize. Add the required antibiotics ampicillin (0.1 mg/mL) and chloramphenicol (0.035 mg/mL) before inoculation.
PMSF stock solution
Dissolve PMSF (mol. wt. 174 g/mol) in absolute ethanol (0.174 g solid in 10 mL) to prepare a 100 mM stock solution. The stock solution can be stored at -20 °C.
Buffers
Prepare buffers without glycerol for full-length Amph1 and with 10% glycerol for the N-BAR. Use diluted sodium hydroxide or HCl solutions to adjust the buffer pH.
GST lysis buffer (1 L)
300 mM NaCl 17.532 g
50 mM Tris 6.06 g
1 mM EDTA 4 mL of 0.25 M stock
2 mM DTT 0.308 g
pH = 8, filter and store at 4 °C
GST wash buffer (0.25 L)
150 mM NaCl 2.19 g
50 mM Tris 1.515 g
1 mM EDTA 1 mL of 0.25 M stock
2 mM DTT 0.077 g
pH = 8, filter and store at 4 °C
GST elution buffer (0.25 L)
150 mM NaCl 2.19 g
50 mM Tris 1.515 g
20 mM GSH 1.536 g
1 mM EDTA 1 mL of 0.25 M stock
2 mM DTT 0.077 g
pH = 8, filter and store at 4 °C
Cation exchange buffer A (1 L)
150 mM NaCl 8.77 g
20 mM sodium phosphate (including both dibasic and monobasic salts)
Sodium dihydrogen phosphate 1 g
Disodium hydrogen phosphate 3.11g
1 mM EDTA 4 mL of 0.25 M stock
1 mM DTT 0.154 g
pH = 7, filter and store at 4 °C
Cation exchange buffer B (0.5 L)
1 M NaCl 29.22 g
20 mM sodium phosphate (including both dibasic and monobasic salts)
Sodium dihydrogen phosphate 0.5 g
Disodium hydrogen phosphate 1.56 g
1 mM EDTA 2 mL of 0.25 M
1 mM DTT 0.08 g
pH = 7, filter and store at 4 °C
Size exclusion buffer (1 L)
150 mM NaCl 8.766 g
20 mM HEPES 4.766 g
1 mM TCEP 0.287 g
pH = 7.4, filter and store at 4 °C
SDS-PAGE running buffer (1 L)
Tris 3.0 g
Glycine 14.4 g
SDS 1 g
pH 8.3
Acknowledgments
The authors acknowledge Pietro DeCamilli’s lab for providing us with plasmids for full-length amphiphysin, N-BAR domain, and PreScission protease. This research was financially supported by the National Institutes of Health, grant No GM 097552.
Competing interests
The authors declare no competing interests.
Ethics
No animal/human subject is used in this protocol.
References
- Ambroso, M. R., Hegde, B. G. and Langen, R. (2014). Endophilin A1 induces different membrane shapes using a conformational switch that is regulated by phosphorylation. Proc Natl Acad Sci U S A 111(19): 6982-6987.
- Capraro, B. R., Shi, Z., Wu, T., Chen, Z., Dunn, J. M., Rhoades, E. and Baumgart, T. (2013). Kinetics of endophilin N-BAR domain dimerization and membrane interactions. J Biol Chem 288(18): 12533-12543.
- David, C., McPherson, P. S., Mundigl, O. and de Camilli, P. (1996). A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci U S A 93(1): 331-335.
- Frost, A., Unger, V. M. and De Camilli, P. (2009). The BAR domain superfamily: membrane-molding macromolecules. Cell 137(2): 191-196.
- Gallop, J. L., Butler, P. J. and McMahon, H. T. (2005). Endophilin and CtBP/BARS are not acyl transferases in endocytosis or Golgi fission. Nature 438(7068): 675-678.
- Gallop, J. L., Jao, C. C., Kent, H. M., Butler, P. J., Evans, P. R., Langen, R. and McMahon, H. T. (2006). Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J 25(12): 2898-2910.
- Milosevic, I., Giovedi, S., Lou, X., Raimondi, A., Collesi, C., Shen, H., Paradise, S., O'Toole, E., Ferguson, S., Cremona, O., et al. (2011). Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron 72(4): 587-601.
- Mondal, S., Narayan, K., Botterbusch, S., Powers, I., Zheng, J., James, H. P., Jin, R. and Baumgart, T. (2022). Multivalent interactions between molecular components involved in fast endophilin mediated endocytosis drive protein phase separation. Nat Commun 13(1): 5017.
- Ringstad, N., Nemoto, Y. and De Camilli, P. (1997). The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc Natl Acad Sci U S A 94(16): 8569-8574.
- Smith, D. B. and Johnson, K. S. (1988). Single-Step Purification of Polypeptides Expressed in Escherichia coli as Fusions with Glutathione S-Transferase. Gene 67(1): 31-40.
- Takei, K., Slepnev, V. I., Haucke, V. and De Camilli, P. (1999). Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol 1(1): 33-39.
- Vagenende, V., Yap, M. G. S. and Trout, B. L. (2009). Mechanisms of Protein Stabilization and Prevention of Protein Aggregation by Glycerol. Biochemistry 48(46): 11084-11096.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Mondal, S., James, H. P., Milano, F., Jin, R. and Baumgart, T. (2023). Purification of Recombinant Human Amphiphysin 1 and its N-BAR Domain. Bio-protocol 13(12): e4699. DOI: 10.21769/BioProtoc.4699.
Category
Biochemistry > Protein > Expression
Molecular Biology > Protein > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









